Adaptation of Droplet Digital PCR-Based HIV Transcription Profiling to Digital PCR and Association of HIV Transcription and Total or Intact HIV DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Nucleic Acid Extractions
2.3. Preparation of HIV-1 RNA Standards
2.4. Reverse Transcription for ‘HIV Transcription Profiling’ Assays
2.5. Droplet Digital PCR
2.6. QIAcuity dPCR
2.7. Calculations for Limit of Blank (LoB), Limit of Detection (LoD), Limit of Quantitation (LoQ), and Intra- and Inter-Assay Variability (%CV)
triplicate mean × 100)
2.8. Intact Proviral DNA Assay
2.9. HIV Transcription Profiling of CD4+ T Cells from PLWH
2.10. Statistical Analyses
3. Results
3.1. Assay Optimization for dPCR
3.2. Fluorescence Signal-to-Noise Ratio and Primer/Probe Efficiency
3.3. Assessment of Linear Dynamic Range and Precision of ddPCR and qPCR Technologies for Low Target Concentration
3.4. Intra- and Inter-Assay Variability
3.5. Concordance between ddPCR and dPCR Platforms
3.6. Levels of HIV Transcription in People Living with HIV Are Not Influenced by Size of the Intact Reservoir, Duration of Suppressive ART, or HLA Allele Carriage
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018, 10, eaap9927. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS—Mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Telwatte, S.; Lee, S.; Somsouk, M.; Hatano, H.; Baker, C.; Kaiser, P.; Kim, P.; Chen, T.-H.; Milush, J.; Hunt, P.W.; et al. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog. 2018, 14, e1007357. [Google Scholar] [CrossRef] [PubMed]
- Dreo, T.; Pirc, M.; Ramšak, Z.; Pavšič, J.; Milavec, M.; Žel, J.; Gruden, K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014, 406, 6513–6528. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar]
- Porco, D.; Hermant, S.; Purnomo, C.A.; Horn, M.; Marson, G.; Colling, G. Getting rid of ‘rain’ and ‘stars’: Mitigating inhibition effects on ddPCR data analysis, the case study of the invasive crayfish Pacifastacus leniusculus in the streams of Luxembourg. PLoS ONE 2022, 17, e0275363. [Google Scholar] [CrossRef]
- Tumpach, C.; Cochrane, C.R.; Kim, Y.; Ong, J.; Rhodes, A.; Angelovich, T.A.; Churchill, M.J.; Lewin, S.R.; Telwatte, S.; Roche, M. Adaptation of the intact proviral DNA assay to a nanowell-based digital PCR platform. J. Virus Erad. 2023; in press. [Google Scholar] [CrossRef]
- Bruner, K.M.; Wang, Z.; Simonetti, F.R.; Bender, A.M.; Kwon, K.J.; Sengupta, S.; Fray, E.J.; Beg, S.A.; Antar, A.A.R.; Jenike, K.M.; et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019, 566, 120–125. [Google Scholar] [CrossRef]
- Kinloch, N.N.; Ren, Y.; Alberto, W.D.C.; Dong, W.; Khadka, P.; Huang, S.H.; Mota, T.M.; Wilson, A.; Shahid, A.; Kirkby, D.; et al. HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA). Nat. Commun. 2021, 12, 165. [Google Scholar] [CrossRef]
- Ma, H.; Bell, K.N.; Loker, R.N. qPCR and qRT-PCR analysis: Regulatory points to consider when conducting biodistribution and vector shedding studies. Mol. Ther. Methods Clin. Dev. 2021, 20, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Muccini, C.; Guffanti, M.; Spagnuolo, V.; Cernuschi, M.; Galli, L.; Bigoloni, A.; Galli, A.; Poli, A.; Racca, S.; Castagna, A. Association between low levels of HIV-1 DNA and HLA class I molecules in chronic HIV-1 infection. PLoS ONE 2022, 17, e0265348. [Google Scholar] [CrossRef]
- Dorak, M.T.; Tang, J.; Tang, S.; Penman-Aguilar, A.; Coutinho, R.A.; Goedert, J.J.; Detels, R.; Kaslow, R.A. Influence of Human Leukocyte Antigen–B22 Alleles on the Course of Human Immunodeficiency Virus Type 1 Infection in 3 Cohorts of White Men. J. Infect. Dis. 2003, 188, 856–863. [Google Scholar] [CrossRef]
- QIAGEN. QIAcuity User Manual; QIAGEN: Hilden, Germany, 2021; pp. 23–233. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=ad681197-cdf1-401c-89c7-2db518129e86&lang=en (accessed on 17 July 2023).
