Nicotinamide Efficiently Suppresses Porcine Epidemic Diarrhea Virus and Porcine Deltacoronavirus Replication
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Viruses and Drugs
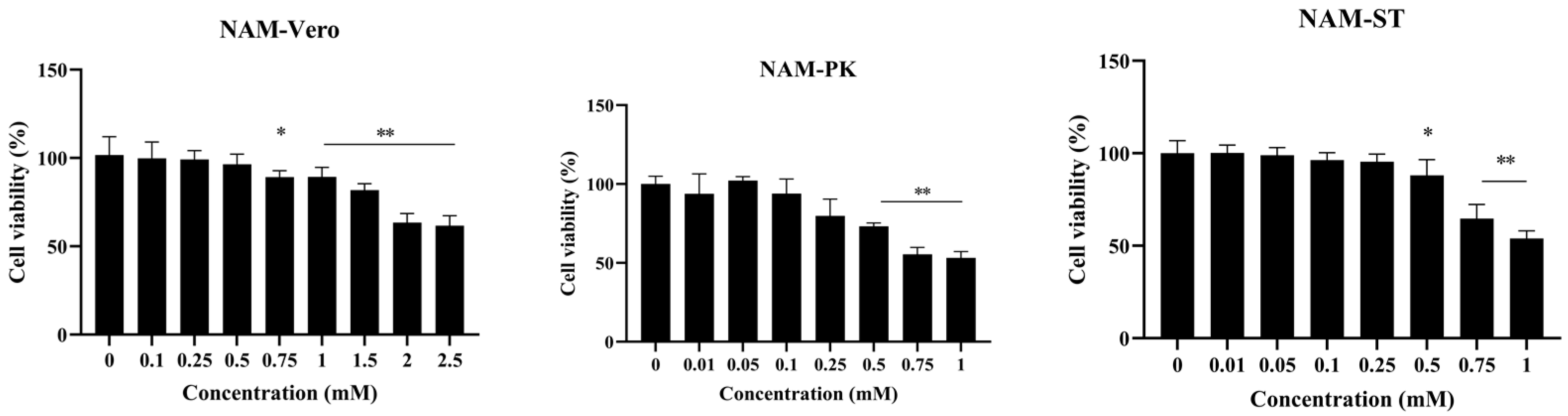
2.2. Cell Viability Assay (Cytotoxicity)
2.3. Quantitative Real-Time Reverse-Transcription PCR (RT-qPCR)
2.4. Western Blotting
2.5. Indirect Immunofluorescence Assay (IFA)
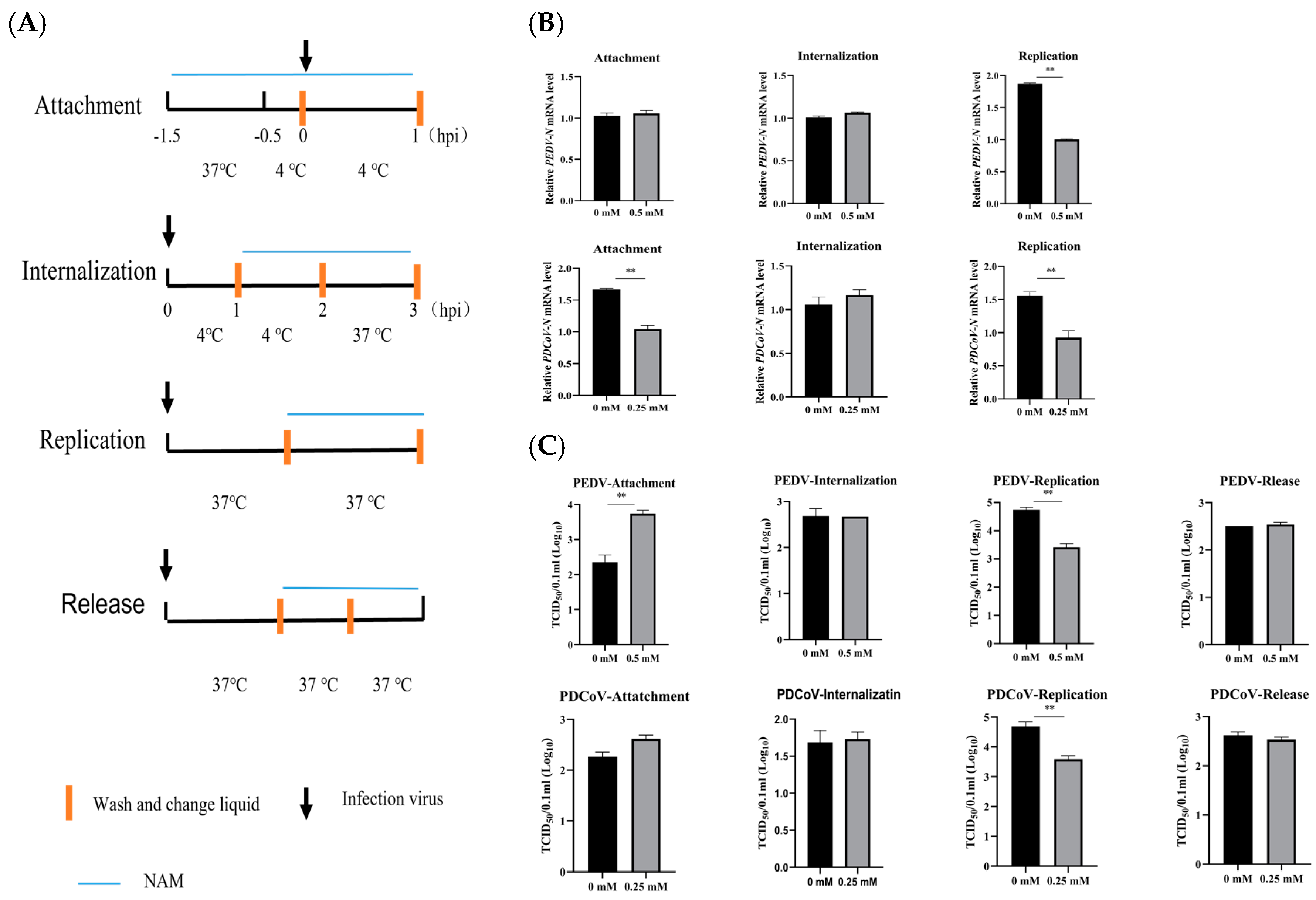
2.6. Investigation of the Effects of NAM on the Viral Replication Cycle
2.7. NAM Antiviral Activity Assay
2.8. Statistical Analyses
3. Results
3.1. Minimal Cytotoxicity of NAM
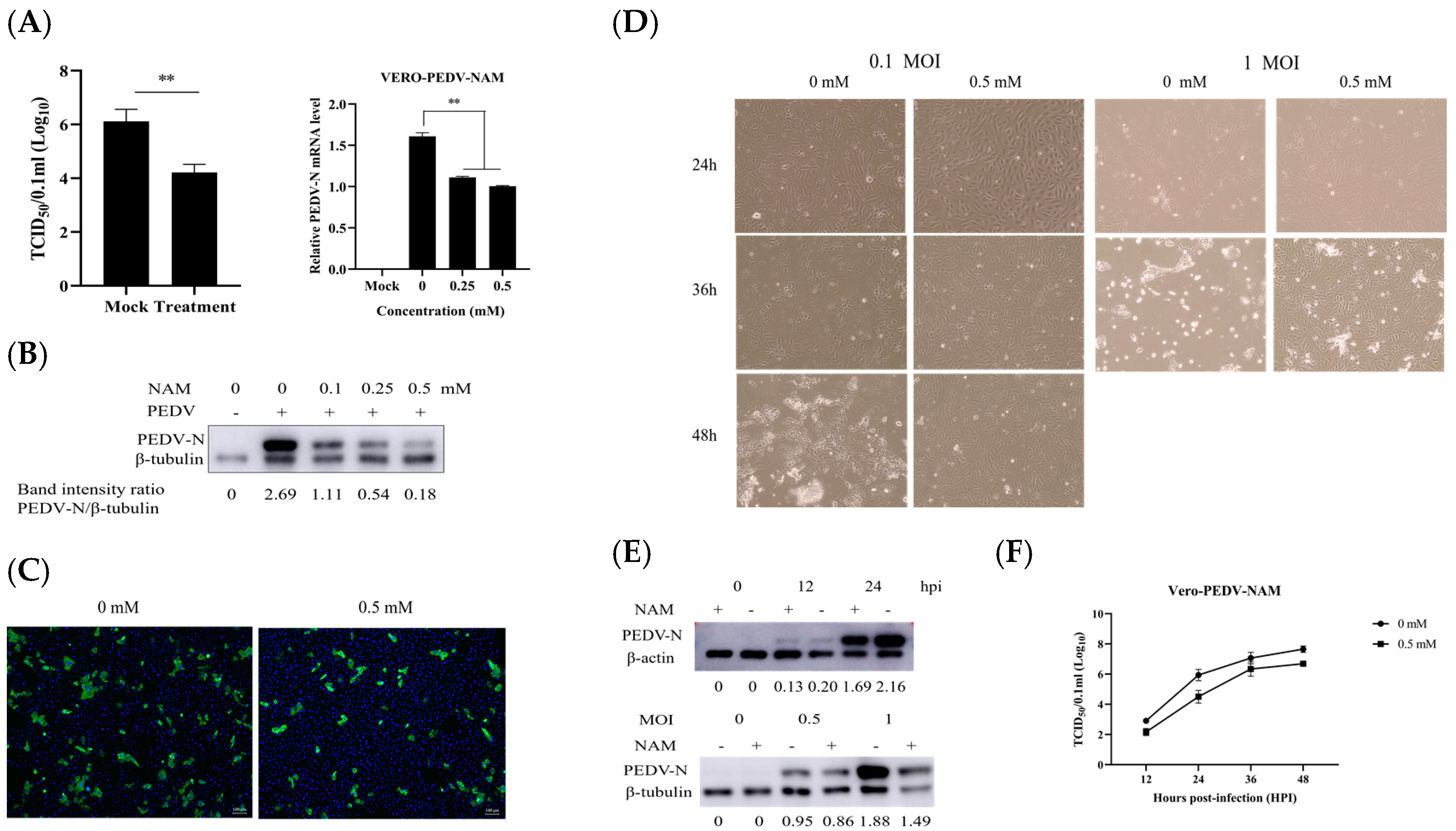
3.2. NAM Exhibits Antiviral Activity against PEDV
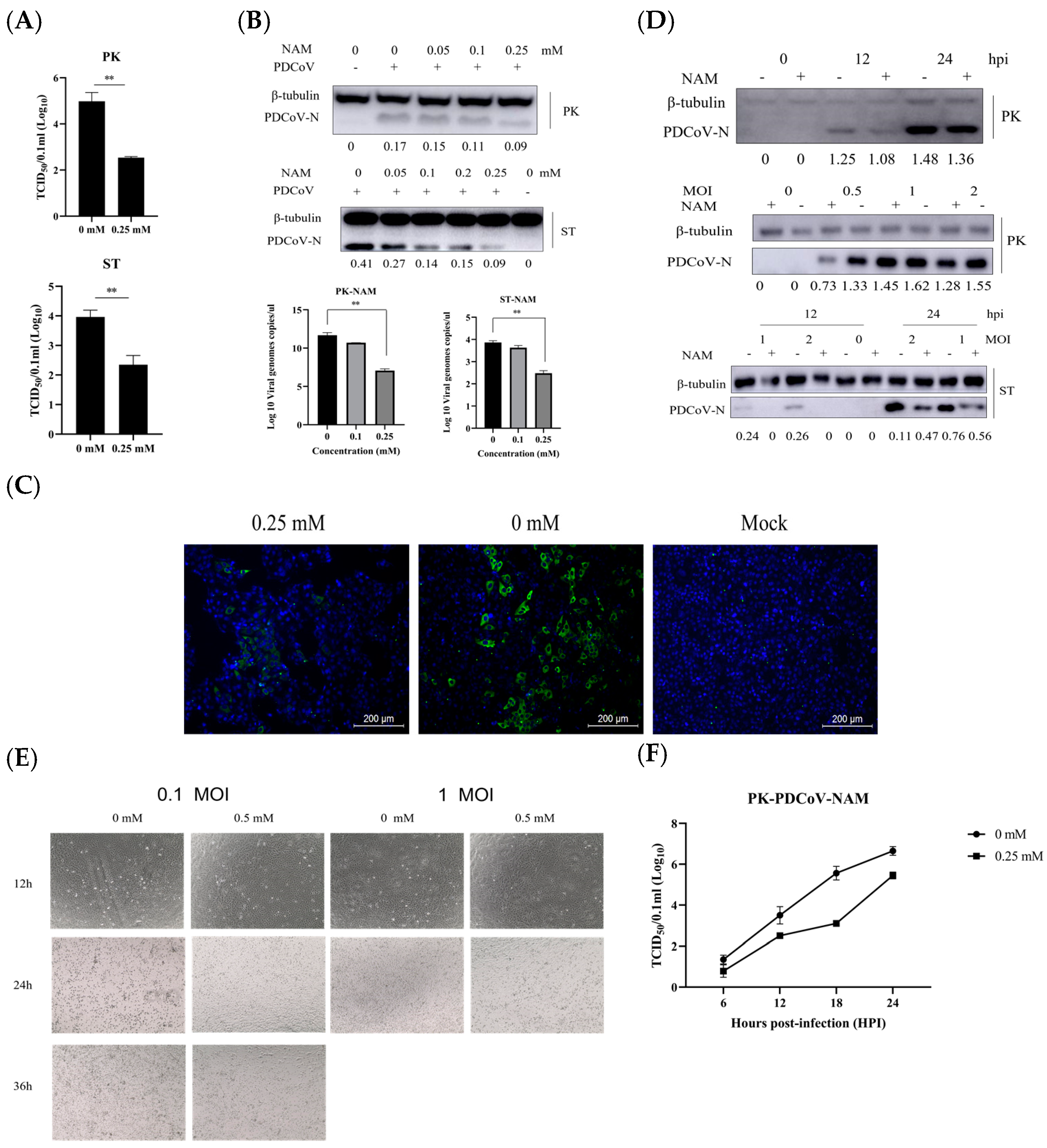
3.3. NAM Exhibits Antiviral Activity against PDCoV Infection
3.4. NAM Inhibits the Synthesis of Viral RNA
3.5. Evaluation of the Therapeutic Effect of NAM
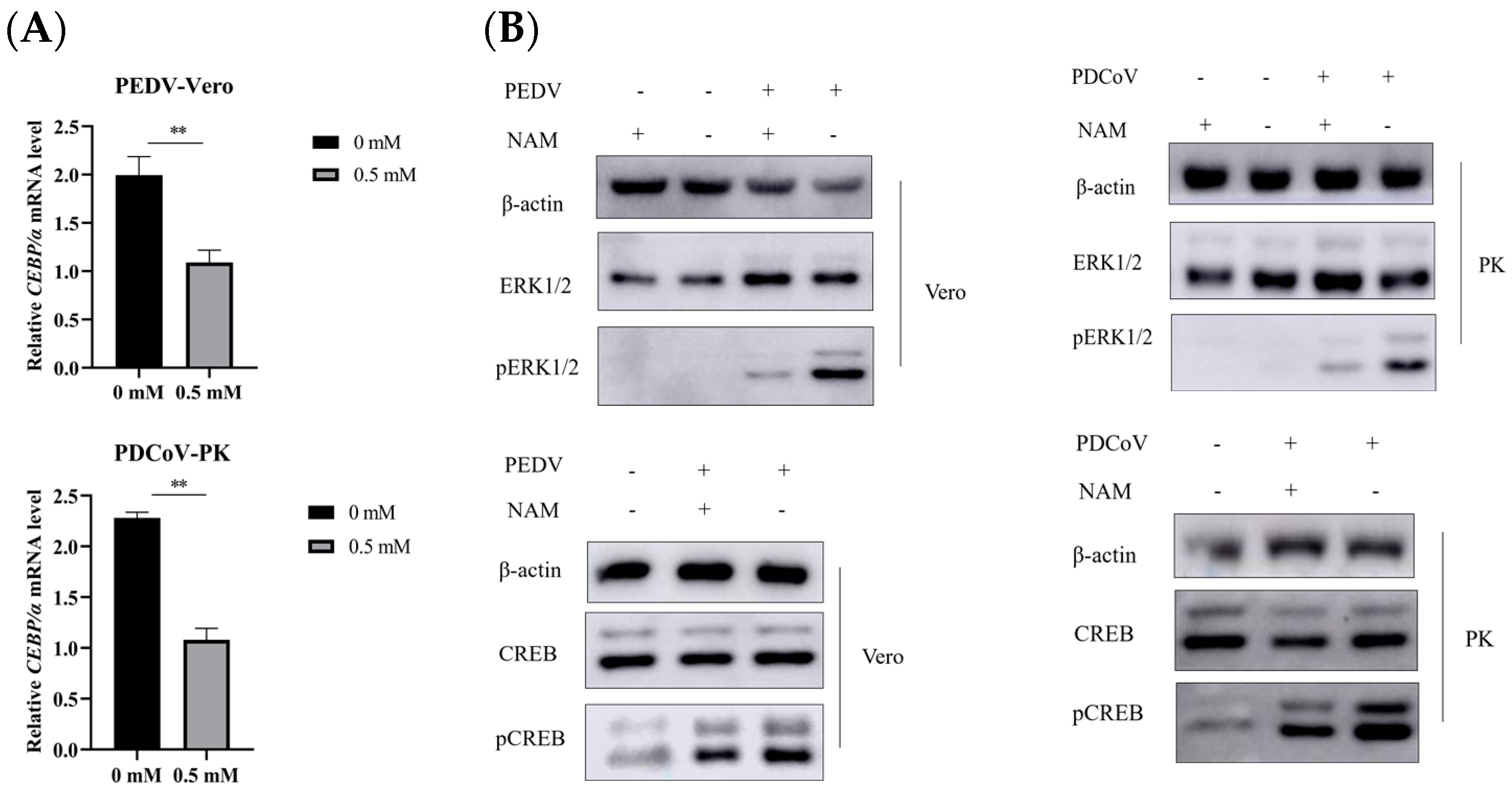
3.6. Suppression of ERK1/2/MAPK Activity by NAM
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef]
- Wood, E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977, 100, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Liang, X.; Oglesbee, M.; Krakowka, S.; Niehaus, A.; Wang, G.; Jia, A.; Song, H.; Li, J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet. Microbiol. 2016, 186, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Wang, N.; Jiao, H.; Zhang, J.; Li, C.; Ren, W.; Reiter, R.J.; Su, S. Melatonin and other indoles show antiviral activities against swine coronaviruses in vitro at pharmacological concentrations. J. Pineal Res. 2021, 71, e12754. [Google Scholar] [CrossRef]
