Circulation of Bluetongue Virus Serotypes 1, 4, 8, 10 and 16 and Epizootic Hemorrhagic Disease Virus in the Sultanate of Oman in 2020–2021
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Serological Analyses
2.3. RT-qPCR Analyses
2.4. Virus Isolation
2.5. Serotype Determination
2.6. Full Genome Sequencing
2.7. Genome Annotation and Phylogenetic Analyses
3. Results
3.1. Serological Results
- The seroprevalence for BT in Oman is high (74%) in cattle and illustrates a significant circulation of BTV in the country (Table 1). It seems less important in small ruminants with 50 and 30.4% of goats and sheep, respectively, showing antibodies against BTV (Table 2). Seroprevalence is also high for EHDV: more than one out of two cattle has antibodies against this virus (Table 1). In total, 46 (90.2%) of the 51 cattle with antibodies to EHDV were also positive in BTV ELISA.
3.2. Pan rt-qPCR Results and Virus Isolation
- In total, 15 of the 61 animals tested were positive for BTV rt-qPCR (Ct value range: 22 to 34), and 11 of the 56 cattle tested were positive for pan EHDV rt-qPCR (Ct value range: 30 to 35). Three cattle were positive for both viruses. Viral isolation assays on embryonated eggs and KC cells were tested several times on each rt-qPCR-positive blood sample. Only one isolate of BTV was obtained, and no EHDV isolate was recovered.
3.3. Typing and Sequencing
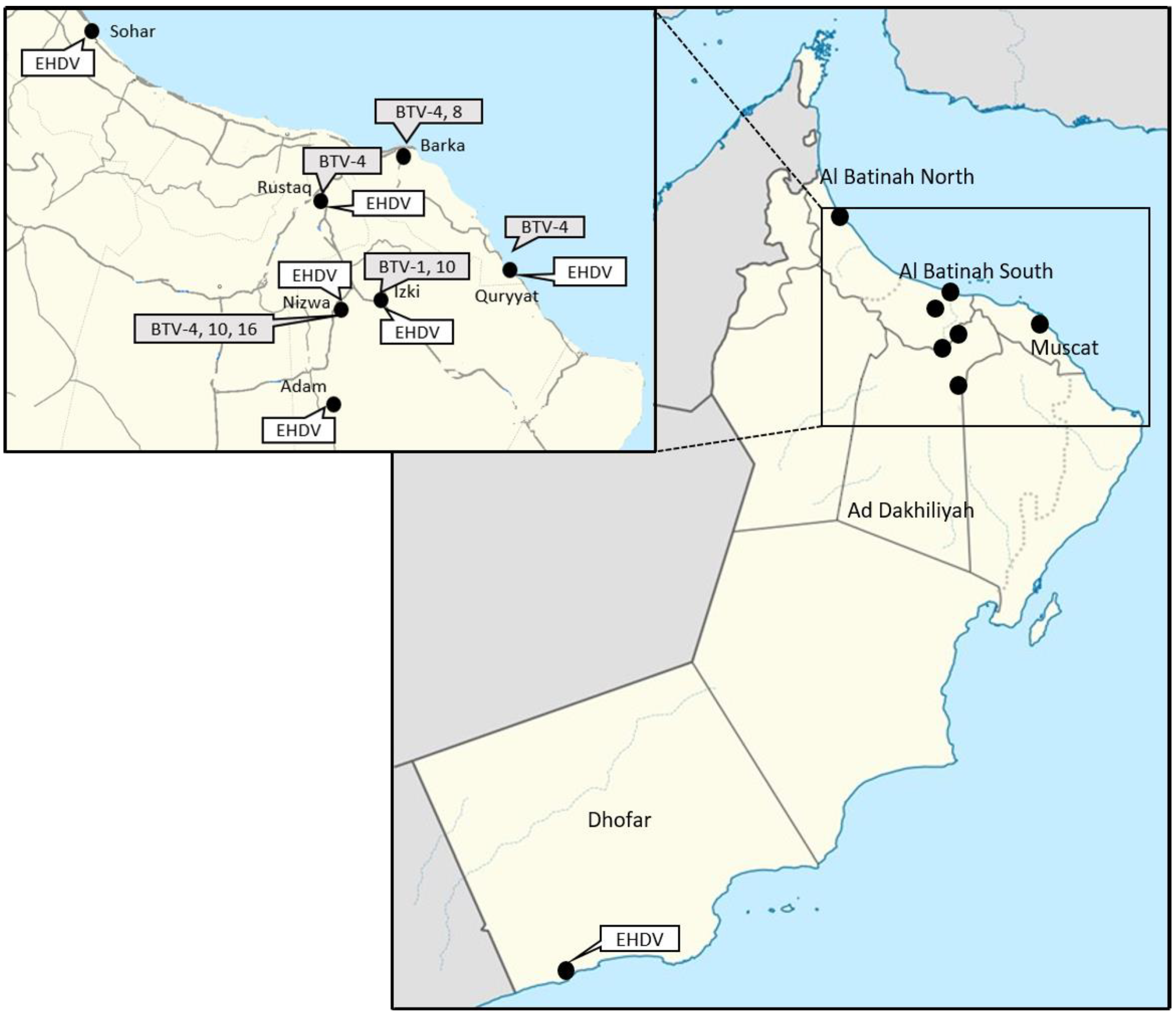