- Bio-Rad Laboratories Inc. QX200™ Droplet Digital™ PCR System. 2022. Available online: https://www.bio-rad.com/sites/default/files/webroot/web/pdf/lsr/literature/Bulletin_6311.pdf (accessed on 17 July 2023).
- Bio-Rad Laboratories Inc. QX200 Droplet Reader and QX Manager Software Standard Edition. 2022. Available online: https://www.bio-rad.com/sites/default/files/webroot/web/pdf/lsr/literature/10000107223.pdf (accessed on 17 July 2023).
- Foster, J.L.; Garcia, J.V. Role of Nef in HIV-1 Replication and Pathogenesis. Adv. Pharmacol. 2007, 55, 389–409. [Google Scholar] [CrossRef]
- Hiener, B.; Horsburgh, B.A.; Eden, J.-S.; Barton, K.; Schlub, T.E.; Lee, E.; von Stockenstrom, S.; Odevall, L.; Milush, J.M.; Liegler, T.; et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4 + T Cells from Effectively Treated Participants. Cell Rep. 2017, 21, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Sannier, G.; Dubé, M.; Dufour, C.; Richard, C.; Brassard, N.; Delgado, G.-G.; Pagliuzza, A.; Baxter, A.E.; Niessl, J.; Brunet-Ratnasingham, E.; et al. Combined single-cell transcriptional, translational, and genomic profiling reveals HIV-1 reservoir diversity. Cell Rep. 2021, 36, 109643. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.-C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell 2013, 155, 540–551. [Google Scholar] [CrossRef]
- Lee, G.Q.; Orlova-Fink, N.; Einkauf, K.; Chowdhury, F.Z.; Sun, X.; Harrington, S.; Kuo, H.-H.; Hua, S.; Chen, H.-R.; Ouyang, Z.; et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Investig. 2017, 127, 2689–2696. [Google Scholar] [CrossRef] [PubMed]
