A Whole-Genome Analysis of the African Swine Fever Virus That Circulated during the First Outbreak in Vietnam in 2019 and Subsequently in 2022
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Library Preparation
2.3. Clustering and Sequencing
2.4. Genome Sequencing and Annotation
2.5. Comparative Genomic Analysis
3. Results
3.1. Preparation of ASFV-Infected Samples
3.2. Complete Genome Sequence Analysis of VN/HY/2019-ASFV1 and VN/QP/2019-ASFV1
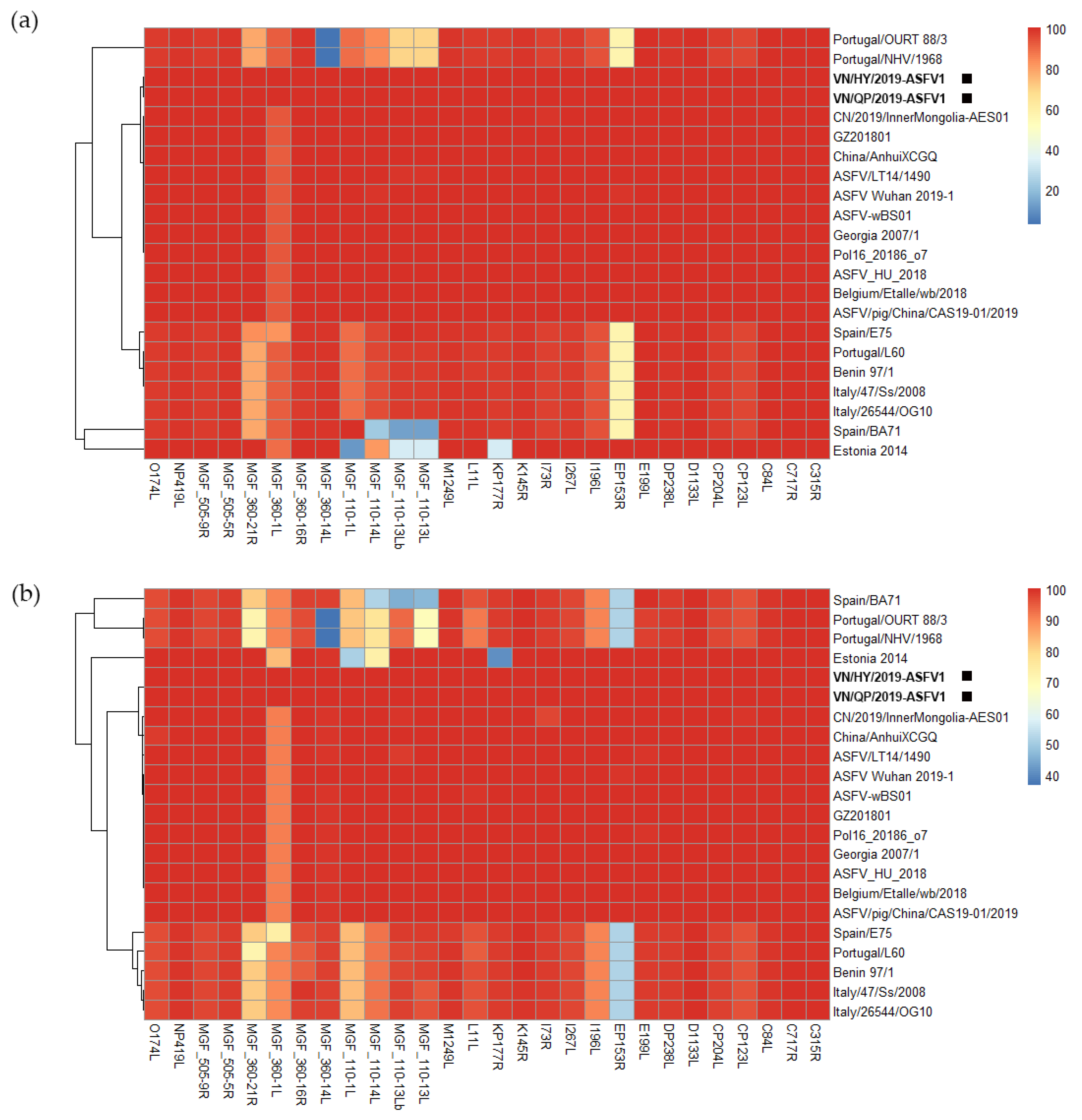
3.3. Genomic Comparison of VN/HY/2019-ASFV1 and VN/QP/2019-ASFV1 with Genotype I and II ASFV Strains
3.4. Genome Comparison of VN/HY/2022-ASFV2 with VN/HY/2019-ASFV1 and VN/QP/2019-ASFV1
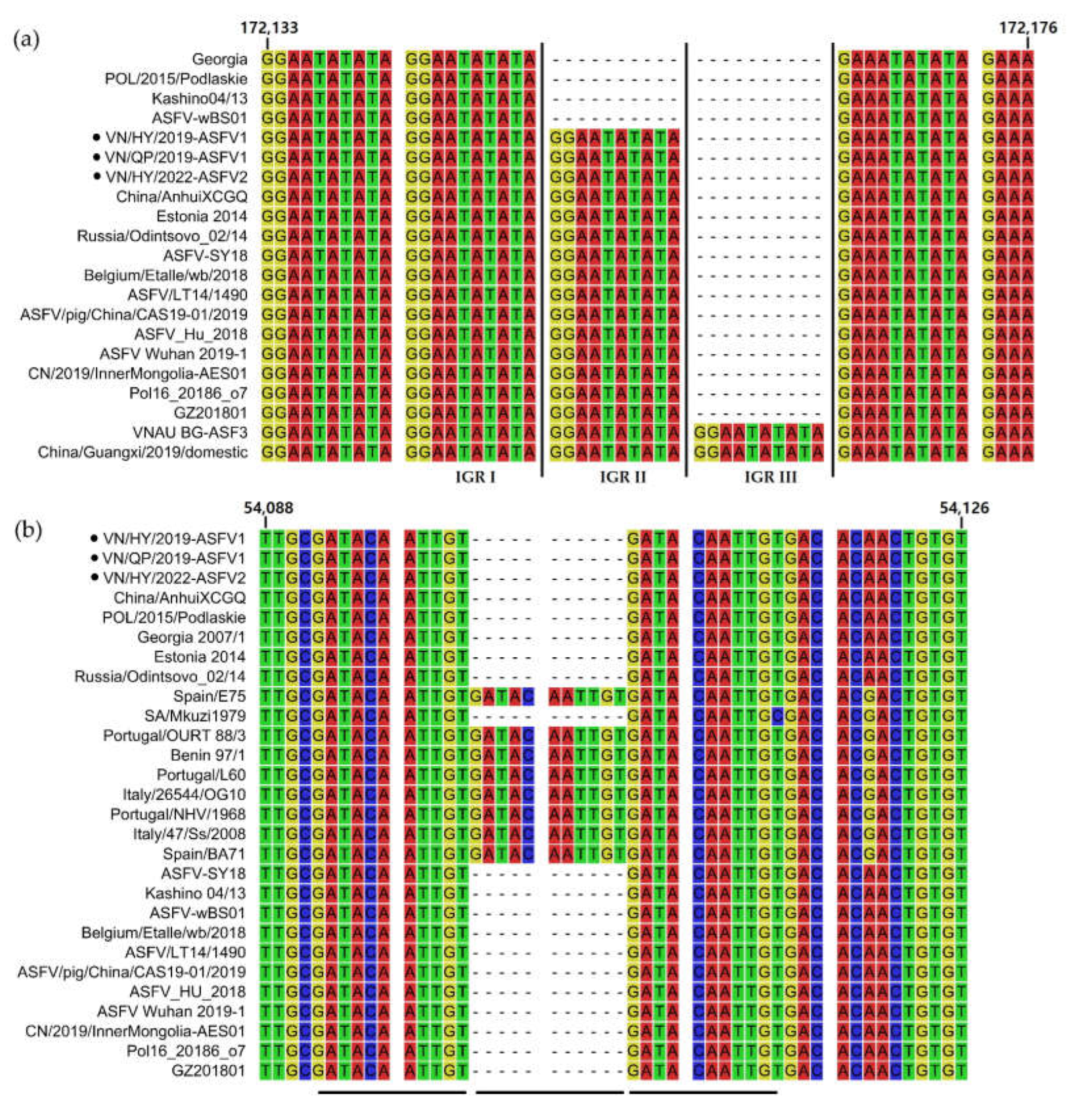
3.5. Sequence Alignment Analysis of the IGR in ASFV Genomes
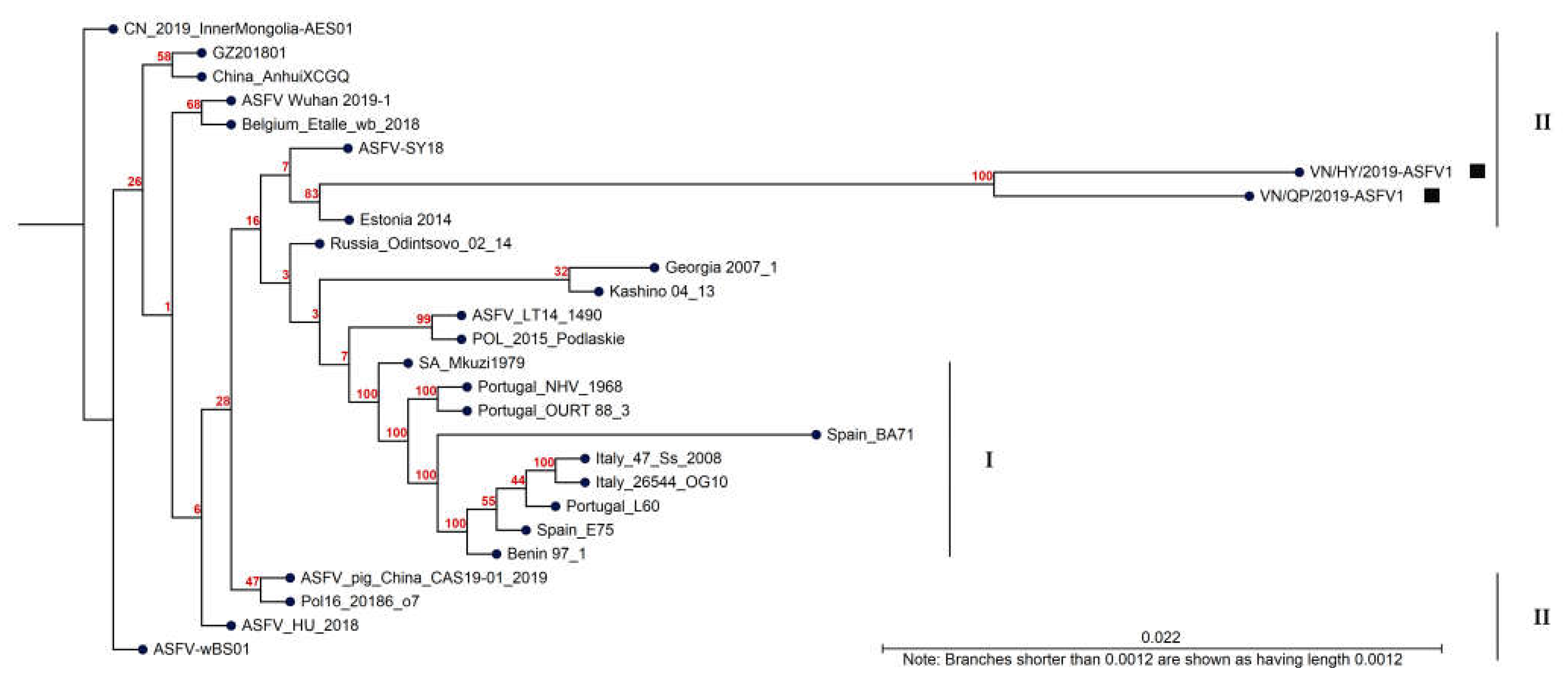
3.6. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sánchez-Cordón, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Urbano, A.C.; Ferreira, F. African swine fever control and prevention: An update on vaccine development. Emerg. Microbes Infect. 2022, 11, 2021–2033. [Google Scholar] [CrossRef]
- Galindo, I.; Alonso, C. African swine fever virus: A review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Bastos, A.D.; Penrith, M.L.; Crucière, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef]
- De Kock, G.; Robinson, E.M.; Keppel, J.J.G. Swine fever in South Africa. Onderstepoort J. Vet. Sci. Anim. Ind. 1940, 14, 31–93. [Google Scholar]