- The Figure 1 illustrates the location of the herds where BTV and EHDV genome detection occurred. The rt-qPCRs from Maan et al. [18,19], performed on the RNA extracts from the blood, have shown the presence of the BTV-1, 4, 8, 10 and 16. The end-point RT-PCRs [16] performed on the RNAs from the bloods allowed the determination of partial sequences of S2 of BTV-1, 4 and 10. No S2 sequence could be obtained for BTV-16-positive blood by classical RT-PCRs. The complete genome sequences of the BTV-8 strain were obtained by NGS.
- The EHDV type specific rt-qPCRs as well as the EHDV end-point RT-PCRs did not yield positive result.
3.4. BTV Genome Annotation and Phylogenetic Analyses
- The complete length of the S1, 2, 3, 4, 6, 7, 8, 9 and 10 were obtained for the BTV-8 genome, with the usual 5′ and 3′ extremities sequences (5′- GTTAAA … ACTTAC-3′). Regarding S5, only three nucleotides were missing at the 3′ extremity. Thus, all expected CDS were recovered (Table 4).
- Table 4 shows the sequence homologies obtained between the 10 segments of the BTV-8 Oman strain and 3 other BTV-8 strains: one isolated in France in 2015 (AN: MN495893 to MN495902), the second isolated in Mayotte (Indian Ocean) in 2016 (AN: OQ860834 to OQ860843) and a 3rd isolated in SA in 1937 (AN: MT078269 to MT078278). Table 4 also shows the best homology found between BTV-8 Oman sequences and homologous sequences deposited on GenBank. Sequence alignments (in AA) of the VP3, NS1, VP5, VP7 and NS3 of the four BTV-8 strains (Table 4) show more than 99% for all these VPs.