- Einkauf, K.B.; Lee, G.Q.; Gao, C.; Sharaf, R.; Sun, X.; Hua, S.; Chen, S.M.; Jiang, C.; Lian, X.; Chowdhury, F.Z.; et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Investig. 2019, 129, 988–998. [Google Scholar] [CrossRef]
- Peluso, M.J.; Bacchetti, P.; Ritter, K.D.; Beg, S.; Lai, J.; Martin, J.N.; Hunt, P.W.; Henrich, T.J.; Siliciano, J.D.; Siliciano, R.F.; et al. Differential decay of intact and defective proviral DNA in HIV-1–infected individuals on suppressive antiretroviral therapy. J. Clin. Investig. 2020, 5, e132997. [Google Scholar] [CrossRef]
- Einkauf, K.B.; Osborn, M.R.; Gao, C.; Sun, W.; Sun, X.; Lian, X.; Parsons, E.M.; Gladkov, G.T.; Seiger, K.W.; Blackmer, J.E.; et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 2022, 185, 266–282.e215. [Google Scholar] [CrossRef]
- Trachtenberg, E.; Korber, B.; Sollars, C.; Kepler, T.B.; Hraber, P.T.; Hayes, E.; Funkhouser, R.; Fugate, M.; Theiler, J.; Hsu, Y.S.; et al. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 2003, 9, 928–935. [Google Scholar] [CrossRef]
- Altfeld, M.; Kalife, E.T.; Qi, Y.; Streeck, H.; Lichterfeld, M.; Johnston, M.N.; Burgett, N.; Swartz, M.E.; Yang, A.; Alter, G.; et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8+ T Cell Response against HIV-1. PLoS Med. 2006, 3, e403. [Google Scholar] [CrossRef] [PubMed]
- Kaslow, R.; Carrington, M.; Apple, R.; Park, L.; Muñoz, A.; Saah, A.; Goedert, J.; Winkler, C.; O’Brien, S.; Rinaldo, C.; et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV–1 infection. Nat. Med. 1996, 2, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Hendel, H.; Caillat-Zucman, S.; Lebuanec, H.; Carrington, M.; O’brien, S.; Andrieu, J.-M.; Schächter, F.; Zagury, D.; Rappaport, J.; Winkler, C.; et al. New Class I and II HLA Alleles Strongly Associated with Opposite Patterns of Progression to AIDS. J. Immunol. 1999, 162, 6942–6946. [Google Scholar] [CrossRef] [PubMed]