- Eustace Montgomery, R.E. On a form of swine fever occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Thomson, G.R. The epidemiology of African swine fever: The role of free-living hosts in Africa. Onderstepoort J. Vet. Res. 1985, 52, 201–209. [Google Scholar] [PubMed]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Gomez-Villamandos, J.C.; Carrasco, L. An update on the epidemiology and pathology of African swine fever. J. Comp. Pathol. 2015, 152, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Cwynar, P.; Stojkov, J.; Wlazlak, K. African swine fever status in Europe. Viruses 2019, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Gogin, A.; Gerasimov, V.; Malogolovkin, A.; Kolbasov, D. African Swine Fever in the North Caucasus region and the Russian Federation in years 2007–2012. Virus Res. 2013, 173, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Linden, A.; Licoppe, A.; Volpe, R.; Paternostre, J.; Lesenfants, C.; Cassart, D.; Garigliany, M.; Tignon, M.; van den Berg, T.; Desmecht, D.; et al. Summer 2018: African swine fever virus hits north-western Europe. Transbound. Emerg. Dis. 2019, 66, 54–55. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. GLOBAL SITUATION OF AFRICAN SWINE FEVER. African swine fever (ASF) Report N° 47: 2016–2020. 2020. Available online: https://www.woah.org/app/uploads/2022/01/asf-situation-report-18062020.pdf (accessed on 1 September 2023).