- Maiese, K.; Chong, Z.Z.; Hou, J.; Shang, Y.C. The Vitamin Nicotinamide: Translating Nutrition into Clinical Care. Molecules 2009, 14, 3446–3485. [Google Scholar] [CrossRef]
- Jackson, T.M.; Rawling, J.M.; Roebuck, B.D.; Kirkland, J.B. Large supplements of nicotinic acid and nicotinamide increase tissue NAD+ and poly(ADP-ribose) levels but do not affect diethylnitrosamine-induced altered hepatic foci in Fischer-344 rats. J. Nutr. 1995, 125, 1455–1461. [Google Scholar]
- Li, F.; Chong, Z.Z.; Maiese, K. Navigating novel mechanisms of cellular plasticity with the NAD+ precursor and nutrient nicotinamide. Front. Biosci. 2004, 9, 2500–2520. [Google Scholar] [CrossRef]
- Magni, G.; Amici, A.; Emanuelli, M.; Orsomando, G.; Raffaelli, N.; Ruggieri, S. Enzymology of NAD+ homeostasis in man. Cell Mol. Life Sci. 2004, 61, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Lautrup, S.; Hou, Y.; Demarest, T.G.; Croteau, D.L.; Mattson, M.P.; Bohr, V.A. NAD+ in Aging: Molecular Mechanisms and Translational Implications. Trends Mol. Med. 2017, 23, 899–916. [Google Scholar] [CrossRef] [PubMed]
- Fania, L.; Mazzanti, C.; Campione, E.; Candi, E.; Abeni, D.; Dellambra, E. Role of Nicotinamide in Genomic Stability and Skin Cancer Chemoprevention. Int. J. Mol. Sci. 2019, 20, 5946. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Kudo, N.; Thelen, J.N.; Ito, A.; Yoshida, M.; Denu, J.M. Kinetic and Structural Basis for Acyl-Group Selectivity and NAD+ Dependence in Sirtuin-Catalyzed Deacylation. Biochemistry 2015, 54, 3037–3050. [Google Scholar] [CrossRef] [PubMed]