- Migueles, S.A.; Sabbaghian, M.S.; Shupert, W.L.; Bettinotti, M.P.; Marincola, F.M.; Martino, L.; Hallahan, C.W.; Selig, S.M.; Schwartz, D.; Sullivan, J.; et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 2000, 97, 2709–2714. [Google Scholar] [CrossRef]
- Altfeld, M.; Addo, M.M.; Rosenberg, E.S.; Hecht, F.M.; Lee, P.K.; Vogel, M.; Yu, X.G.; Draenert, R.; Johnston, M.N.; Strick, D.; et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. Aids 2003, 17, 2581–2591. [Google Scholar] [CrossRef]
- Li, S.S.; Hickey, A.; Shangguan, S.; Ehrenberg, P.K.; Geretz, A.; Butler, L.; Kundu, G.; Apps, R.; Creegan, M.; Clifford, R.J.; et al. HLA-B∗46 associates with rapid HIV disease progression in Asian cohorts and prominent differences in NK cell phenotype. Cell Host Microbe 2022, 30, 1173–1185.e1178. [Google Scholar] [CrossRef]
- Klein, M.R.; Keet, I.P.M.; D’Amaro, J.; Bende, R.J.; Hekman, A.; Mesman, B.; Koot, M.; de Waal, L.P.; Coutinho, R.A.; Miedema, F. Associations between HLA Frequencies and Pathogenic Features of Human Immunodeficiency Virus Type 1 Infection in Seroconverters from the Amsterdam Cohort of Homosexual Men. J. Infect. Dis. 1994, 169, 1244–1249. [Google Scholar] [CrossRef]
- Gao, X.; Nelson, G.W.; Karacki, P.; Martin, M.P.; Phair, J.; Kaslow, R.; Goedert, J.J.; Buchbinder, S.; Hoots, K.; Vlahov, D.; et al. Effect of a Single Amino Acid Change in MHC Class I Molecules on the Rate of Progression to AIDS. N. Engl. J. Med. 2001, 344, 1668–1675. [Google Scholar] [CrossRef]