| % nt and (AA) Homology between BTV8 Oman and | ||||||||
|---|---|---|---|---|---|---|---|---|
| Seg. | AN | Length bp | CDS (nt Position/Length AA) | Product | BTV-8 France | BTV-8 Mayotte | BTV-8 SA | (%)—Better Homology in GenBank (AN) |
| 1 | OQ860824 | 3944 | 12–3920/1302 | VP1 | 93.6 (98.8) | 92.5 (98.6) | 96 (99.1) | 97.8—BTV-1 Isr 2019 (OM502362) |
| 2 | OQ860825 | 2939 | 18–2903/961 | VP2 | 94.6 (97.2) | 98.7 (99) | 92.5 (97.1) | 95.9—BTV-8 Nig 1982 (AJ585184) |
| 3 | OQ860826 | 2772 | 18–2723/901 | VP3 | 94.5 (99.7) | 94.3 (99.7) | 95.5 (99.8) | 97.1—BTV-24 SA 2017 (MG255591) |
| 4 | OQ860827 | 1982 | 9–1943/644 | VP4 | 94.2 (97.8) | 94 (98.4) | 95.2 (98.8) | 97.5—BTV-3 Isr 2016 (MG344993) |
| 5 | OQ860828 | 1773 | 35–1693/552 | NS1 | 93.7 (99.3) | 96.9 (99.3) | 93 (99.1) | 97.7—BTV-4 Fr 2020 (OK018214) |