- Maiese, K. Nicotinamide as a Foundation for Treating Neurodegenerative Disease and Metabolic Disorders. Curr. Neurovascular Res. 2021, 18, 134–149. [Google Scholar] [CrossRef]
- Hwang, E.S.; Song, S.B. Possible Adverse Effects of High-Dose Nicotinamide: Mechanisms and Safety Assessment. Biomolecules 2020, 10, 687. [Google Scholar] [CrossRef]
- Feily, A.; Omidian, M.; Khazanee, A.; Yaghoobi, R.; Ghorbani, A.R.; Pazyar, N.; Beladimousavi, S.S.; Ghadimi, M.; Mohebbipour, A. Therapeutic effect of oral nicotinamide on refractory uremic pruritus: A randomized, double-blind study. Saudi J. Kidney Dis. Transplant. 2013, 24, 995–999. [Google Scholar] [CrossRef]
- Murray, M.F. Nicotinamide: An Oral Antimicrobial Agent with Activity against Both Mycobacterium tuberculosis and Human Immunodeficiency Virus. Clin. Infect. Dis. 2003, 36, 453–460. [Google Scholar] [CrossRef]
- Krasil’nikov, I.I.; Kalnina, L.B.; Korneeva, M.N.; Nosik, D.N.; Zlobin, A.; Vladimirov, V.G.; L’Vov, D.K. Inhibitors of ADP ribosylation as antiviral agents: An experimental study in a model of HIV infection. Vopr. Virusol. 1991, 36, 216–218. [Google Scholar]
- Li, W.-Y.; Ren, J.-H.; Tao, N.-N.; Ran, L.-K.; Chen, X.; Zhou, H.-Z.; Liu, B.; Li, X.-S.; Huang, A.-L.; Chen, J. The SIRT1 inhibitor, nicotinamide, inhibits hepatitis B virus replication in vitro and in vivo. Arch. Virol. 2016, 161, 621–630. [Google Scholar] [CrossRef]
