SARS-CoV-2 Serological Investigation of White-Tailed Deer in Northeastern Ohio
Abstract
1. Introduction
2. Materials and Methods
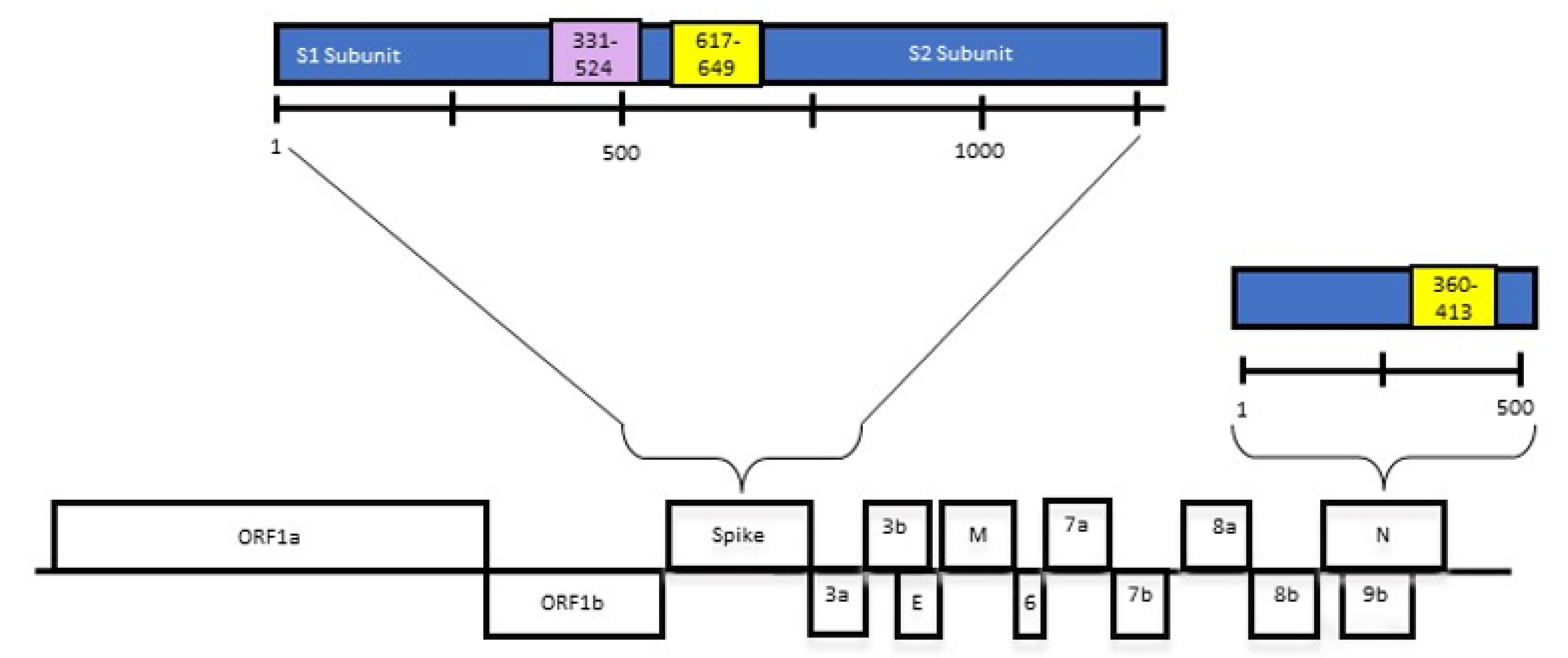
2.1. Recombinant Proteins
2.2. Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.3. Western Blotting
2.4. ELISA
2.5. Reference Viruses, Sera, Anti-Sera, and Antibodies
2.6. 50%. Plaque-Reduction Neutralization Test (PRNT50)
2.7. Virus Neutralization Test (VNT)
2.8. Deer Sample Collection
3. Results
3.1. Expression Constructs and Expression of Recombinant Proteins
3.2. ELISA Development and Optimization
3.3. ELISA Testing for CoV S and N Protein Cross-Reactivity
3.4. ELISA Results in Deer Samples
3.5. ELISA Epitope Sequence Stability
3.6. Neutralizing Antibody Titer Results in Deer Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medicine, J.H.U. Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/us-map (accessed on 1 July 2023).
