Abstract
Worldwide, acute gastroenteritis (AGE) is a major cause of morbidity and mortality in children under 5 years of age. Viruses, including norovirus, rotavirus, and enteric adenovirus, are the leading causes of pediatric AGE. In this prospective cohort study, we investigated the viral load and duration of shedding of norovirus, rotavirus, and adenovirus in stool samples collected from 173 children (median age: 15 months) with AGE who presented to emergency departments (EDs) across Canada on Day 0 (day of enrollment), and 5 and 28 days after enrollment. Quantitative RT-qPCR was performed to assess the viral load. On Day 0, norovirus viral load was significantly lower compared to that of rotavirus and adenovirus (p < 0.001). However, on Days 5 and 28, the viral load of norovirus was higher than that of adenovirus and rotavirus (p < 0.05). On Day 28, norovirus was detected in 70% (35/50) of children who submitted stool specimens, while rotavirus and adenovirus were detected in 52.4% (11/24) and 13.6% (3/22) of children (p < 0.001), respectively. Overall, in stool samples of children with AGE who presented to EDs, rotavirus and adenovirus had higher viral loads at presentation compared to norovirus; however, norovirus was shed in stool for the longest duration.
Keywords:
acute gastroenteritis; norovirus; rotavirus; adenovirus; viral load; fecal shedding; children 1. Introduction
Worldwide, acute gastroenteritis (AGE) is one of the leading causes of morbidity and mortality of children younger than 5 years [1,2]. AGE cases are predominantly caused by viruses, most commonly norovirus, rotavirus, and enteric adenovirus [3]. Despite the fact that the majority of virus-related AGE infections are self-limiting, they still significantly contribute to the burden of disease in many countries [1].
Human norovirus is the leading etiology of non-bacterial AGE among individuals of all ages globally [1,2,3], and is responsible for 60% of AGE cases in the United States, accounting for 400,000 emergency department (ED) visits and 71,000 hospitalizations each year [4]. Norovirus is highly contagious due to its low infective dose, various transmission routes, and persistence in the environment. Noroviruses were recently grouped into ten genogroups (GI-GX), with GII being the most predominant [5]. Infected individuals can shed over 109 copies of viral particles in one gram of feces [6]. In immunocompetent individuals, norovirus infection is usually self-limiting, with clinical symptoms lasting for 2–3 days [6,7].
Rotavirus was the most common cause of AGE in infants and young children worldwide before the introduction of the rotavirus vaccine. According to the Global Burden of Disease report, rotavirus infection caused 128,500 deaths and more than 250 million episodes of diarrhea among children younger than 5 years in 2016 [8]. Although the rotavirus vaccine has substantially reduced its incidence in countries with high vaccine coverage [9,10], rotavirus remains a major cause of AGE in children. Although most children with rotavirus gastroenteritis have high amounts of virus in their feces, shedding usually ceases within 10 days of symptom onset [10].
Adenovirus consists of non-enveloped, double-stranded DNA viruses, and belongs to the family of Adenoviridae. In humans, there are 88 serotypes in seven species (species A–G) [11]. Enteric adenovirus, which classically refers to serotypes 40 and 41 of specie F, was estimated to be responsible for over 52,000 deaths globally among children younger than 5 years in 2016, behind only rotavirus and Shigella [1]. The incubation period for adenovirus, typically 8–10 days, is longer than that of norovirus and rotavirus, and the illness can last as long as two weeks [12,13].
Although it is known that viruses can be shed for an extended period after the resolution of acute symptoms, the duration of virus shedding in stool varied among different studies [14,15]. Few studies have reported the kinetics and duration of viral shedding in stool samples of children with AGE infected with norovirus, rotavirus, and adenovirus [16]. In this prospective cohort study, we evaluated the viral load and virus shedding in stool samples collected from children with AGE at an index ED visit and during follow-up 5 and 28 days after the visit using real-time reverse transcription quantitative PCR (RT-qPCR).