- Child, S.J.; Franke, C.A.; Hruby, D.E. Inhibition of vaccinia virus replication by nicotinamide: Evidence for ADP-ribosylation of viral proteins. Virus Res. 1988, 9, 119–132. [Google Scholar] [CrossRef]
- Déry, C.V.; de Murcia, G.; Lamarre, D.; Morin, N.; Poirier, G.G.; Weber, J. Possible role of ADP-ribosylation of adenovirus core proteins in virus infection. Virus Res. 1986, 4, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Deng, Y.; Pang, H.; Ma, T.; Ye, Q.; Chen, Q.; Chen, H.; Hu, Z.; Qin, C.-F.; Xu, Z. Treatment of SARS-CoV-2-induced pneumonia with NAD+ and NMN in two mouse models. Cell Discov. 2022, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Ge, X.; Wang, J.; Wei, Z.; Feng, W.-H.; Wang, J. Ergosterol peroxide exhibits antiviral and immunomodulatory abilities against porcine deltacoronavirus (PDCoV) via suppression of NF-κB and p38/MAPK signaling pathways in vitro. Int. Immunopharmacol. 2021, 93, 107317. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, C. Extracellular signal-regulated kinase (ERK) activation is required for porcine epidemic diarrhea virus replication. Virology 2015, 484, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhao, D.; Zhou, P.; Zhang, L.; Li, M.; Li, W.; Zhang, Y.; Wang, Y.; Liu, X. Characterization, pathogenicity and protective efficacy of a cell culture-derived porcine deltacoronavirus. Virus Res. 2020, 282, 197955. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, L.; Zhang, Q.; Zhou, P.; Fang, Y.; Zhao, D.; Feng, J.; Li, W.; Zhang, Y.; Wang, Y. Evaluation and comparison of immunogenicity and cross-protective efficacy of two inactivated cell culture-derived GIIa- and GIIb-genotype porcine epidemic diarrhea virus vaccines in suckling piglets. Vet. Microbiol. 2019, 230, 278–282. [Google Scholar] [CrossRef]
- Sun, R.-Q.; Cai, R.-J.; Chen, Y.-Q.; Liang, P.-S.; Chen, D.-K.; Song, C.-X. Outbreak of Porcine Epidemic Diarrhea in Suckling Piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, P.; Guo, L.; Liu, Y.; Du, Y.; Ren, S.; Li, J.; Zhang, Y.; Fan, Y.; Huang, B.; et al. Porcine Epidemic Diarrhea Virus Variants with High Pathogenicity, China. Emerg. Infect. Dis. 2013, 19, 2048–2049. [Google Scholar] [CrossRef]
- Zhu, T.; Du, S.; Cao, D.; Pei, Z.; Guo, Y.; Shao, H.; Wang, H.; Wang, K.; Hu, G. Isolation and identification of a variant subtype G 2b porcine epidemic diarrhea virus and S gene sequence characteristic. Infect. Genet. Evol. 2019, 71, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, L.; Yuan, Y.; Li, Q.; Han, L.; Yang, G.; Hu, H. Rhodanine derivative LJ001 inhibits TGEV and PDCoV replication in vitro. Virus Res. 2020, 289, 198167. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Song, Y.; Li, Z.; Xue, Z.; Cao, W.; Liu, X.; Zheng, H. African Swine Fever Virus Regulates Host Energy and Amino Acid Metabolism To Promote Viral Replication. J. Virol. 2022, 96, e0191921. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Niu, C.; Wu, F.; Zhou, S.; Han, T.; Zhang, Z.; Li, E.; Zhang, X.; Xu, S.; Wang, J.; et al. DNA damage contributes to age-associated differences in SARS-CoV-2 infection. Aging Cell 2022, 21, e13729. [Google Scholar] [CrossRef]