- WHO. World Health Organization Coronavirus (COVID-19) Dashboard. 2023. Available online: https://covid19.who.int (accessed on 1 July 2023).
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [PubMed]
- Geographic, N. More Animal Species are Getting COVID-19 for the First Time. Available online: https://www.nationalgeographic.com/animals/article/more-animal-species-are-getting-covid-19-for-the-first-time (accessed on 1 July 2023).
- Murphy, H.L.; Ly, H. Understanding the prevalence of SARS-CoV-2 (COVID-19) exposure in companion, captive, wild, and farmed animals. Virulence 2021, 12, 2777–2786. [Google Scholar] [CrossRef]
- APHIS, U. Confirmed Cases of SARS-CoV-2 in Animals in the United States. Available online: https://www.aphis.usda.gov/aphis/dashboards/tableau/sars-dashboard (accessed on 1 July 2023).
- Francisco, R.; Hernandez, S.M.; Mead, D.G.; Adcock, K.G.; Burke, S.C.; Nemeth, N.M.; Yabsley, M.J. Experimental Susceptibility of North American Raccoons (Procyon lotor) and Striped Skunks (Mephitis mephitis) to SARS-CoV-2. Front. Vet. Sci. 2021, 8, 715307. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Lu, S.; Zhao, Y.; Yu, W.; Yang, Y.; Gao, J.; Wang, J.; Kuang, D.; Yang, M.; Yang, J.; et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct. Target. Ther. 2020, 5, 157. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Lamers, M.M.; Okba, N.M.; Breugem, T.I.; Schipper, D.; van den Doel, P.B.; van Run, P.; van Amerongen, G.; de Waal, L.; Koopmans, M.P.; et al. Susceptibility of rabbits to SARS-CoV-2. Emerg. Microbes Infect. 2021, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.V.; Martins, M.; Falkenberg, S.; Buckley, A.; Caserta, L.C.; Mitchell, P.K.; Cassmann, E.D.; Rollins, A.; Zylich, N.C.; Renshaw, R.W.; et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 2021, 95, 10–1128. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.M.; Chin, A.W.; Fung, K.; Choy, K.T.; Wong, A.Y.; Kaewpreedee, P.; Perera, R.A.; Poon, L.L.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020. 583, 834–838. [CrossRef]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Penaranda, S.; Bankamp, B.; Maher, K.; Chen, M.H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–13999. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Drosten, C.; Muller, M.A. Serological assays for emerging coronaviruses: Challenges and pitfalls. Virus Res. 2014, 194, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Infantino, M.; Damiani, A.; Gobbi, F.L.; Grossi, V.; Lari, B.; Macchia, D.; Casprini, P.; Veneziani, F.; Villalta, D.; Bizzaro, N.; et al. Serological Assays for SARS-CoV-2 Infectious Disease: Benefits, Limitations and Perspectives. Isr. Med. Assoc. J. 2020, 22, 203–210. [Google Scholar]
- Krahling, V.; Halwe, S.; Rohde, C.; Becker, D.; Berghoefer, S.; Dahlke, C.; Eickmann, M.; Ercanoglu, M.S.; Gieselmann, L.; Herwig, A.; et al. Development and characterization of an indirect ELISA to detect SARS-CoV-2 spike protein-specific antibodies. J. Immunol. Methods 2021, 490, 112958. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2021, 602, 481–486. [Google Scholar] [CrossRef]