| 6 | OQ860829 | 1637 | 28–1608/526 | VP5 | 96.8 (99.8) | 96.6 (99.8) | 96.2 (99.6) | 96.9—BTV-8 Nig 1982 (AJ586706) |
| 7 | OQ860830 | 1156 | 18–1067/349 | VP7 | 97.3 (100) | 94.7 (99.4) | 95.1 (100) | 97.7—BTV-1 Sud 1987 (KP821626) |
| 8 | OQ860831 | 1125 | 20–1084/354 | NS2 | 92.9 (96.9) | 92.4 (97.7) | 94.7 (97.5) | 97.5—BTV-1 Isr 2019 (OM502359) |
| 9 | OQ860832 | 1049 | 16–1005/329 | VP6 | 95.9 (94.8) | 95 (93.0) | 95.3 (95.5) | 96.8—BTV-24 SA 2020 (MT090653) |
| 182–415/77 | NS4 | 98.7 (98.7) | 98.7 (100) | 98.7 (100) | ||||
| 10 | OQ860833 | 822 | 20–709/229 | NS3 | 95.7 (100) | 98.6 (99.6) | 92.3 (99.1) | 98.8—BTV-15 Isr 2006 (JX272377) |
| 108–287/59 | NS5 | 93.9 (100) | 93.3 (99.6) | 93.3 (99.1) | ||||
- The phylogenetic tree of S2 (Figure 2) was performed with selected sequences of BTV-8, 18 and 23 that constitute the nucleotype D of the BTV S2 [24]. The tree shows two clusters of S2 of BTV-8: the Oman and Mayotte S2, which are very similar, form one of these clusters also consisting of S2s of European, Mediterranean and Nigerian BTV-8 strains. The second cluster includes the S2 of the SA and Kenia strains.
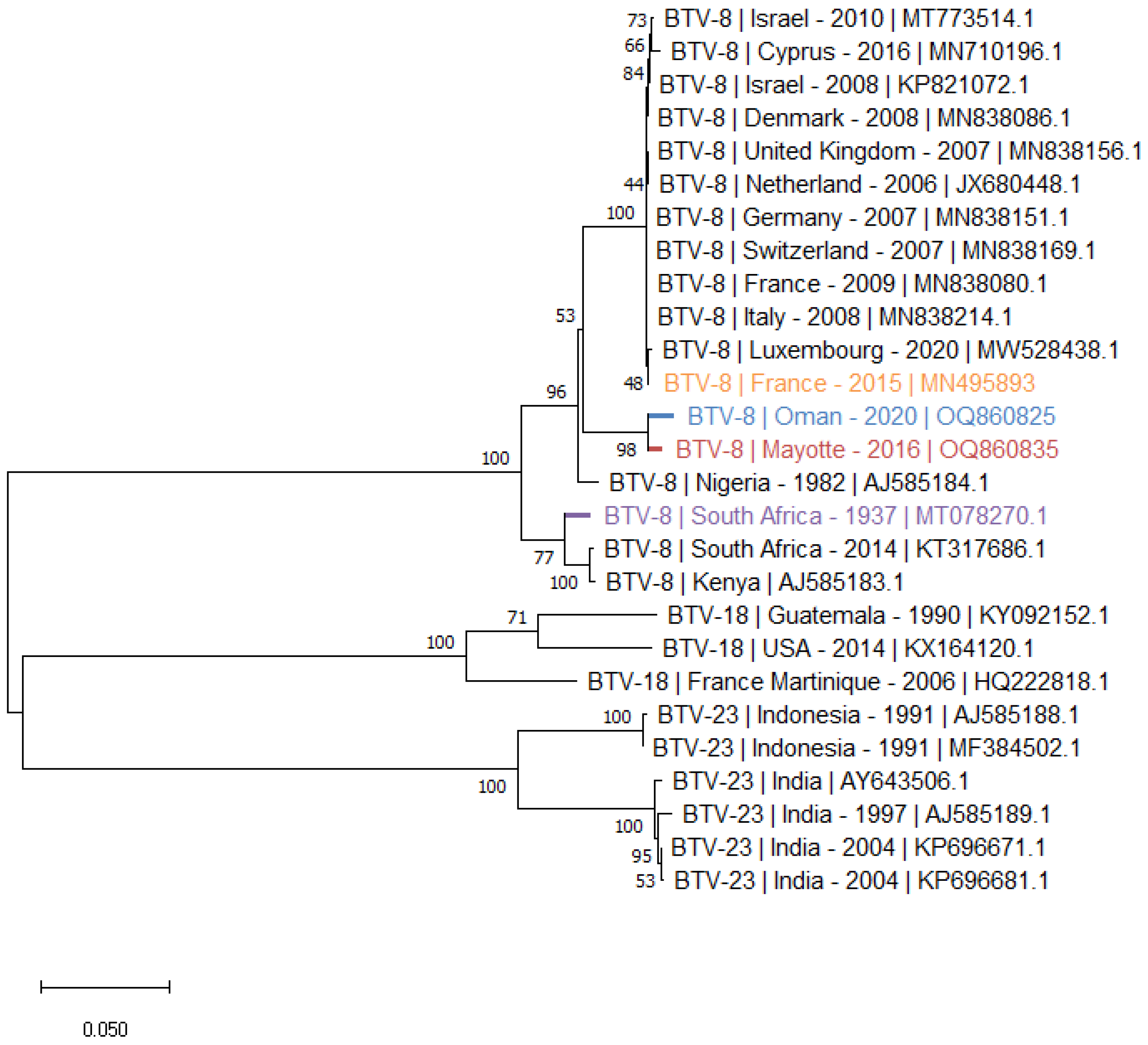
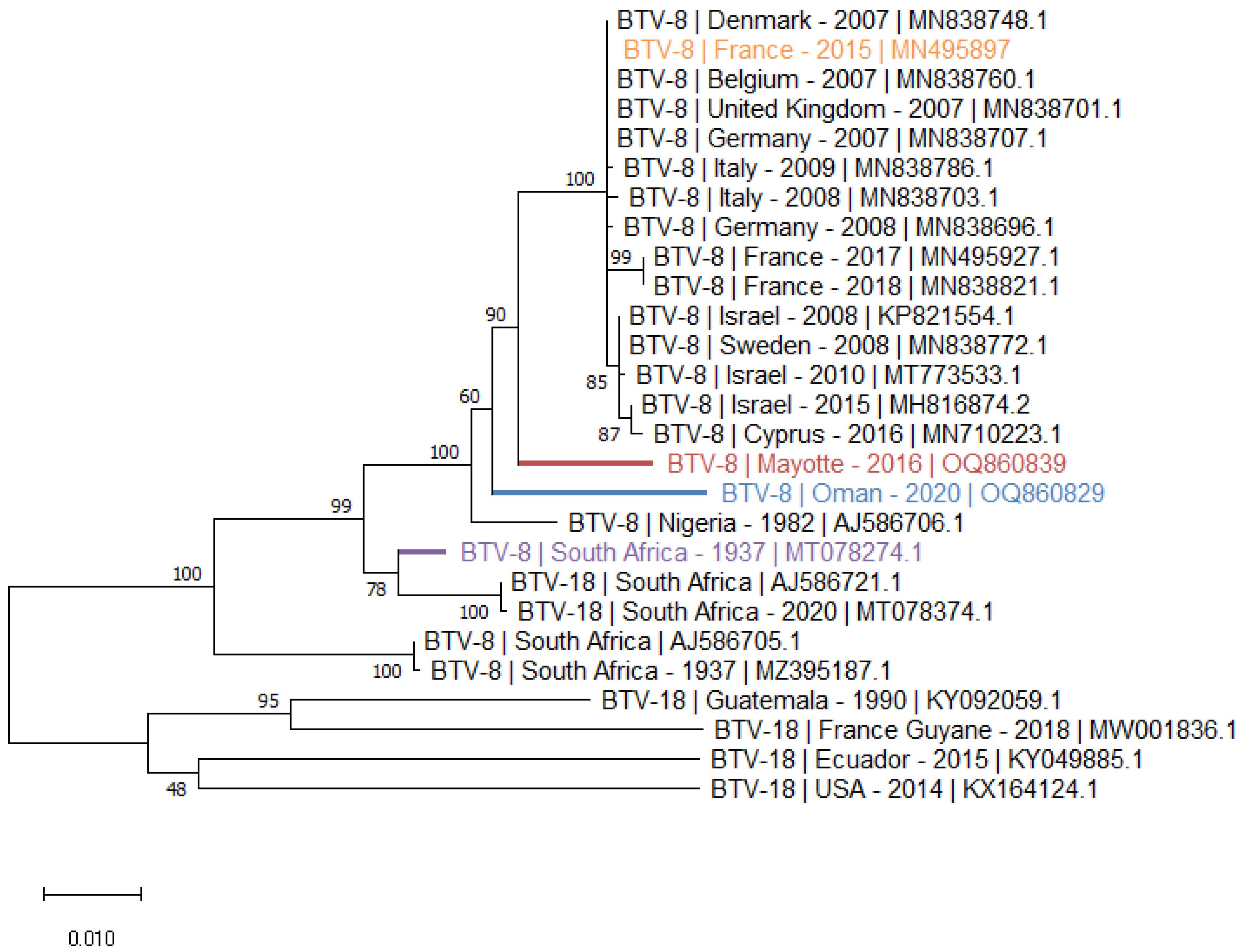
- The S6 phylogenetic tree (Figure 3) illustrates the homologies between S6 of BTV-8 and BTV-18, constituting the G nucleotype of BTV VP5 [25]. As observed with the S2 phylogenetic tree, the S6 of the Oman and Mayotte strains are closely related and form a cluster with BTV strains S6 isolated in Europe, in the Mediterranean basin and in Nigeria. In the cluster of the BTV-8 S6, two BTV-18 S6 from SA were found having a strong homology with BTV-8 S6 from the same area. These two SA BTV-18 S6 are clearly distinct from those isolated from American strains (Figure 3).