2. Materials and Methods
2.1. Study Design
This study was part of the Probiotic Regimen for Outpatient Gastroenteritis Utility of Treatment (PROGUT) study, which found no overall or virus-specific clinical benefits associated with probiotic therapy in children who presented to the ED with AGE as compared to a placebo [17,18]. In addition, probiotic therapy was not associated with any reduction in viral shedding in stool [18]. We performed a secondary analysis focused on stool virus shedding to assess and compare the kinetics and duration of norovirus, rotavirus, and adenovirus shedding in children with AGE.
Five Canadian tertiary pediatric centers participated in this study. Children aged 3 to 48 months who presented to the ED with ≥3 diarrheal stools in a 24 h period and had a duration of illness of less than 72 h were eligible to participate. Attempts were made to collect stool specimens from all participants on Day 0 (ED enrollment), and 5 and 28 days later. Only participants who provided stool specimens at multiple time points in this study and tested positive for norovirus, adenovirus, or rotavirus were included in this analysis.
2.2. Study Objective and Outcome Measures
We sought to evaluate the viral load and duration of norovirus, rotavirus, and adenovirus shedding in children who presented to the ED with acute diarrhea. The study outcome was to compare the viral load and detection rate of norovirus with rotavirus and adenovirus on Days 0, 5, and 28. The Modified Vesikari Scale (MVS), with a maximum 20-point score, was used to quantify disease severity over a broad range of symptoms and interventions [19,20]. This scale has been employed in several studies, and effectively measured overall disease severity in children with AGE [17,18,21]. In this study, MVS scores were only available for Day 0.
2.3. Specimen Collection
Participants were enrolled into the trial between 5 November 2013, and 7 April 2017. Study participants were asked to provide a stool sample on Day 0 prior to ED discharge. If a specimen was not provided prior to ED discharge, caregivers were instructed to collect a stool sample at home, which was retrieved by a study-funded courier service.
Day 5 and Day 28 stool samples were requested from all study participants who provided a Day 0 stool sample, either while in the ED or at home. Caregivers were provided with collection instructions along with containers. Specimens were labeled with the date and time of collection and the subject’s study identification number. They were returned to the research team by a study-funded courier service within 12 h of collection. All specimens were placed in coolers with ice packs while in transit to the laboratory. Upon receipt, each sample was split and frozen for future testing. Sites then batch-shipped all frozen stool samples to the Alberta Public Laboratories-ProvLab (Edmonton, AB, Canada) bi-annually to enable interim laboratory analyses to verify collection and processing procedures.
2.4. Detection of Gastroenteritis Viruses
Total nucleic acid was extracted from 100–150 mg solid or 100 µL liquid stool using NucliSENS® easyMag®. Nucleic acid extracts were tested using the Luminex xTAG® Gastrointestinal Pathogen Panel (GPP) (Toronto, ON, Canada) that detects norovirus GI and GII, rotavirus, adenovirus 40/41, and 11 non-viral pathogens, as per the manufacturer’s instructions. Day 5 and 28 specimens were tested only if the Day 0 specimen tested positive for a pathogen.
2.5. Virus Quantification
The serial Day 0, 5, and 28 stool samples with norovirus, rotavirus, and/or adenovirus detected using the Luminex xTAG GPP assay were further tested using RT-qPCR to quantify the viral load in the sample. A 10% (weight/volume) stool solution was prepared with phosphate-buffered saline (PBS) and centrifuged at 12,000 rpm for 5 min at 4 °C. Two hundred µL of the supernatant was used for nucleic acid extraction using a MagaZorb RNA extraction kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions; the volume of the final elution was 50 µL. Two-step reactions, including reverse transcription and qPCR, were performed to detect norovirus, rotavirus, and adenovirus using an ABI 7500 fast sequence detection system (Applied Biosystems, Foster City, CA, USA), as previously described [22,23,24]. An individual qPCR reaction was performed for each virus using probes all labeled with FAM and TAMRA dyes. Ten-fold serial dilutions from 100 copies to 1 × 106 copies of DNA fragments of each virus were used to establish the external standard curve for viral quantification [23,24]. Virus load was expressed as genome equivalent (GE) copies/gram of stool.