- Cool, K.; Gaudreault, N.N.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; McDowell, C.; Carossino, M.; Bold, D.; Mitzel, D.; Kwon, T.; et al. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg. Microbes Infect. 2022, 11, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Palermo, P.M.; Orbegozo, J.; Watts, D.M.; Morrill, J.C. SARS-CoV-2 Neutralizing Antibodies in White-Tailed Deer from Texas. Vector Borne Zoonotic Dis. 2022, 22, 62–64. [Google Scholar] [CrossRef]
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl. Acad. Sci. USA 2021, 118, e2114828118. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Surendran-Nair, M.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; Jayarao, B.M.; Maranas, C.D.; et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc. Natl. Acad. Sci. 2022, 119, e2121644119. [Google Scholar] [CrossRef]
- Pickering, B.; Lung, O.; Maguire, F.; Kruczkiewicz, P.; Kotwa, J.D.; Buchanan, T.; Gagnier, M.; Guthrie, J.L.; Jardine, C.M.; Marchand-Austin, A.; et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat. Microbiol. 2022, 7, 2011–2024. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Zhang, X.; Hasoksuz, M.; Nagesha, H.S.; Haynes, L.M.; Fang, Y.; Lu, S.; Saif, L.J. Two-way antigenic cross-reactivity between severe acute respiratory syndrome coronavirus (SARS-CoV) and group 1 animal CoVs is mediated through an antigenic site in the N-terminal region of the SARS-CoV nucleoprotein. J. Virol. 2007, 81, 13365–13377. [Google Scholar] [CrossRef]
- Frey, A.; Di Canzio, J.; Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 1998, 221, 35–41. [Google Scholar] [CrossRef]
- Case, J.B.; Bailey, A.L.; Kim, A.S.; Chen, R.E.; Diamond, M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 2020, 548, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Che, X.Y.; Qiu, L.W.; Liao, Z.Y.; Wang, Y.D.; Wen, K.; Pan, Y.X.; Hao, W.; Mei, Y.B.; Cheng, V.C.; Yuen, K.Y. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005, 191, 2033–2037. [Google Scholar] [CrossRef]
- Sun, Z.F.; Meng, X.J. Antigenic cross-reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group I animal coronaviruses: Implication for SARS diagnosis. J. Clin. Microbiol. 2004, 42, 2351–2352. [Google Scholar] [CrossRef] [PubMed]
- Xiang, F.; Wang, X.; He, X.; Peng, Z.; Yang, B.; Zhang, J.; Zhou, Q.; Ye, H.; Ma, Y.; Li, H.; et al. Antibody Detection and Dynamic Characteristics in Patients with COVID-19. Clin. Infect. Dis. 2020, 71, 1930–1934. [Google Scholar] [CrossRef]
- Maache, M.; Komurian-Pradel, F.; Rajoharison, A.; Perret, M.; Berland, J.L.; Pouzol, S.; Bagnaud, A.; Duverger, B.; Xu, J.; Osuna, A.; et al. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid-based western blot assay were rectified by the use of two subunits (S1 and S2) of spike for detection of antibody to SARS-CoV. Clin. Vaccine Immunol. 2006, 13, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; Shen, H.; Jiang, L.; Zhang, M.; Xiao, D.; Liu, Y.; Ma, X.; Zhang, Y.; Guo, N.; et al. Cross-reaction of SARS-CoV antigen with autoantibodies in autoimmune diseases. Cell Mol. Immunol. 2004, 1, 304–307. [Google Scholar]