- S1, 3 to 5 and 7 to 10 of the Oman strain have the strongest homologies with segments all originating from BTV of many serotypes strains circulating further west: in the Mediterranean basin (Israel, Lebanon, Cyprus, Maghreb...), in the European countries, in Central Africa (Sudan, Nigeria, Cameroon) or SA (See Supplementary Figures S1–S8 for phylogenetic analysis of each BTV segment). Note the presence of a S4 of a Chinese BTV-7 strain in the cluster otherwise presenting only BTV S4 from strains isolated in Africa, Europe or the Arabian Peninsula (Supplementary Figure S4), suggesting for this Chinese BTV-7 segment 4 a “western” origin.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gibbs, E.P.; Greiner, E.C. The epidemiology of bluetongue. Comp. Immunol. Microbiol. Infect. Dis. 1994, 17, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Maclachlan, N.J.; Zientara, S.; Savini, G.; Daniels, P.W. Epizootic haemorrhagic disease. Rev. Sci. Tech. 2015, 34, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Savini, G.; Afonso, A.; Mellor, P.; Aradaib, I.; Yadin, H.; Sanaa, M.; Wilson, W.; Monaco, F.; Domingo, M. Epizootic heamorragic disease. Res. Vet. Sci. 2011, 91, 1–17. [Google Scholar] [PubMed]
- Hulten, C.; Frossling, J.; Chenais, E.; Sternberg Lewerin, S. Seroprevalence after vaccination of cattle and sheep against Bluetongue Virus (BTV) serotype 8 in Sweden. Transbound. Emerg. Dis. 2013, 60, 438–447. [Google Scholar] [CrossRef]
- Mertens, P.P.; Brown, F.; Sangar, D.V. Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology 1984, 135, 207–217. [Google Scholar] [CrossRef]
- Mohd Jaafar, F.; Monsion, B.; Mertens, P.P.C.; Attoui, H. Identification of Orbivirus Non-Structural Protein 5 (NS5), Its Role and Interaction with RNA/DNA in Infected Cells. Int. J. Mol. Sci. 2023, 24, 6845. [Google Scholar] [CrossRef]
- al-Busaidy, S.M.; Mellor, P.S. Isolation and identification of arboviruses from the Sultanate of Oman. Epidemiol. Infect. 1991, 106, 403–413. [Google Scholar]
- al-Busaidy, S.M.; Mellor, P.S. Epidemiology of bluetongue and related orbiviruses in the Sultanate of Oman. Epidemiol. Infect. 1991, 106, 167–178. [Google Scholar] [CrossRef]
- Frolich, K.; Hamblin, C.; Jung, S.; Ostrowski, S.; Mwanzia, J.; Streich, W.J.; Anderson, J.; Armstrong, R.M.; Anajariyah, S. Serologic surveillance for selected viral agents in captive and free-ranging populations of Arabian oryx (Oryx leucoryx) from Saudi Arabia and the United Arab Emirates. J. Wildl. Dis. 2005, 41, 67–79. [Google Scholar] [CrossRef]
- Bosnic, S.; Beck, R.; Listes, E.; Lojkic, I.; Savini, G.; Roic, B. Bluetongue virus in Oryx antelope (Oryx leucoryx) during the quarantine period in 2010 in Croatia. Vet. Ital. 2015, 51, 139–143. [Google Scholar]
- Anthony, S.J.; Maan, S.; Maan, N.; Kgosana, L.; Bachanek-Bankowska, K.; Batten, C.; Darpel, K.E.; Sutton, G.; Attoui, H.; Mertens, P.P. Genetic and phylogenetic analysis of the outer-coat proteins VP2 and VP5 of epizootic haemorrhagic disease virus (EHDV): Comparison of genetic and serological data to characterise the EHDV serogroup. Virus Res. 2009, 145, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Sghaier, S.; Sailleau, C.; Marcacci, M.; Thabet, S.; Curini, V.; Ben Hassine, T.; Teodori, L.; Portanti, O.; Hammami, S.; Jurisic, L.; et al. Epizootic Haemorrhagic Disease Virus Serotype 8 in Tunisia, 2021. Viruses 2022, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Cappai, S.; Loi, F.; Pinna, L.; Ruiu, A.; Puggioni, G.; Guercio, A.; Purpari, G.; Vicari, D.; Sghaier, S.; et al. Epizootic Hemorrhagic Disease Virus Serotype 8, Italy, 2022. Emerg. Infect. Dis. 2023, 29, 1063–1065. [Google Scholar] [CrossRef]