2.6. Statistical Analysis
Viral load quantification values were log10 transformed to make data conform to normality. To compare viral load among viruses collected on Day 5, we conducted a multivariable linear regression model using viral load on Day 5 as the dependent variable. Covariates in the model included the type of virus infected (with norovirus being the reference group), age, baseline duration of symptoms, baseline MVS score, and Day 0 viral load. To compare viral load between the viruses collected on Day 28, we conducted the same regression model, with norovirus viral load on Day 28 being the dependent variable. All analyses were specified a priori and were two-sided; statistical significance was set at p < 0.05. We included data from all participants who met study eligibility criteria and provided the required specimens. Analyses were performed using SPSS 26.0 (Armonk, NY, USA: IBM Corp).
The Pearson correlation coefficient (r) was calculated to measure the correlation between viral load and disease severity (MVS score) on Day 0.
3. Results
3.1. Norovirus Shedding in Stool Samples
Data for children who tested positive for norovirus, rotavirus, or adenovirus and provided serial stool samples are summarized in Table 1. Seventy-nine children tested positive for norovirus, with a median age of 13 months (IQR: 9.5, 20.5). Norovirus GII was detected in 78 patients, and norovirus GI in 1 patient. The median viral load of norovirus was 9.9 log GE copies/g stool on Day 0 and 9.7 log GE copies/g stool on Day 5. On Day 28, norovirus was detected in 70.0% (35/50) of specimens, with a median viral load of 5.1 log GE copies/g stool (Table 2, Figure 1 and Figure S1A).

Table 1.
Number of serial stool samples collected on different days that were norovirus-, rotavirus-, and adenovirus-positive.

Table 2.
Norovirus, rotavirus, and adenovirus viral loads in different groups of serial stool samples based on availability of samples for qPCR tests.
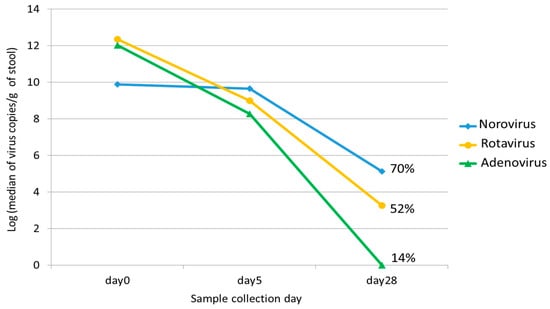
Figure 1.
The median viral loads of norovirus, rotavirus, and adenovirus on different collection days (Days 0, 5, and 28). The percentage on Day 28 represents the positive detection rate for each virus in stool.
3.2. Rotavirus Shedding in Stool Samples
The median age of the 52 children who tested positive for rotavirus was 20 months (IQR: 12.8, 31.8). The median viral load of rotavirus was 12.3 log GE copies/g stool on Day 0 and 8.8 log GE copies/g stool on Day 5. On Day 28, rotavirus was detected in 52.4% (11/21) of specimens with a median viral load of 3.3 log GE copies/g stool (Table 2 and Figure 1 and Figure S1B).
3.3. Adenovirus Shedding in Stool Samples
Forty-two children with a median age of 13 months (IQR: 9, 20.5) tested positive for enteric adenovirus 40/41. The median viral load of adenovirus was 12.1 and 8.4 log GE copies/g stool on Days 0 and 5, respectively. Adenovirus was detected in 13.6% (3/22) of specimens on Day 28 (Table 2 and Figure 1 and Figure S1C).