- Gao, X.; O’Brien, T.R.; Welzel, T.M.; Marti, D.; Qi, Y.; Goedert, J.J.; Phair, J.; Pfeiffer, R.; Carrington, M. HLA-B alleles associate consistently with HIV heterosexual transmission, viral load, and progression to AIDS, but not susceptibility to infection. Aids 2010, 24, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Bruner, K.M.; Murray, A.J.; Pollack, R.A.; Soliman, M.G.; Laskey, S.B.; Capoferri, A.A.; Lai, J.; Strain, M.C.; Lada, S.M.; Hoh, R.; et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016, 22, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.A.; Kadiyala, G.N.; Telwatte, S.; Wedrychowski, A.; Chen, T.-H.; Moron-Lopez, S.; Arneson, D.; Hoh, R.; Deeks, S.; Wong, J.; et al. New Assay Reveals Vast Excess of Defective over Intact HIV-1 Transcripts in Antiretroviral Therapy-Suppressed Individuals. J. Virol. 2022, 96, e0160522. [Google Scholar] [CrossRef]
- Deeks, S.G.; Archin, N.; Cannon, P.; Collins, S.; Jones, R.B.; de Jong, M.A.W.P.; Lambotte, O.; Lamplough, R.; Ndung’u, T.; Sugarman, J.; et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 2021, 27, 2085–2098. [Google Scholar] [CrossRef] [PubMed]

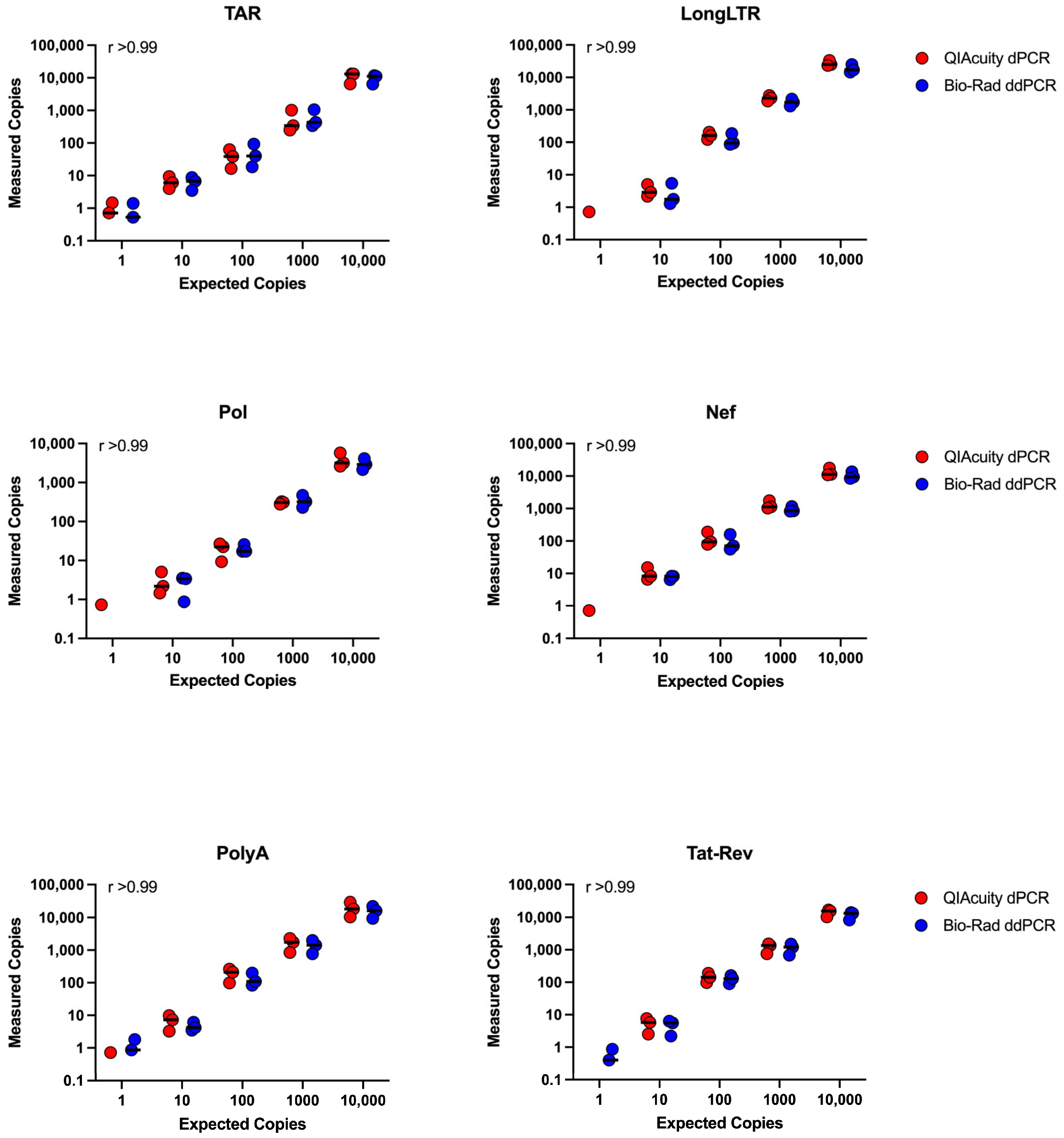
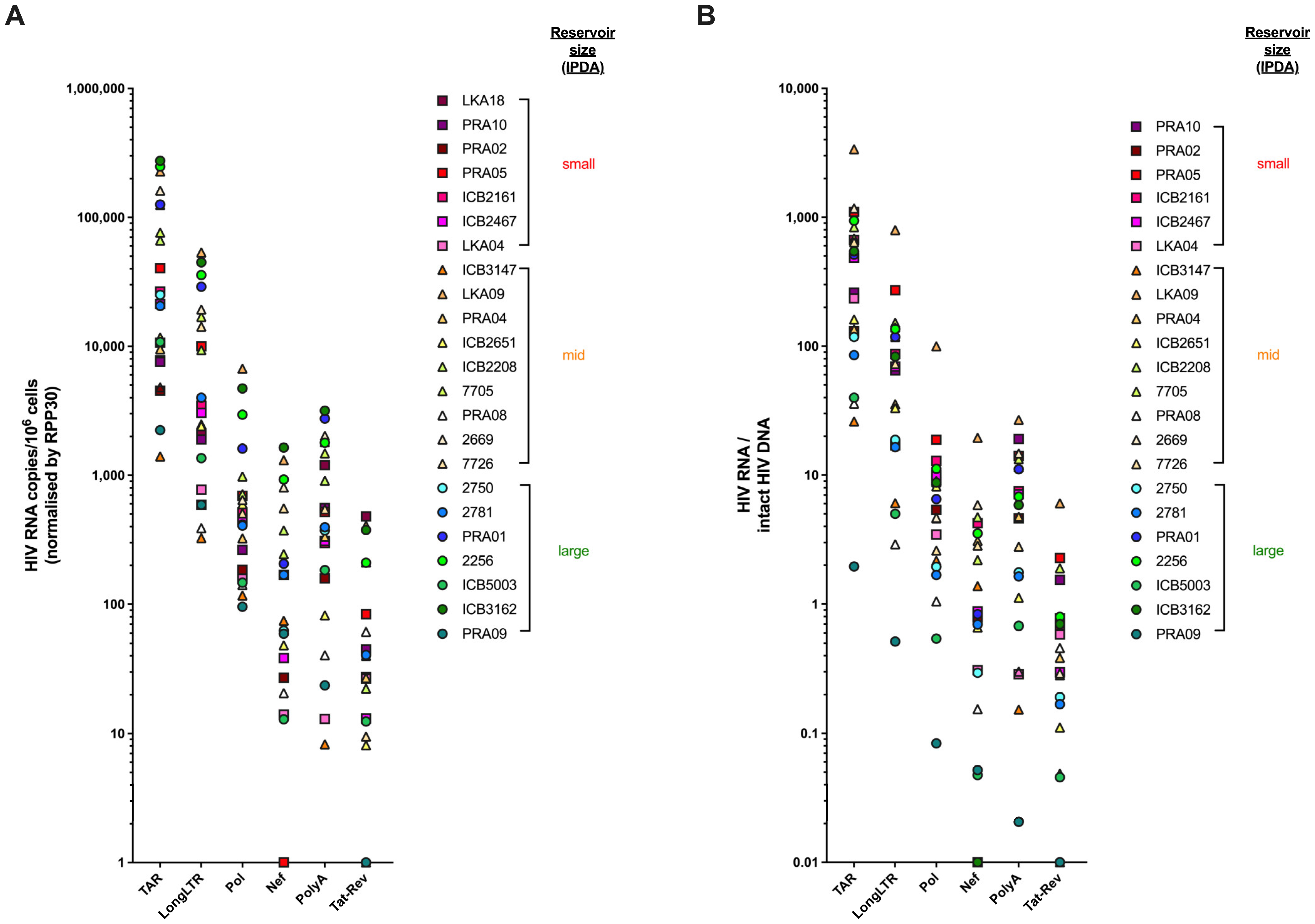

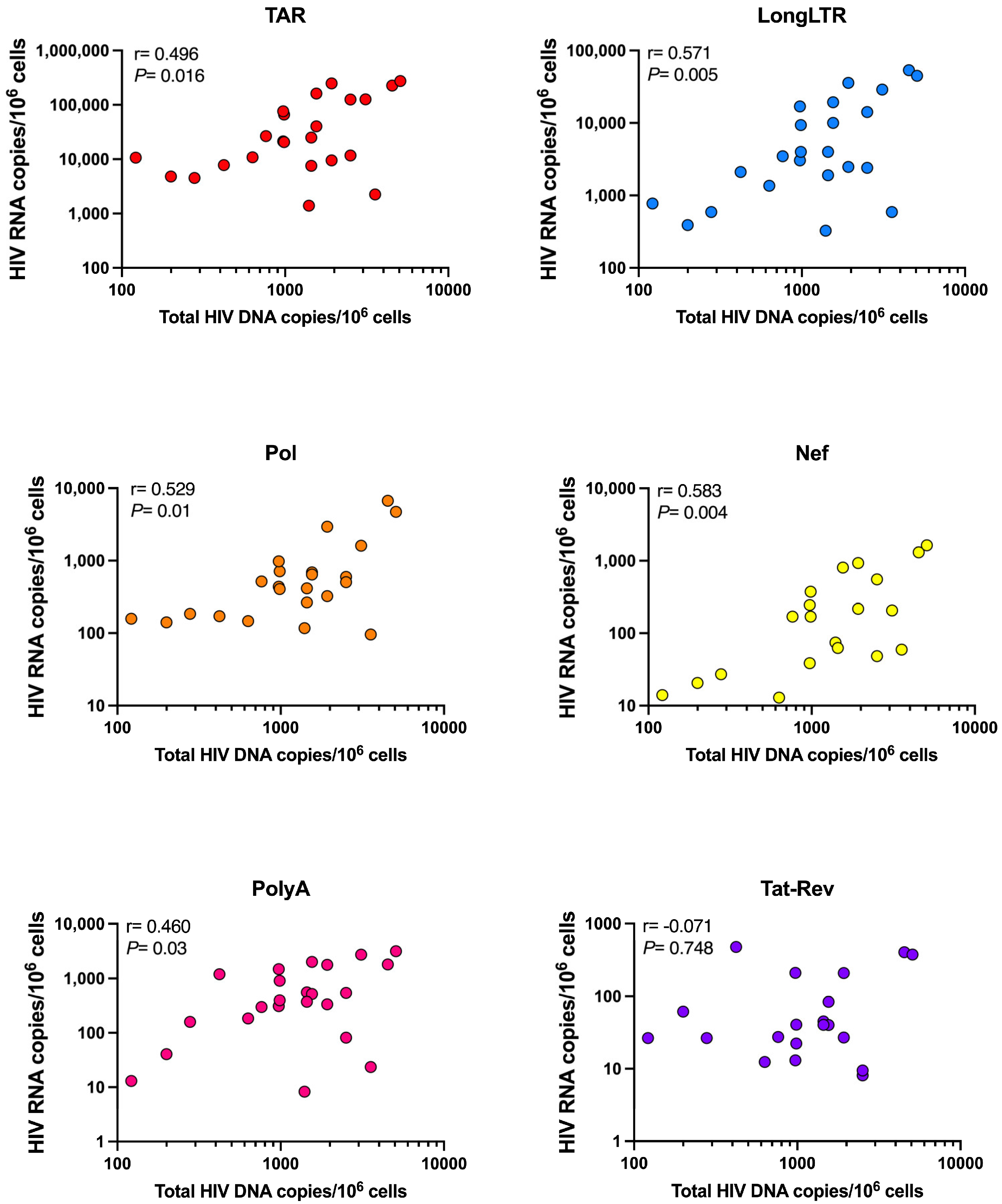
| Assay (Primer/Probe Set) | Intra-Assay 1 %CV | Inter-Assay 2 %CV (High Copy) | Inter-Assay %CV (Low Copy) | Frequency of 10 Copies Detected (%) 3 | ||||
|---|---|---|---|---|---|---|---|---|
| dPCR | ddPCR | dPCR | ddPCR | dPCR | ddPCR | dPCR | ddPCR | |
| TAR | 8.59 | 7.7 | 8.33 | 4.24 | 22.67 | 18.56 | 95 | 95 |
| LongLTR | 4.97 | 4.1 | 4.4 | 3.68 | 39.2 | 33.01 | 90 | 90 |
| Pol | 9.93 | 13.0 | 4.53 | 4.27 | 26.59 | 9.56 | 81 | 83 |
| Nef | 3.38 | 4.32 | 2.97 | 4.67 | 18.63 | 28.14 | 100 | 100 |
| PolyA | 7.02 | 3.3 | 2.26 | 1.59 | 8.72 | 8.54 | 100 | 100 |