- Kundlacz, C.; Caignard, G.; Sailleau, C.; Viarouge, C.; Postic, L.; Vitour, D.; Zientara, S.; Breard, E. Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses 2019, 11, 672. [Google Scholar] [CrossRef] [PubMed]
- Viarouge, C.; Breard, E.; Zientara, S.; Vitour, D.; Sailleau, C. Duplex Real-Time RT-PCR Assays for the Detection and Typing of Epizootic Haemorrhagic Disease Virus. PLoS ONE 2015, 10, e0132540. [Google Scholar] [CrossRef]
- Viarouge, C.; Lancelot, R.; Rives, G.; Breard, E.; Miller, M.; Baudrimont, X.; Doceul, V.; Vitour, D.; Zientara, S.; Sailleau, C. Identification of bluetongue virus and epizootic hemorrhagic disease virus serotypes in French Guiana in 2011 and 2012. Vet. Microbiol. 2014, 174, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Zanella, G.; Breard, E.; Viarouge, C.; Desprat, A.; Vitour, D.; Adam, M.; Lasne, L.; Martrenchar, A.; Bakkali-Kassimi, L.; et al. Co-circulation of bluetongue and epizootic haemorrhagic disease viruses in cattle in Reunion Island. Vet. Microbiol. 2012, 155, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Maan, N.S.; Maan, S.; Belaganahalli, M.N.; Ostlund, E.N.; Johnson, D.J.; Nomikou, K.; Mertens, P.P. Identification and differentiation of the twenty six bluetongue virus serotypes by RT-PCR amplification of the serotype-specific genome segment 2. PLoS ONE 2012, 7, e32601. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Belaganahalli, M.N.; Potgieter, A.C.; Kumar, V.; Batra, K.; Wright, I.M.; Kirkland, P.D.; Mertens, P.P. Development and Evaluation of Real Time RT-PCR Assays for Detection and Typing of Bluetongue Virus. PLoS ONE 2016, 11, e0163014. [Google Scholar] [CrossRef]
- Briand, F.X.; Schmitz, A.; Ogor, K.; Le Prioux, A.; Guillou-Cloarec, C.; Guillemoto, C.; Allee, C.; Le Bras, M.O.; Hirchaud, E.; Quenault, H.; et al. Emerging highly pathogenic H5 avian influenza viruses in France during winter 2015/16: Phylogenetic analyses and markers for zoonotic potential. Euro Surveill. 2017, 22, 30473. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Maan, S.; Maan, N.S.; Samuel, A.R.; O’Hara, R.; Meyer, A.J.; Rao, S.; Mertens, P.P. Completion of the sequence analysis and comparisons of genome segment 2 (encoding outer capsid protein VP2) from representative isolates of the 24 bluetongue virus serotypes. Vet. Ital. 2004, 40, 484–488. [Google Scholar] [PubMed]
- Maan, S.; Maan, N.S.; Nomikou, K.; Veronesi, E.; Bachanek-Bankowska, K.; Belaganahalli, M.N.; Attoui, H.; Mertens, P.P. Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS ONE 2011, 6, e26147. [Google Scholar] [CrossRef]
- Elbers, A.R.; Meiswinkel, R. Culicoides (Diptera: Ceratopogonidae) host preferences and biting rates in the Netherlands: Comparing cattle, sheep and the black-light suction trap. Vet. Parasitol. 2014, 205, 330–337. [Google Scholar] [CrossRef]
- Ayllon, T.; Nijhof, A.M.; Weiher, W.; Bauer, B.; Allene, X.; Clausen, P.H. Feeding behaviour of Culicoides spp. (Diptera: Ceratopogonidae) on cattle and sheep in northeast Germany. Parasit. Vectors 2014, 7, 34. [Google Scholar] [CrossRef]