3.4. Comparison of Viral Load and Detection Rate among the Three Viruses
Multivariable linear regression showed a lower viral load of norovirus on Day 0 compared to rotavirus and adenovirus (p < 0.001) (Table 3). However, the median viral loads of norovirus were higher than rotavirus and adenovirus on Days 5 and 28 (p < 0.001) (Table 3). In terms of detection rate on Day 28, norovirus was detected in more children compared to rotavirus and adenovirus (p < 0.001). Adenovirus had the lowest detection rate on Day 28 among the three viruses (p < 0.05). The Pearson correlation coefficient showed no association between the Day 0 viral load and the MVS score for norovirus (r = 0.18, p = 0.13), a low correlation for rotavirus (r = 0.29, p = 0.05), and a moderate correlation for adenovirus (r = 0.4, p = 0.01).

Table 3.
Multivariable linear regression model comparing viral loads between norovirus and rotavirus, and between norovirus and adenovirus.
3.5. Co-Infection of Mixed Viruses in Children with AGE
Stool samples collected from 10 children had more than one virus detected, including four with adenovirus/rotavirus, four with norovirus/rotavirus, and two with adenovirus/norovirus (Table 4). Nine of the ten (90%) specimens collected from children with mixed viral infection had a dominant viral pathogen which had a higher viral load on Day 0 and Day 5.

Table 4.
The viral load of stool samples in each patient with co-infection of mixed viruses.
4. Discussion
In this prospective study of children who presented to the ED with acute infectious diarrhea, we examined the kinetics of viral shedding in stool samples of three major gastroenteritis viruses—norovirus, adenovirus, and rotavirus. All three viruses were shed in high levels at the time of presentation for ED care when the first specimen was obtained, and demonstrated unique fecal shedding kinetics. Norovirus had the lowest viral load on Day 0 among the three viruses, but it shed longer and at a higher viral load compared to rotavirus and adenovirus on Days 5 and 28.
Among the children recruited with acute diarrhea who tested positive for one of the three viruses, norovirus was detected in 46%, followed by rotavirus (30%) and adenovirus (24%). The peak shedding of norovirus has been reported to occur between symptom onset to 5 days after infection [25], with over 109 copies of viral particles shed in one gram of feces [6]. Cheng et al. reported that among hospitalized pediatric patients, norovirus viral load increased between days 2 and 9 following illness onset, and most norovirus shedding in feces ceased by day 15, with an average viral load of 7.25 log genome copies/mL for all samples [26]. In our study, we did not observe an increase in viral load after illness onset, but the median viral load of norovirus on Day 0 was 9.9 log GE copies/g stool, and it remained persistently high on Day 5 (9.7 log GE copies/g stool). On Day 28, 70% of children still tested positive for norovirus, with a median viral load of 5.1 log GE copies/g stool. This is consistent with previous reports that younger children shed norovirus in stool for a longer period, especially infants less than 6 months of age, who have been reported to shed detectable virus for a median of 42 days compared to just 10 days among patients older than 1 year of age [27]. Human challenge studies also report that norovirus shedding can continue for up to 60 days after symptoms disappear [6,28,29]. Norovirus reinfection is often defined as a positive result 14 days after a previous positive test. Our results support the notion that use of a 14-day interval to define reinfection is inadequate given the longer duration of shedding in young infants. Further studies of norovirus shedding with a larger sample size and among different age groups are needed to characterize the optimal time frame to define incidence of norovirus infection.
In recent years, RT-qPCR has become the gold standard for rapid, sensitive, and specific detection of norovirus in clinical samples [30]. However, a limitation of molecular assays is the inability to differentiate between infectious and non-infectious virus (i.e., to determine transmissibility); thus the communicable period of norovirus is unclear, and this has major implications for infection prevention and control practices. Chan et al. reported that a RT-qPCR cycle threshold (Ct) cut-off of 30 might define the lower limit of transmissibility, based on experiments conducted with human intestinal enteroid cultures inoculated with GII.4 Sydney[P31]-infected fecal samples [31]. However, due to the lower sensitivity of viral culture systems for norovirus, the variable replication efficiencies of different norovirus genotypes in the enteroid system, and inter-laboratory variation in RT-qPCR assays, it is possible that norovirus in stool samples with a Ct value >30 might still be transmissible.