- Malgras, M.; Garcia, M.; Jousselin, C.; Bodet, C.; Lévêque, N. The Antiviral Activities of Poly-ADP-Ribose Polymerases. Viruses 2021, 13, 582. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, M.E.; Chen, Y.; Kuny, C.; Maejima, T.; Lease, R.; Ferraris, D.; Aikawa, M.; Sullivan, C.S.; Perlman, S.; Fehr, A.R. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019, 15, e1007756. [Google Scholar] [CrossRef]
- Puthanveetil, P. Metabolic Activation of PARP as a SARS-CoV-2 Therapeutic Target—Is It a Bait for the Virus or the Best Deal We Could Ever Make with the Virus? Is AMBICA the Potential Cure? Biomolecules 2023, 13, 374. [Google Scholar] [CrossRef]
- Heer, C.D.; Sanderson, D.J.; Voth, L.S.; Alhammad, Y.M.O.; Schmidt, M.S.; Trammell, S.A.J.; Perlman, S.; Cohen, M.S.; Fehr, A.R.; Brenner, C. Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity. J. Biol. Chem. 2020, 295, 17986–17996. [Google Scholar] [CrossRef]
- Shoji, S.; Yamaji, T.; Makino, H.; Ishii, J.; Kondo, A. Metabolic design for selective production of nicotinamide mononucleotide from glucose and nicotinamide. Metab. Eng. 2021, 65, 167–177. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Zhong, H.; Xu, Y. The mechanism of nicotinamide on reducing acute lung injury by inhibiting MAPK and NF-κB signal pathway. Mol. Med. 2021, 27, 115. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Lee, Y.J.; Lee, C. Porcine deltacoronavirus activates the Raf/MEK/ERK pathway to promote its replication. Virus Res. 2020, 283, 197961. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Ma, X.; Yuan, M.; Yi, Y.; Liu, G.; Wen, M.; Jiang, W.; Ji, R.; Zhu, L.; Tang, Z.; et al. E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF. Nat. Commun. 2021, 12, 1194. [Google Scholar] [CrossRef] [PubMed]
- Raines, L.N.; Zhao, H.; Wang, Y.; Chen, H.-Y.; Gallart-Ayala, H.; Hsueh, P.-C.; Cao, W.; Koh, Y.; Alamonte-Loya, A.; Liu, P.-S.; et al. PERK is a critical metabolic hub for immunosuppressive function in macrophages. Nat. Immunol. 2022, 23, 431–445. [Google Scholar] [CrossRef]
- Luo, X.; Chen, X.-X.; Qiao, S.; Li, R.; Lu, Q.; Geng, R.; Wang, L.; Zhou, E.-M.; Zhang, G. Porcine reproductive and respiratory syndrome virus increases SOCS3 production via activation of p38/AP-1 signaling pathway to promote viral replication. Vet. Microbiol. 2021, 257, 109075. [Google Scholar] [CrossRef]
- Luo, X.; Chen, X.-X.; Qiao, S.; Li, R.; Xie, S.; Zhou, X.; Deng, R.; Zhou, E.-M.; Zhang, G. Porcine Reproductive and Respiratory Syndrome Virus Enhances Self-Replication via AP-1–Dependent Induction of SOCS1. J. Immunol. 2020, 204, 394–407. [Google Scholar] [CrossRef]
- Hu, W.; Wang, X.; Ding, X.; Li, Y.; Zhang, X.; Xie, P.; Yang, J.; Wang, S. MicroRNA-141 Represses HBV Replication by Targeting PPARA. PLoS ONE 2012, 7, e34165. [Google Scholar] [CrossRef]
- Malesu, R.; Martin, A.J.; Lyons, J.G.; Scolyer, R.A.; Chen, A.C.; McKenzie, C.A.; Madore, J.; Halliday, G.M.; Damian, D.L. Nicotinamide for skin cancer chemoprevention: Effects of nicotinamide on melanoma in vitro and in vivo. Photochem. Photobiol. Sci. 2020, 19, 171–179. [Google Scholar] [CrossRef]