| Tat-Rev | 4.86 | 5.30 | 2.88 | 3.34 | 46.16 | 33.59 | 90 | 94 |
| Assay | RNA Standard 2 | dPCR | ddPCR | |
|---|---|---|---|---|
| Average Measured/Expected Copies | Average Measured/Expected Copies | Difference (%) | ||
| TAR | IVT | 0.78 | 0.71 | 6.6 |
| LongLTR | IVT | 1.44 | 1.20 | 24.29 |
| Pol 1 | VIR | 0.273 | 0.259 | 1.4 |
| Nef | IVT | 1.13 | 0.79 | 34.43 |
| PolyA | IVT | 1.51 | 1.25 | 26.51 |
| Tat-Rev | IVT | 0.8 | 0.76 | 3.95 |
| Participant ID | Age (Years) | Sex | Race | HIV Diagnosis | CD4+ Count (Cells/μL) | CD4 (%) | CD8+ Count (Cells/μL) | CD8 (%) | Nadir CD4+ Count (Cells/μL) | ART Regimen 1 | VL (Copies/mL) | Peak VL (Copies/mL) | Duration HIV RNA < 50 Copies (Years) | HLA-B Alleles 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICB2161 | 69 | M | Caucasian | 1985 | 800 | 42 | 647 | 34 | 98 | 3TC, DRV, RTV, DTG | <40 | 80,410 | 7 | 14:01:01G + 27:05:02G |
| ICB2208 | 66 | M | Caucasian | 1984 | 466 | 31 | 546 | 36 | 54 | FTC/TAF, DRV/COBI | <40 | 50,000 | 9.8 | 07:02:01G + 57:01:01G |
| ICB2467 | 46 | M | Hispanic/Latino | 2006 | 429 | 43 | 316 | 32 | 324 | RPV/TAF/FTC | <40 | 47,100 | 10.4 | 39:05:01G + 48:01:01G |
| ICB2651 | 52 | M | Caucasian | 2001 | 655 | 37 | 681 | 39 | 275 | ABC/DTG/3TC | <40 | 45,069 | 14 | 14:02:01G + 14:02:01G |
| ICB3147 | 61 | M | Hispanic/Latino | 1993 | 837 | 44 | 522 | 27 | 4 | ABC/DTG/3TC | <40 | 119,870 | 11 | 44:02:01G + 52:01:01G |
| ICB3162 | 56 | M | Caucasian | 1987 | 586 | 37 | 471 | 30 | 200 | DRV, RTV, ABC/DTG/3TC | <40 | 171,000 | 11.5 | 07:02:01G + 51:09:01G |
| ICB5003 | 47 | M | Caucasian | 1993 | 279 | 25 | 385 | 35 | 56 | ATV, ABC/DTG/3TC | <40 | 171,000 | 6.7 | 39:01:01G + 52:01:01G |