The median rotavirus viral load of 12.3 log GE copies/g stool detected on Day 0 in our study aligns with a previous report [32]. We also found that compared to norovirus, rotavirus had a higher viral load on Day 0, but lower viral loads on Days 5 and 28. Moreover, we found that 52% of children tested positive for rotavirus on Day 28, which was lower than the 70% detection rate of norovirus on Day 28. However, prolonged rotavirus shedding of up to 57 days after symptom resolution has been reported [33,34]. Although the ability to culture rotavirus was established in the 1980s [35], no studies examining the infectivity of rotavirus in stools had been undertaken to define the period of transmissibility.
Few studies have reported the kinetics of viral load and shedding of adenovirus in stool samples [36,37], especially for comparison with other gastroenteritis viruses. The available literature reports that adenovirus 40/41 had higher viral loads compared to non-enteric adenovirus in children with AGE [36]. We report a high viral load of adenovirus on Day 0, with a rapid decline in virus level on Days 5 and 28. Compared to norovirus and rotavirus, adenovirus had the lowest detection rate on Day 28 (13.6%), indicating that the duration of adenovirus 40/41 shedding in stool is shorter than norovirus and rotavirus.
Although previous studies reported that a higher stool viral load was associated with severe clinical symptoms for norovirus infection in cancer patients at the time of diagnosis [38], we did not identify a significant correlation between norovirus viral load and disease severity on Day 0. A moderate correlation between adenovirus viral load and MVS score was observed in this study, which was in agreement with a previous finding that children infected with adenovirus with higher viral loads had more severe disease [37]. For rotavirus, it was reported that a higher level of fecal viral load was positively associated with disease severity in children [34]. We also identified a low correlation between the Day 0 rotavirus viral load and MVS score. As MVS scores were not available for Days 5 and 28 in this study, it was not clear whether there was an association between viral load and disease severity on those days.
Some limitations merit mention. We did not sample specimens between Day 5 and Day 28; therefore, the kinetic of viral load and shedding during this time period was not clear. In addition, this study enrolled a cohort of otherwise healthy young children [18]; thus, the findings could not be extrapolated to the kinetics of virus shedding among immunocompromised children and those > 48 months of age.
5. Conclusions
In conclusion, rotavirus and adenovirus had higher viral loads at the illness onset compared to norovirus; however, norovirus demonstrated the longest duration of virus shedding, with moderately high viral loads in the stools of young children up to Day 28 after presenting to EDs due to acute infectious diarrhea. The long duration of detection of the gastroenteritis virus in stool samples highlights the importance of further studies regarding the infectivity of qPCR positive samples and the need to revisit the definition of incident infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15071541/s1, Figure S1: Viral load of norovirus, rotavirus and adenovirus in each individual child who had Day 0, 5 and 28 serial samples: (A) Norovirus; (B) Rotavirus; (C) Adenovirus.
Author Contributions
Y.Q.: investigation, data analysis, writing—original draft preparation, writing—review and editing; S.B.F.: conceptualization, methodology, investigation, funding acquisition, writing—review and editing; S.W.-U.: project administration; K.J.F.: resources; S.G.: resources; N.P.: resources; S.S.: resources; Y.F.: resources, writing—review and editing; J.X.: data analysis, writing—review and editing; B.E.L.: investigation, writing—review and editing; L.C.: investigation, writing—review and editing; X.P.: conceptualization, methodology, investigation, supervision, writing—review and editing. Other members from the PROGUT Trial Group: resources. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Canadian Institutes of Health Research (grants 286384 and 325412).
Institutional Review Board Statement
Study protocols were approved by Institutional Review Boards at the University of Calgary and the University of Alberta.