- Scatozza, F.; Moschella, F.; D’arcangelo, D.; Rossi, S.; Tabolacci, C.; Giampietri, C.; Proietti, E.; Facchiano, F.; Facchiano, A. Nicotinamide inhibits melanoma in vitro and in vivo. J. Exp. Clin. Cancer Res. 2020, 39, 211. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhang, L.; Pan, L.; Zhou, P.; Yu, R.; Zhang, Z.; Lv, J.; Guo, H.; Wang, Y.; Xiao, S.; et al. Nicotinamide Efficiently Suppresses Porcine Epidemic Diarrhea Virus and Porcine Deltacoronavirus Replication. Viruses 2023, 15, 1591. https://doi.org/10.3390/v15071591
Li M, Zhang L, Pan L, Zhou P, Yu R, Zhang Z, Lv J, Guo H, Wang Y, Xiao S, et al. Nicotinamide Efficiently Suppresses Porcine Epidemic Diarrhea Virus and Porcine Deltacoronavirus Replication. Viruses. 2023; 15(7):1591. https://doi.org/10.3390/v15071591
Chicago/Turabian StyleLi, Mingxia, Liping Zhang, Li Pan, Peng Zhou, Ruiming Yu, Zhongwang Zhang, Jianliang Lv, Huichen Guo, Yonglu Wang, Sa Xiao, and et al. 2023. "Nicotinamide Efficiently Suppresses Porcine Epidemic Diarrhea Virus and Porcine Deltacoronavirus Replication" Viruses 15, no. 7: 1591. https://doi.org/10.3390/v15071591
APA StyleLi, M., Zhang, L., Pan, L., Zhou, P., Yu, R., Zhang, Z., Lv, J., Guo, H., Wang, Y., Xiao, S., & Liu, X. (2023). Nicotinamide Efficiently Suppresses Porcine Epidemic Diarrhea Virus and Porcine Deltacoronavirus Replication. Viruses, 15(7), 1591. https://doi.org/10.3390/v15071591