- Mazur-Panasiuk, N.; Woźniakowski, G.; Niemczuk, K. The first complete genomic sequences of African swine fever virus isolated in Poland. Sci. Rep. 2019, 9, 4556. [Google Scholar] [CrossRef] [PubMed]
- OIE. OIE-WAHIS. Available online: https://wahis.oie.int/#/home (accessed on 1 September 2023).
- Bao, J.; Wang, Q.; Lin, P.; Liu, C.; Li, L.; Wu, X.; Chi, T.; Xu, T.; Ge, S.; Liu, Y.; et al. Genome comparison of African swine fever virus China/2018/AnhuiXCGQ strain and related European p72 Genotype II strains. Transbound. Emerg. Dis. 2019, 66, 1167–1176. [Google Scholar] [CrossRef]
- Le, V.P.; Jeong, D.G.; Yoon, S.W.; Kwon, H.M.; Trinh, T.B.N.; Nguyen, T.L.; Bui, T.T.N.; Oh, J.; Kim, J.B.; Cheong, K.M.; et al. Outbreak of African swine fever, Vietnam, 2019. Emerg. Infect. Dis. 2019, 25, 1433–1435. [Google Scholar] [CrossRef]
- Liu, J.; Liu, B.; Shan, B.; Wei, S.; An, T.; Shen, G.; Chen, Z. Prevalence of African swine fever in China, 2018–2019. J. Med. Virol. 2020, 92, 1023–1034. [Google Scholar] [CrossRef]
- Mai, N.T.A.; Vu, X.D.; Nguyen, T.T.H.; Nguyen, V.T.; Trinh, T.B.N.; Kim, Y.J.; Kim, H.J.; Cho, K.H.; Nguyen, T.L.; Bui, T.T.N.; et al. Molecular profile of African swine fever virus (ASFV) circulating in Vietnam during 2019–2020 outbreaks. Arch. Virol. 2021, 166, 885–890. [Google Scholar] [CrossRef]
- Hien, N.D.; Nguyen, L.T.; Hoang, L.T.; Bich, N.N.; Quyen, T.M.; Isoda, N.; Sakoda, Y. First report of a complete genome sequence of a variant African swine fever virus in the Mekong delta, Vietnam. Pathogens 2022, 11, 797. [Google Scholar] [CrossRef]