- Liu, W.; Liu, L.; Kou, G.; Zheng, Y.; Ding, Y.; Ni, W.; Wang, Q.; Tan, L.; Wu, W.; Tang, S.; et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef]
- Okba, N.M.A.; Raj, V.S.; Widjaja, I.V.Y.; GeurtsvanKessel, C.H.; De Bruin, E.; Chandler, F.D.; Park, W.B.; Kim, N.J.; Farag, E.A.; Al-Hajri, M.; et al. Sensitive and Specific Detection of Low-Level Antibody Responses in Mild Middle East Respiratory Syndrome Coronavirus Infections. Emerg. Infect. Dis. 2019, 25, 1868–1877. [Google Scholar] [CrossRef]
- Perera, R.A.; Mok, C.K.; Tsang, O.T.; Lv, H.; Ko, R.L.; Wu, N.C.; Yuan, M.; Leung, W.S.; Chan, J.M.; Chik, T.S.; et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill 2020, 25, 2000421. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Sidney, J.; Zhang, Y.; Scheuermann, R.H.; Peters, B.; Sette, A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe 2020, 27, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Muller, K.; Girl, P.; von Buttlar, H.; Dobler, G.; Wölfel, R. Comparison of two commercial surrogate ELISAs to detect a neutralising antibody response to SARS-CoV-2. J. Virol. Methods 2021, 292, 114122. [Google Scholar] [CrossRef] [PubMed]
- Robbiani, D.F.; Gaebler, C.; Muecksch, F.; Lorenzi, J.C.; Wang, Z.; Cho, A.; Agudelo, M.; Barnes, C.O.; Gazumyan, A.; Finkin, S.; et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020, 584, 437–442. [Google Scholar] [CrossRef]
- Nayak, K.; Gottimukkala, K.; Kumar, S.; Reddy, E.S.; Edara, V.V.; Kauffman, R.; Floyd, K.; Mantus, G.; Savargaonkar, D.; Goel, P.K.; et al. Characterization of neutralizing versus binding antibodies and memory B cells in COVID-19 recovered individuals from India. Virology 2021, 558, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; West, A.P.; Huey-Tubman, K.E.; Hoffmann, M.A.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842. [Google Scholar] [CrossRef]
- Marques, A.D.; Sherrill-Mix, S.; Everett, J.K.; Adhikari, H.; Reddy, S.; Ellis, J.C.; Zeliff, H.; Greening, S.S.; Cannuscio, C.C.; Strelau, K.M.; et al. Multiple Introductions of SARS-CoV-2 Alpha and Delta Variants into White-Tailed Deer in Pennsylvania. medRxiv 2022, 13, e02101-22. [Google Scholar] [CrossRef]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488. [Google Scholar] [CrossRef]
- Agarwal, V.; Venkatakrishnan, A.J.; Puranik, A.; Kirkup, C.; Lopez-Marquez, A.; Challener, D.W.; Theel, E.S.; O’Horo, J.C.; Binnicker, M.J.; Kremers, W.K.; et al. Long-term SARS-CoV-2 RNA shedding and its temporal association to IgG seropositivity. Cell Death Discov. 2020, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Zhao, J.; Zeng, H.L.; Lu, Q.B.; Fang, L.Q.; Wang, Q.; Wu, Q.M.; Liu, W. Prolonged viral shedding of SARS-CoV-2 and related factors in symptomatic COVID-19 patients: A prospective study. BMC Infect. Dis. 2021, 21, 1282. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Waterborne Disease & Outbreak Surveillance Reporting. 2020. Available online: https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html#:~:text=While%20SARS%2DCoV%2D2%20can,to%20treated%20or%20untreated%20wastewater (accessed on 1 July 2023).