Informed Consent Statement
Written informed consent was obtained from the guardians of all participants involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the participants and their families for participating this study, and the investigators, support staff, and site coordinators across all sites for their support. The Pediatric Emergency Research Canada Probiotic Regimen for Outpatient Gastroenteritis Utility of Treatment (PROGUT) Trial Group: Yuanyuan Qiu 1, Stephen B. Freedman 2, Sarah Williamson-Urquhart 3, Ken J. Farion 4, Serge Gouin 5, Naveen Poonai 6, Suzanne Schuh 7, Yaron Finkelstein 8, Jianling Xie 9, Bonita E. Lee 10, Linda Chui 1, Xiaoli Pang 1,11, David Schnadower 12, Philip M. Sherman 13, Katrina F. Hurley 14, Karen Black 15. 12 Division of Emergency Medicine, Cincinnati Children’s Hospital Medical Center, Department of Pediatrics, University of Cincinnati, Cincinnati, OH 45229, USA. 13 Division of Gastroenterology, Hepatology, and Nutrition, The Hospital for Sick Children, University of Toronto, Toronto, ON M5G 1X8, Canada. 14 Division of Pediatric Emergency Medicine, IWK Health Centre, Dalhousie University, Halifax, NS B3K 6R8, Canada. 15 Department of Pediatrics, University of British Columbia, Vancouver, BC V6H 3N1, Canada.
Conflicts of Interest
The authors declare no conflict of interest.
References
- GBD Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Aguilar, J.; Berkley, J.; Broekaert, I.; Vazquez-Frias, R.; Holtz, L.; Lo Vecchio, A.; Meskini, T.; Moore, S.; Rivera Medina, J.F.; et al. Acute Gastroenteritis in Children of the World: What Needs to Be Done? J. Pediatr. Gastroenterol. Nutr. 2020, 70, 694–701. [Google Scholar] [CrossRef]
- Zweigart, M.R.; Becker-Dreps, S.; Bucardo, F.; Gonzalez, F.; Baric, R.S.; Lindesmith, L.C. Serological Humoral Immunity Following Natural Infection of Children with High Burden Gastrointestinal Viruses. Viruses 2021, 13, 2033. [Google Scholar] [CrossRef]
- Hall, A.J.; Lopman, B.A.; Payne, D.C.; Patel, M.M.; Gastanaduy, P.A.; Vinje, J.; Parashar, U.D. Norovirus disease in the United States. Emerg. Infect. Dis. 2013, 19, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553–1557. [Google Scholar] [CrossRef] [PubMed]
- Sukhrie, F.H.; Teunis, P.; Vennema, H.; Copra, C.; Thijs Beersma, M.F.; Bogerman, J.; Koopmans, M. Nosocomial transmission of norovirus is mainly caused by symptomatic cases. Clin. Infect. Dis. 2012, 54, 931–937. [Google Scholar] [CrossRef]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization-Coordinated Global Rotavirus Surveillance Network. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin. Infect. Dis. 2016, 62 (Suppl. S2), S96–S105. [Google Scholar] [CrossRef]
- Burnett, E.; Parashar, U.D.; Tate, J.E. Real-world effectiveness of rotavirus vaccines, 2006–2019: A literature review and meta-analysis. Lancet Glob. Health 2020, 8, e1195–e1202. [Google Scholar] [CrossRef]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Bottcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T.; et al. Molecular Evolution of Human Adenovirus (HAdV) Species C. Sci. Rep. 2019, 9, 1039. [Google Scholar] [CrossRef]