- Truong, Q.L.; Nguyen, T.L.; Nguyen, T.H.; Shi, J.; Vu, H.L.X.; Lai, T.L.H.; Nguyen, V.G. Genome sequence of a virulent African swine fever virus isolated in 2020 from a domestic pig in Northern Vietnam. Microbiol. Resour. Announc. 2021, 10, 1110–1128. [Google Scholar] [CrossRef]
- Tran, H.T.T.; Truong, A.D.; Dang, A.K.; Ly, D.V.; Nguyen, C.T.; Chu, N.T.; Hoang, T.V.; Nguyen, H.T.; Dang, H.V. Circulation of two different variants of intergenic region (IGR) located between the I73R and I329L genes of African swine fever virus strains in Vietnam. Transbound. Emerg. Dis. 2021, 68, 2693–2695. [Google Scholar] [CrossRef]
- Nga, B.T.T.; Dao, B.T.A.; Thi, L.N.; Osaki, M.; Kawashima, K.; Song, D.; Salguero, F.J.; Le, V.P. Clinical and Pathological Study of the First Outbreak Cases of African Swine Fever in Vietnam, 2019. Front. Vet. Sci. 2020, 7, 392. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Ravarapu, K.; Apparao, D.S.K. Genomics, proteomics and drug designing approaches on avian leukemia virus. J. Adv. Bioinform. Appl. Res. 2011, 2, 149–154. [Google Scholar]
- Arahal, D.R. Whole-Genome Analyses: Average Nucleotide Identity. In Methods in Microbiology; Michael Goodfellow, I.S., Chun, J., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 41. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing; Vienna, Austria. 2019. Available online: https://www.R-project.org/ (accessed on 1 September 2023).