| Disease | Full Name | ID | Host | Source | Type | Assay |
|---|---|---|---|---|---|---|
| BCoV | Bovine coronavirus | B-12272 B574 | Calf | LJS | HS | S and N |
| BCoV | Bovine coronavirus | Gp#99-10 Mebus | GP | LJS | HS | S and N |
| BtCoV | Bat coronavirus | HKU5.5N | Mouse | LJS | HS | S and N |
| CCoV | Canine coronavirus | 2CoV UDC1 | GP | LJS | HS | S and N |
| FCoV | Feline coronavirus | 79-1146 | GP | LJS | HS | S and N |
| HCoV | Human coronavirus | NL-63 | GP | LJS | HS | S and N |
| IBV | Infectious bronchitis virus | M41 | Chicken | LJS | HS | S and N |
| MERS | Middle East respiratory syndrome | PA5-81778 | Rabbit | TF | Ab | N |
| PDCoV | Porcine deltacoronavirus | DC97 | GP | LJS | HS | S and N |
| PDCoV | Porcine deltacoronavirus | DC163 | Pig (Gn) | LJS | Serum | S and N |
| PDCoV | Porcine deltacoronavirus | DC173 | Pig (Gn) | LJS | Serum | S and N |
| PEDV | Porcine epidemic diarrhea virus | PV1610 | Pig (Gn) | QW | Serum | S and N |
| PEDV | Porcine epidemic diarrhea virus | PV1736 | Pig (Gn) | QW | Serum | S and N |
| PRCV | Porcine respiratory coronavirus | ISU-1 PP12 | Pig (Gn) | LJS | HS | S and N |
| SARS | Severe acute respiratory syndrome | VCR830L | Mouse | LJS | HS | S |
| SARS | Severe acute respiratory syndrome | Anti-S COV50 | Mouse | LJS | HS | S |
| SARS | Severe acute respiratory syndrome | Urbani | Mouse | LJS | HS | S |
| SARS | Severe acute respiratory syndrome | NR-5469 | Rabbit | BEI | HS | S |
| SARS | Severe acute respiratory syndrome | NR-10361 | GP | BEI | HS | N |
| SARS | Severe acute respiratory syndrome | NRC-2146 | Calf | LJS | HS | S and N |
| SARS | Severe acute respiratory syndrome | 40150-T62 | Rabbit | Sino | Ab | S |
| SARS 2 | Severe acute respiratory syndrome 2 | 40588-T62 | Rabbit | Sino | Ab | N |
| TGEV | Transmissible gastroenteritis virus | Purdue-ATCC | GP | LJS | HS | S and N |
| TGEV | Transmissible gastroenteritis virus | Purdue-ATCC | Pig (Gn) | LJS | HS | S and N |
| TGEV | Transmissible gastroenteritis virus | M2 H5 | Pig (Gn) | LJS | HS | S and N |
| TGEV | Transmissible gastroenteritis virus | MM973 | Pig (Gn) | LJS | HS | S and N |
| Sample # | Deer Tag # | ELISA Titer | PRNT50 Titer | pVNT | PCR+ |
|---|---|---|---|---|---|
| 1109 | 009 | 1:640 | Neg | Neg | NT |
| 1110 | 010 | 1:320 | Neg | Neg | NT |
| 1111 | 011 | 1:10 | Neg | Neg | NT |
| 1113 | 013 | 1:40 | Neg | Neg | NT |
| 1115 | 015 | 1:20 | 16 | 723.1 | NT |
| 1116 | 016 | 1:10 | 16 | 1021.9 | NT |
| 1118 | 018 | 1:10 | 64 | 1047.3 | NT |
| 1142 | 042 | 1:640 | 4 | 91.8 | NT |
| 1148 | 048 | 1:10 | Neg | <80 | NT |
| 1150 | 050 | 1:320 | 16; 40 b | 285.3 | NT |
| 1162 | 062 | 1:640 | 4 | 252.9 | NT |
| 1288 | 188 | Neg | Neg | Neg | Neg |
| 1325 | 225 | 1:2560 | Neg | <80 | Neg |