- Grimwood, K.; Carzino, R.; Barnes, G.L.; Bishop, R.F. Patients with enteric adenovirus gastroenteritis admitted to an Australian pediatric teaching hospital from 1981 to 1992. J. Clin. Microbiol. 1995, 33, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Uhnoo, I.; Wadell, G.; Svensson, L.; Johansson, M.E. Importance of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children. J. Clin. Microbiol. 1984, 20, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, L.; Norden, R.; Westin, J.; Lindh, M.; Andersson, L.M. Slow Clearance of Norovirus following Infection with Emerging Variants of Genotype GII.4 Strains. J. Clin. Microbiol. 2017, 55, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Chiu, C.H.; Lee, H.Y.; Tsai, C.N.; Chen, C.L.; Chen, S.Y. Clinical and virological characteristics of viral shedding in children with norovirus gastroenteritis. J. Microbiol. Immunol. Infect. 2022, 55, 1188–1194. [Google Scholar] [CrossRef] [PubMed]
- Akihara, S.; Phan, T.G.; Nguyen, T.A.; Hansman, G.; Okitsu, S.; Ushijima, H. Existence of multiple outbreaks of viral gastroenteritis among infants in a day care center in Japan. Arch. Virol. 2005, 150, 2061–2075. [Google Scholar] [CrossRef]
- Freedman, S.B.; Williamson-Urquhart, S.; Farion, K.J.; Gouin, S.; Willan, A.R.; Poonai, N.; Hurley, K.; Sherman, P.M.; Finkelstein, Y.; Lee, B.E.; et al. Multicenter Trial of a Combination Probiotic for Children with Gastroenteritis. N. Engl. J. Med. 2018, 379, 2015–2026. [Google Scholar] [CrossRef]
- Freedman, S.B.; Xie, J.; Nettel-Aguirre, A.; Pang, X.L.; Chui, L.; Williamson-Urquhart, S.; Schnadower, D.; Schuh, S.; Sherman, P.M.; Lee, B.E.; et al. A randomized trial evaluating virus-specific effects of a combination probiotic in children with acute gastroenteritis. Nat. Commun. 2020, 11, 2533. [Google Scholar] [CrossRef]
- Freedman, S.B.; Eltorky, M.; Gorelick, M.; Pediatric Emergency Research Canada Gastroenteritis Study Group. Evaluation of a gastroenteritis severity score for use in outpatient settings. Pediatrics 2010, 125, e1278–e1285. [Google Scholar] [CrossRef]
- Schnadower, D.; Tarr, P.I.; Gorelick, M.H.; O’Connell, K.; Roskind, C.G.; Powell, E.C.; Rao, J.; Bhatt, S.; Freedman, S.B. Validation of the modified Vesikari score in children with gastroenteritis in 5 US emergency departments. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 514–519. [Google Scholar] [CrossRef]
- Schnadower, D.; Tarr, P.I.; Casper, T.C.; Gorelick, M.H.; Dean, J.M.; O’Connell, K.J.; Mahajan, P.; Levine, A.C.; Bhatt, S.R.; Roskind, C.G.; et al. Lactobacillus rhamnosus GG versus Placebo for Acute Gastroenteritis in Children. N. Engl. J. Med. 2018, 379, 2002–2014. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Pabbaraju, K.; Pang, X.L.; Lee, B.E.; Fox, J.D. Detection of a broad range of human adenoviruses in respiratory tract samples using a sensitive multiplex real-time PCR assay. J. Med. Virol. 2008, 80, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.L.; Lee, B.E.; Pabbaraju, K.; Gabos, S.; Craik, S.; Payment, P.; Neumann, N. Pre-analytical and analytical procedures for the detection of enteric viruses and enterovirus in water samples. J. Virol. Methods 2012, 184, 77–83. [Google Scholar] [CrossRef]