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Dhar, A.; Minin, V.N. Maximum likelihood phylogenetic inference. Encycl. Evol. Biol. 2016, 499–506. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Cho, K.H.; Mai, N.T.A.; Park, J.Y.; Trinh, T.B.N.; Jang, M.K.; Nguyen, T.T.H.; Vu, X.D.; Nguyen, T.L.; Nguyen, V.D.; et al. Multiple variants of African swine fever virus circulating in Vietnam. Arch. Virol. 2022, 167, 1137–1140. [Google Scholar] [CrossRef]
- Tran, H.T.T.; Truong, A.D.; Dang, A.K.; Ly, D.V.; Chu, N.T.; Van Hoang, T.; Nguyen, H.T.; Netherton, C.L.; Dang, H.V. Novel method for sub-grouping of genotype II African swine fever viruses based on the intergenic region between the A179L and A137R genes. Vet. Med. Sci. 2022, 8, 607–609. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.H.; Zhu, Z.Y.; Wang, S.J.; Tu, S.Y.; Zhang, Y.Q.; Zou, Y.L.; Liu, Y.T.; Liu, C.J.; Ren, W.J.; et al. Tracing the Origin of Genotype II African Swine Fever Virus in China by Genomic Epidemiology Analysis. Transbound. Emerg. Dis. 2023, 2023, 4820809. [Google Scholar] [CrossRef]
- Kreindel, S.; Pittiglio, C.; Pinto, J.; Lockhart, C.; Calistri, P.; Lubroth, J.; Correa, M. African Swine Fever Threatens People’s Republic of China: A Rapid Risk Assessment of ASF Introduction; FAO: Rome, Italy, 2018; p. 20. Available online: https://www.fao.org/3/I8805EN/i8805en.pdf (accessed on 6 March 2018).