| 1392 | 292 | Neg | 64 | 661.8 | Pos |
| 1435 | 335 | 1:640 | 40 | <80 | Pos |
| 1581 | 7 a | Neg | Neg | Neg | NT |
| 1601 | 148 a | Neg | Neg | Neg | NT |
| Date of Collection | Samples | Positive | Estimated Seroprevalence | Upper 95% CI | Lower 95% CI | |
|---|---|---|---|---|---|---|
| Site 1 | 2021-02-02 | 41 | 3 | 7.3% | 19.43% | 2.52% |
| 2021-02-25 | 33 | 2 | 6.1% | 19.61% | 1.07% | |
| Site 2 | 2021-01-27 | 4 | 3 | 75% | 98.71% | 30.06% |
| 2021-03-03 | 16 | 5 | 31.3% | 55.60% | 14.17% | |
| Site 3 | 2021-02-04 | 14 | 3 | 21.4% | 47.59% | 7.57% |
| 2021-03-02 | 24 | 3 | 12.5% | 31.0% | 4.34% | |
| Site 4 | 2021-01-26 | 14 | 5 | 35.7% | 61.24% | 16.35% |
| 2021-02-17 | 16 | 2 | 12.5% | 36.02% | 2.22% | |
| 2021-03-09 | 8 | 2 | 25% | 59.07% | 4.44% | |
| Site 5 | 2021-01-19 | 11 | 3 | 27.3% | 56.57% | 9.75% |
| 2021-01-20 | 10 | 0 | 0% | 27.75% | 0% | |
| 2021-01-21 | 32 | 6 | 18.8% | 35.31% | 8.89% | |
| 2021-01-25 | 20 | 1 | 5% | 23.61% | 0.26% | |
| 2021-02-09 | 8 | 2 | 25% | 59.08% | 4.44% | |
| 2021-03-08 | 25 | 3 | 12% | 29.96% | 4.17% | |
| Site 6 | 2021-01-28 | 33 | 16 | 48.5% | 64.78% | 32.50% |
| 2021-03-04 | 15 | 1 | 6.7% | 29.82% | 0.34% | |
| Site 7 | 2021-02-22 | 20 | 3 | 15% | 36.04% | 5.24% |
| 2021-02-23 | 19 | 3 | 15.8% | 37.57% | 5.52% | |
| 2021-02-24 | 50 | 7 | 14% | 26.19% | 6.95% | |
| Site 8 | 2020-12-09 | 5 | 3 | 60% | 92.89% | 23.07% |
| 2021-02-16 | 13 | 2 | 15.4% | 42.24% | 2.73% | |
| Site 9 | 2021-02-01 | 22 | 2 | 9.1% | 27.82% | 1.62% |
| 2021-03-01 | 10 | 0 | 0% | 27.75% | 0% | |
| Site 10 | 2020-11-24 | 9 | 2 | 22.2% | 54.74% | 3.95% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boley, P.A.; Dennis, P.M.; Faraone, J.N.; Xu, J.; Liu, M.; Niu, X.; Gibson, S.; Hale, V.; Wang, Q.; Liu, S.-L.; et al. SARS-CoV-2 Serological Investigation of White-Tailed Deer in Northeastern Ohio. Viruses 2023, 15, 1603. https://doi.org/10.3390/v15071603
Boley PA, Dennis PM, Faraone JN, Xu J, Liu M, Niu X, Gibson S, Hale V, Wang Q, Liu S-L, et al. SARS-CoV-2 Serological Investigation of White-Tailed Deer in Northeastern Ohio. Viruses. 2023; 15(7):1603. https://doi.org/10.3390/v15071603
Chicago/Turabian StyleBoley, Patricia A., Patricia M. Dennis, Julia N. Faraone, Jiayu Xu, Mingde Liu, Xiaoyu Niu, Stormy Gibson, Vanessa Hale, Qiuhong Wang, Shan-Lu Liu, and et al. 2023. "SARS-CoV-2 Serological Investigation of White-Tailed Deer in Northeastern Ohio" Viruses 15, no. 7: 1603. https://doi.org/10.3390/v15071603
APA StyleBoley, P. A., Dennis, P. M., Faraone, J. N., Xu, J., Liu, M., Niu, X., Gibson, S., Hale, V., Wang, Q., Liu, S.-L., Saif, L. J., & Kenney, S. P. (2023). SARS-CoV-2 Serological Investigation of White-Tailed Deer in Northeastern Ohio. Viruses, 15(7), 1603. https://doi.org/10.3390/v15071603