| LKA04 | 56 | M | Caucasian | 1977 | 769 | 35 | 530 | 24 | 230 | ABC/3TC/DTG | <20 | NA | 28 | 18:01:01G + 51:01:01G |
| LKA09 | 51 | M | Caucasian | 1997 | 372 | 27 | 771 | 56 | 72 | TDF/3TC, ATV, RAL | <20 | 199,100 | 13.49 | 07:02:01G + 50:01:01G |
| LKA18 | 60 | M | Caucasian | 1993 | 312 | 19 | 312 | 34 | 2 | TAF/FTC | <20 | NA | 12 | 41:02:01G + 44:15:01G |
| PRA01 | 64 | M | Caucasian | 1985 | 403 | 24 | 1061 | 63 | 10 | ATV, TDF/FTC | <20 | 148,430 | 14.1 | 08:01:01G + 44:02:01G |
| PRA02 | 48 | M | Caucasian | 2006 | 1460 | 47 | 793 | 26 | 698 | ABC/3TC, EFV | <20 | NA | 12 | 27:05:02G + 39:01:01G |
| PRA04 | 55 | M | Caucasian | 1996 | 1036 | 40 | 1069 | 42 | 266 | ATV, TDF/FTC | <20 | 100,000 | 11.1 | 35:01:01G + 55:01:01G |
| PRA05 | 49 | M | Caucasian | 2003 | 388 | 28 | 717 | 51 | 168 | TAF/FTC, MVC | <20 | 146,000 | 12 | 08:01:01G + 08:01:01G |
| PRA08 | 38 | M | Other (PNG) | 2006 | 281 | 25 | 328 | 30 | 168 | EVG/TAF/FTC/COBI | <20 | 63,300 | 13 | 40:01:01G + 40:02:01G |
| PRA09 | 49 | M | Caucasian | 2010 | 474 | 25 | 1085 | 56 | 42 | EVG/TAF/FTC/COBI | <20 | 211,930 | 7 | 35:01:01G + 51:01:01G |
| PRA10 | 48 | M | Caucasian | 2000 | 484 | 28 | 895 | 52 | 411 | TAF, FTC, RPV | <20 | N/A 2 | N/A | 40:01:01G + 50:01:01G |
| 2256 | 62 | M | Caucasian | 1985 | 310 | 25 | 550 | 45 | 86 | RPV/TAF/FTC, TCV | <40 | 29,900 | 13.2 | 15:02 + 40:01 |
| 2669 | 59 | M | Caucasian | 1989 | 420 | 24 | 672 | 38 | 180 | ABC/TCV/3TC | <40 | 900,000 | 8.39 | 14:02 + 44:03 |
| 2750 | 56 | M | Caucasian | 2005 | 474 | 36 | 398 | 30 | 190 | RPV/TAF/FTC | <40 | 175,000 | 2.98 | 08:01:01G + 13:02 |
| 2781 | 42 | M | Caucasian | 2009 | 433 | 27 | 714 | 44 | 267 | ABC/TCV/3TC | <40 | 187,090 | 2.2 | N/A |