- Qiu, Y.; Lee, B.E.; Ruecker, N.J.; Neumann, N.; Ashbolt, N.; Pang, X. A one-step centrifugal ultrafiltration method to concentrate enteric viruses from wastewater. J. Virol. Methods 2016, 237, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Franco, M.A.; Greenberg, H.B. Rotaviruses, Noroviruses, and Other Gastrointestinal Viruses. In Goldman’s Cecil Medicine, 24th ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Cheng, H.Y.; Lee, C.C.; Chang, Y.C.; Tsai, C.N.; Chao, H.C.; Tsai, Y.T.; Hsieh, C.H.; Su, S.S.; Chen, S.Y. Viral shedding in gastroenteritis in children caused by variants and novel recombinant norovirus infections. Medicine 2021, 100, e25123. [Google Scholar] [CrossRef]
- Murata, T.; Katsushima, N.; Mizuta, K.; Muraki, Y.; Hongo, S.; Matsuzaki, Y. Prolonged norovirus shedding in infants <or=6 months of age with gastroenteritis. Pediatr. Infect. Dis. J. 2007, 26, 46–49. [Google Scholar] [CrossRef]
- Okhuysen, P.C.; Jiang, X.; Ye, L.; Johnson, P.C.; Estes, M.K. Viral shedding and fecal IgA response after Norwalk virus infection. J. Infect. Dis. 1995, 171, 566–569. [Google Scholar] [CrossRef]
- Teunis, P.F.; Sukhrie, F.H.; Vennema, H.; Bogerman, J.; Beersma, M.F.; Koopmans, M.P. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol. Infect. 2015, 143, 1710–1717. [Google Scholar] [CrossRef]
- Vinje, J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015, 53, 373–381. [Google Scholar] [CrossRef]
- Chan, M.C.; Cheung, S.K.C.; Mohammad, K.N.; Chan, J.C.M.; Estes, M.K.; Chan, P.K.S. Use of Human Intestinal Enteroids to Detect Human Norovirus Infectivity. Emerg. Infect. Dis. 2019, 25, 1730–1735. [Google Scholar] [CrossRef]
- Kirkwood, C.D.; Ma, L.F.; Carey, M.E.; Steele, A.D. The rotavirus vaccine development pipeline. Vaccine 2019, 37, 7328–7335. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.; Grimwood, K.; Gorrell, R.; Palombo, E.; Barnes, G.; Bishop, R. Extended excretion of rotavirus after severe diarrhoea in young children. Lancet 1998, 351, 1844–1848. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.; Pollock, L.; Jere, K.C.; Pitzer, V.E.; Lopman, B.; Bar-Zeev, N.; Iturriza-Gomara, M.; Cunliffe, N.A. Duration and Density of Fecal Rotavirus Shedding in Vaccinated Malawian Children With Rotavirus Gastroenteritis. J. Infect. Dis. 2020, 222, 2035–2040. [Google Scholar] [CrossRef] [PubMed]
- Birch, C.J.; Rodger, S.M.; Marshall, J.A.; Gust, I.D. Replication of human rotavirus in cell culture. J. Med. Virol. 1983, 11, 241–250. [Google Scholar] [CrossRef] [PubMed]
- do Nascimento, L.G.; Fialho, A.M.; de Andrade, J.; de Assis, R.M.S.; Fumian, T.M. Human enteric adenovirus F40/41 as a major cause of acute gastroenteritis in children in Brazil, 2018 to 2020. Sci. Rep. 2022, 12, 11220. [Google Scholar] [CrossRef]
- Pabbaraju, K.; Tellier, R.; Pang, X.L.; Xie, J.; Lee, B.E.; Chui, L.; Zhuo, R.; Vanderkooi, O.G.; Ali, S.; Tarr, P.I.; et al. A Clinical Epidemiology and Molecular Attribution Evaluation of Adenoviruses in Pediatric Acute Gastroenteritis: A Case-Control Study. J. Clin. Microbiol. 2020, 59, e02287-20. [Google Scholar] [CrossRef]
- He, T.; McMillen, T.A.; Qiu, Y.; Chen, L.H.; Lu, X.; Pang, X.L.; Kamboj, M.; Tang, Y.W. Norovirus Loads in Stool Specimens of Cancer Patients with Norovirus Gastroenteritis. J. Mol. Diagn. 2017, 19, 836–842. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).