- Duan, Y.; Yang, Z.; Zhu, P.; Xiao, L.; Li, Z.; Li, Z.; Li, L.; Zhu, J. A serologic investigation of epizootic hemorrhagic disease virus in China between 2014 and 2019. Virol. Sin. 2022, 37, 513–520. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Ross-smith, N.; Batten, C.A.; Shaw, A.E.; Anthony, S.J.; Samuel, A.R.; Darpel, K.E.; Veronesi, E.; Oura, C.A.; et al. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 2008, 377, 308–318. [Google Scholar] [CrossRef]
- Nomikou, K.; Hughes, J.; Wash, R.; Kellam, P.; Breard, E.; Zientara, S.; Palmarini, M.; Biek, R.; Mertens, P. Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion. PLoS Pathog. 2015, 11, e1005056. [Google Scholar] [CrossRef]
- Golender, N.; Eldar, A.; Ehrlich, M.; Kenigswald, G.; Shlamovitz, I.; Even-Tov, B.; Zamir, L.; Klement, E.; Bumbarov, V. Genomic Analysis Illustrated a Single Introduction and Evolution of Israeli Bluetongue Serotype 8 Virus Population 2008–2019. Microorganisms 2021, 9, 1955. [Google Scholar] [CrossRef] [PubMed]
| BTV ELISA | ||||
|---|---|---|---|---|
| Negative | Doubtful | Positive | Number Tested | |
| number | 23 | 2 | 71 | 96 |
| % | 24.0 | 2.1 | 74.0 | |
| EHDV ELISA | ||||
| number | 44 | 1 | 51 | 96 |
| % | 45.8 | 1 | 53.1 | |
| BTV ELISA | |||||
|---|---|---|---|---|---|
| Negative | Doubtful | Positive | Number tested | ||
| Goats | number | 13 | 0 | 13 | 26 |
| % | 50 | 0 | 50 | ||
| Sheep | number | 15 | 1 | 7 | 23 |
| % | 65.2 | 4.3 | 30.4 | ||
| AN | Length bp | CDS (nt Position/Length AA) | Product | (% nt/AA)—Better Homology in GenBank (AN) |
|---|---|---|---|---|
| OQ860822 | 342 | 20–335/114 | Partial VP2 BTV-1 | (98.0/98.2)—BTV-1 Spa 2008 (KP821020) |
| OQ860823 | 318 | 1182–1523/105 | Partial VP2 BTV-10 | (93.4/99.0)—BTV-10 SA 2017 (MT078290) |
| OQ860904 | 2874 | 20–2890/956 | Partial VP2 BTV-4 | (97.5/98.6)—BTV-4 Spa 2003 (KP821067) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bréard, E.; Postic, L.; Gondard, M.; Bernelin-Cottet, C.; Le Roux, A.; Turpaud, M.; Lucas, P.; Blanchard, Y.; Vitour, D.; Bakkali-Kassimi, L.; et al. Circulation of Bluetongue Virus Serotypes 1, 4, 8, 10 and 16 and Epizootic Hemorrhagic Disease Virus in the Sultanate of Oman in 2020–2021. Viruses 2023, 15, 1259. https://doi.org/10.3390/v15061259
Bréard E, Postic L, Gondard M, Bernelin-Cottet C, Le Roux A, Turpaud M, Lucas P, Blanchard Y, Vitour D, Bakkali-Kassimi L, et al. Circulation of Bluetongue Virus Serotypes 1, 4, 8, 10 and 16 and Epizootic Hemorrhagic Disease Virus in the Sultanate of Oman in 2020–2021. Viruses. 2023; 15(6):1259. https://doi.org/10.3390/v15061259
Chicago/Turabian StyleBréard, Emmanuel, Lydie Postic, Mathilde Gondard, Cindy Bernelin-Cottet, Aurélie Le Roux, Mathilde Turpaud, Pierrick Lucas, Yannick Blanchard, Damien Vitour, Labib Bakkali-Kassimi, and et al. 2023. "Circulation of Bluetongue Virus Serotypes 1, 4, 8, 10 and 16 and Epizootic Hemorrhagic Disease Virus in the Sultanate of Oman in 2020–2021" Viruses 15, no. 6: 1259. https://doi.org/10.3390/v15061259
APA StyleBréard, E., Postic, L., Gondard, M., Bernelin-Cottet, C., Le Roux, A., Turpaud, M., Lucas, P., Blanchard, Y., Vitour, D., Bakkali-Kassimi, L., Zientara, S., Al Rawahi, W., & Sailleau, C. (2023). Circulation of Bluetongue Virus Serotypes 1, 4, 8, 10 and 16 and Epizootic Hemorrhagic Disease Virus in the Sultanate of Oman in 2020–2021. Viruses, 15(6), 1259. https://doi.org/10.3390/v15061259