| 7705 | 63 | M | African American | 1987 | 594 | 39 | 531 | 35 | 300 | ATV, RTV, FTC/TAF | <40 | N/A | 10 | N/A |
| 7726 | 56 | M | Caucasian | 1986 | 679 | 32 | 938 | 45 | 235 | BIC/FTC/TAF | <40 | N/A | 8.36 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tumpach, C.; Rhodes, A.; Kim, Y.; Ong, J.; Liu, H.; Chibo, D.; Druce, J.; Williamson, D.; Hoh, R.; Deeks, S.G.; et al. Adaptation of Droplet Digital PCR-Based HIV Transcription Profiling to Digital PCR and Association of HIV Transcription and Total or Intact HIV DNA. Viruses 2023, 15, 1606. https://doi.org/10.3390/v15071606
Tumpach C, Rhodes A, Kim Y, Ong J, Liu H, Chibo D, Druce J, Williamson D, Hoh R, Deeks SG, et al. Adaptation of Droplet Digital PCR-Based HIV Transcription Profiling to Digital PCR and Association of HIV Transcription and Total or Intact HIV DNA. Viruses. 2023; 15(7):1606. https://doi.org/10.3390/v15071606
Chicago/Turabian StyleTumpach, Carolin, Ajantha Rhodes, Youry Kim, Jesslyn Ong, Haoming Liu, Doris Chibo, Julian Druce, Deborah Williamson, Rebecca Hoh, Steven G. Deeks, and et al. 2023. "Adaptation of Droplet Digital PCR-Based HIV Transcription Profiling to Digital PCR and Association of HIV Transcription and Total or Intact HIV DNA" Viruses 15, no. 7: 1606. https://doi.org/10.3390/v15071606
APA StyleTumpach, C., Rhodes, A., Kim, Y., Ong, J., Liu, H., Chibo, D., Druce, J., Williamson, D., Hoh, R., Deeks, S. G., Yukl, S. A., Roche, M., Lewin, S. R., & Telwatte, S. (2023). Adaptation of Droplet Digital PCR-Based HIV Transcription Profiling to Digital PCR and Association of HIV Transcription and Total or Intact HIV DNA. Viruses, 15(7), 1606. https://doi.org/10.3390/v15071606