- Petrovan, V.; Rathakrishnan, A.; Islam, M.; Goatley, L.C.; Moffat, K.; Sanchez-Cordon, P.J.; Reis, A.L.; Dixon, L.K. Role of African Swine Fever Virus Proteins EP153R and EP402R in Reducing Viral Persistence in Blood and Virulence in Pigs Infected with BeninΔDP148R. J. Virol. 2022, 96, e01340-21. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zhu, R.; Li, N.; Yang, J.; Yue, H.; Zhang, Y.; Zhou, X.; Ke, J.; Wang, Y.; Li, Q.; et al. African swine fever virus I196L is a virulence determinant and its deletant induces robust protection in Domestic pig. bioRxiv 2023, 2023-06. [Google Scholar] [CrossRef]
- Vuono, E.A.; Ramirez-Medina, E.; Pruitt, S.; Rai, A.; Espinoza, N.; Velazquez-Salinas, L.; Gladue, D.P.; Borca, M.V. Evaluation of the Function of the ASFV KP177R Gene, Encoding for Structural Protein p22, in the Process of Virus Replication and in Swine Virulence. Viruses 2021, 13, 986. [Google Scholar] [CrossRef] [PubMed]
- Zsak, L.; Lu, Z.; Burrage, T.G.; Neilan, J.G.; Kutish, G.F.; Moore, D.M.; Rock, D.L. African swine fever virus multigene family 360 and 530 genes are novel macrophage host range determinants. J. Virol. 2001, 75, 3066–3076. [Google Scholar] [CrossRef]
- Zsak, L.; Sur, J.H.; Burrage, T.G.; Neilan, J.G.; Rock, D.L. African swine fever virus (Asfv) multigene families 360 and 530 genes promote infected macrophage survival. Sci. World J. 2001, 1, 97. [Google Scholar] [CrossRef][Green Version]
- Chapman, D.A.G.; Tcherepanov, V.; Upton, C.; Dixon, L.K. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008, 89, 397–408. [Google Scholar] [CrossRef]
- Farlow, J.; Donduashvili, M.; Kokhreidze, M.; Kotorashvili, A.; Vepkhvadze, N.G.; Kotaria, N.; Gulbani, A. Intra-epidemic genome variation in highly pathogenic African swine fever virus (ASFV) from the country of Georgia. Virol. J. 2018, 15, 190. [Google Scholar] [CrossRef]
- Vietnam, P. Vietnam Halts Import of Pork from Hungary, Poland. 2018. Available online: https://en.vietnamplus.vn/vietnam-halts-import-of-pork-from-hungary-poland/138317.vnp (accessed on 14 September 2018).
- Long, N.V. African Swine Fever in Vietnam Lessons Learnt; MAFF: Tokyo, Japan, 2022.
- Nathan Pitts, T.W. Impact of African Swine Fever on Global Markets. Agric. Commod. 2019, 9, 52–54. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, V.P.; Ahn, M.-J.; Kim, J.-S.; Jung, M.-C.; Yoon, S.-W.; Trinh, T.B.N.; Le, T.N.; Kim, H.K.; Kang, J.-A.; Lim, J.-W.; et al. A Whole-Genome Analysis of the African Swine Fever Virus That Circulated during the First Outbreak in Vietnam in 2019 and Subsequently in 2022. Viruses 2023, 15, 1945. https://doi.org/10.3390/v15091945
Le VP, Ahn M-J, Kim J-S, Jung M-C, Yoon S-W, Trinh TBN, Le TN, Kim HK, Kang J-A, Lim J-W, et al. A Whole-Genome Analysis of the African Swine Fever Virus That Circulated during the First Outbreak in Vietnam in 2019 and Subsequently in 2022. Viruses. 2023; 15(9):1945. https://doi.org/10.3390/v15091945
Chicago/Turabian StyleLe, Van Phan, Min-Ju Ahn, Jun-Seob Kim, Min-Chul Jung, Sun-Woo Yoon, Thi Bich Ngoc Trinh, Thi Ngoc Le, Hye Kwon Kim, Jung-Ah Kang, Jong-Woo Lim, and et al. 2023. "A Whole-Genome Analysis of the African Swine Fever Virus That Circulated during the First Outbreak in Vietnam in 2019 and Subsequently in 2022" Viruses 15, no. 9: 1945. https://doi.org/10.3390/v15091945
APA StyleLe, V. P., Ahn, M.-J., Kim, J.-S., Jung, M.-C., Yoon, S.-W., Trinh, T. B. N., Le, T. N., Kim, H. K., Kang, J.-A., Lim, J.-W., Yeom, M., Na, W., Xie, X., Feng, Z., Song, D., & Jeong, D. G. (2023). A Whole-Genome Analysis of the African Swine Fever Virus That Circulated during the First Outbreak in Vietnam in 2019 and Subsequently in 2022. Viruses, 15(9), 1945. https://doi.org/10.3390/v15091945








