Inference of the Life Cycle of Environmental Phages from Genomic Signature Distances to Their Hosts
Abstract
1. Introduction
2. Results
2.1. Set of Reference Genomes Used for Predicting Phage Life-Cycles
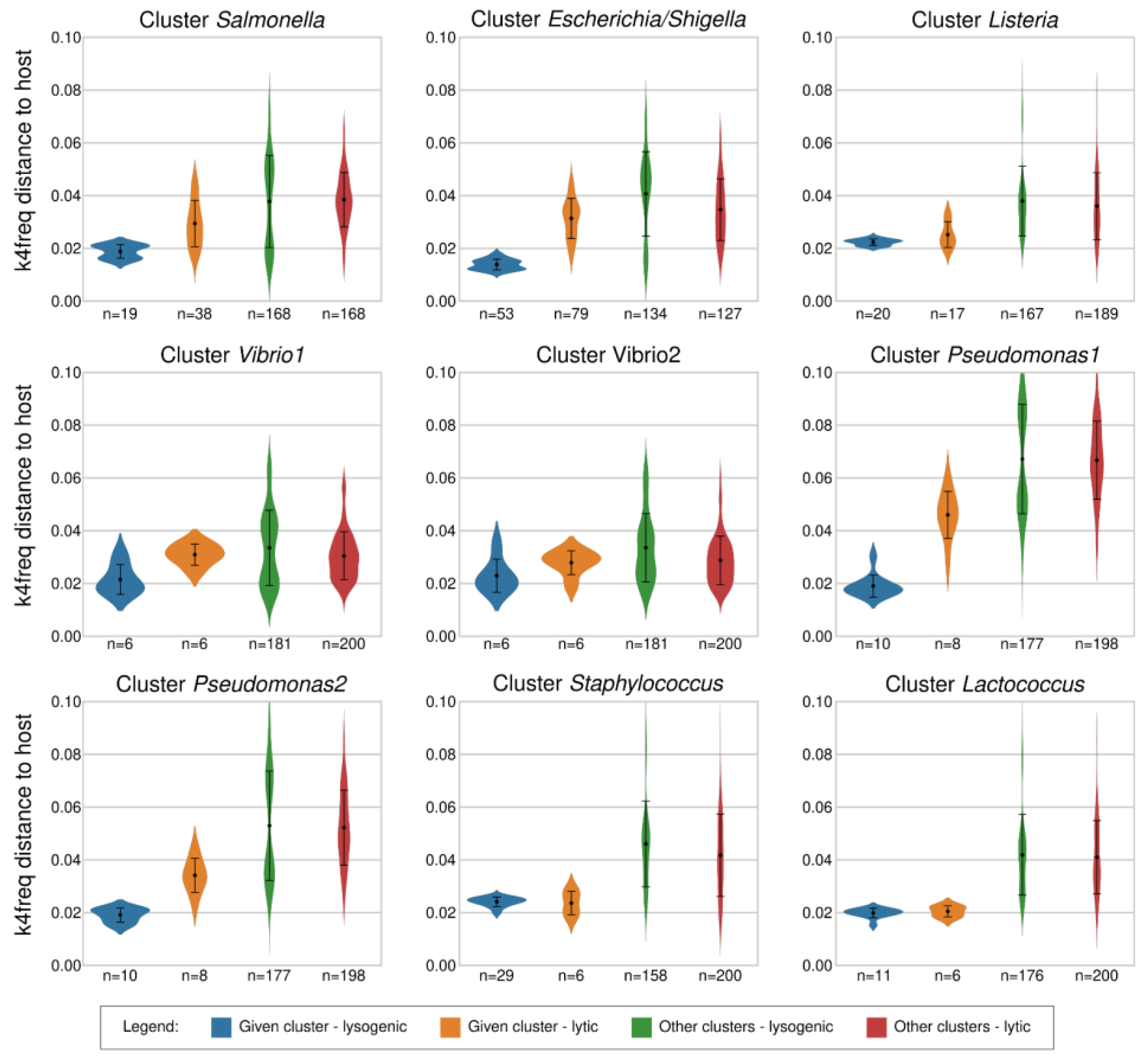
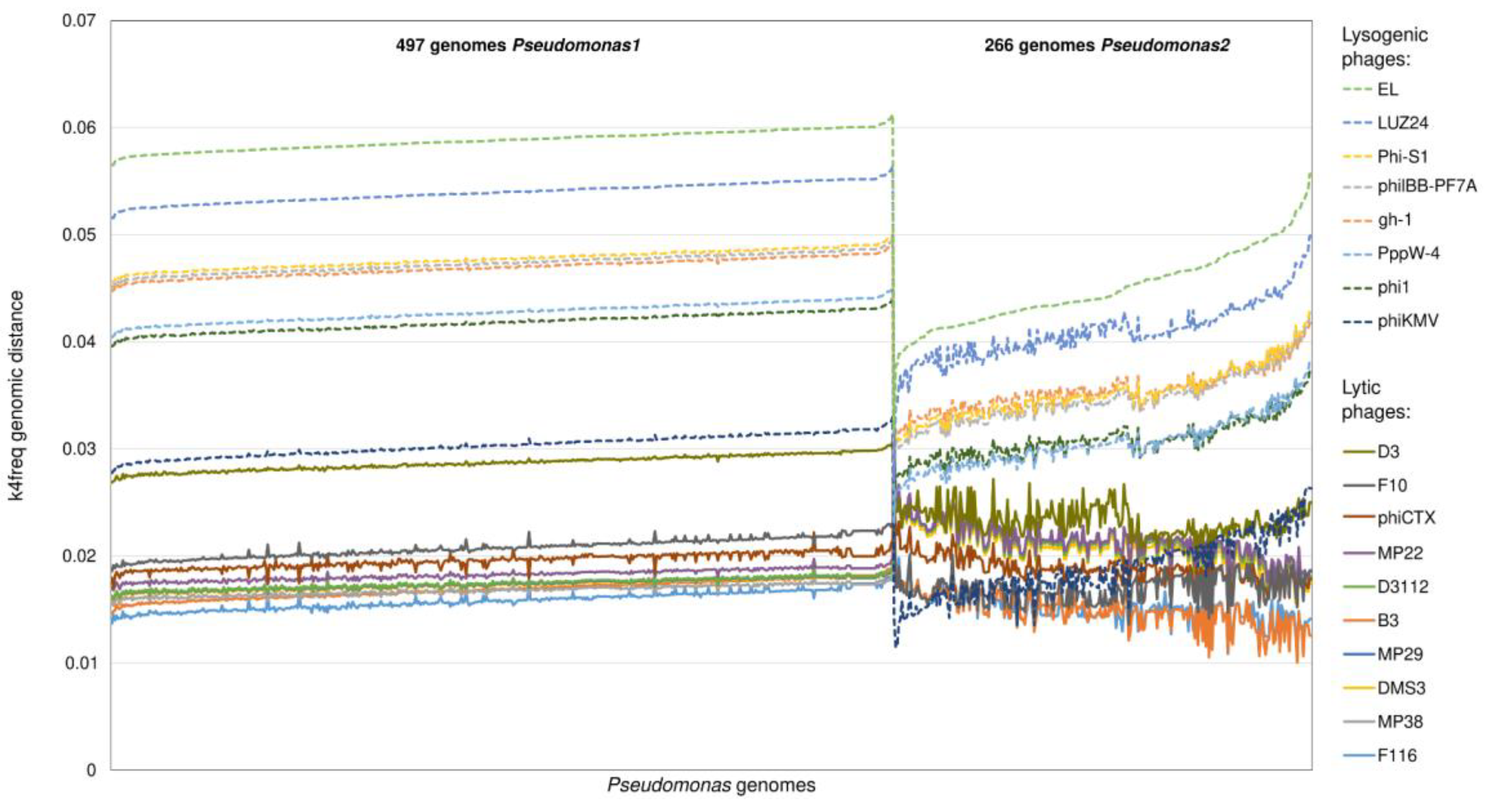
2.2. Distinguishing between Lytic and Lysogenic Phages from the Reference Dataset by the k4freq Method
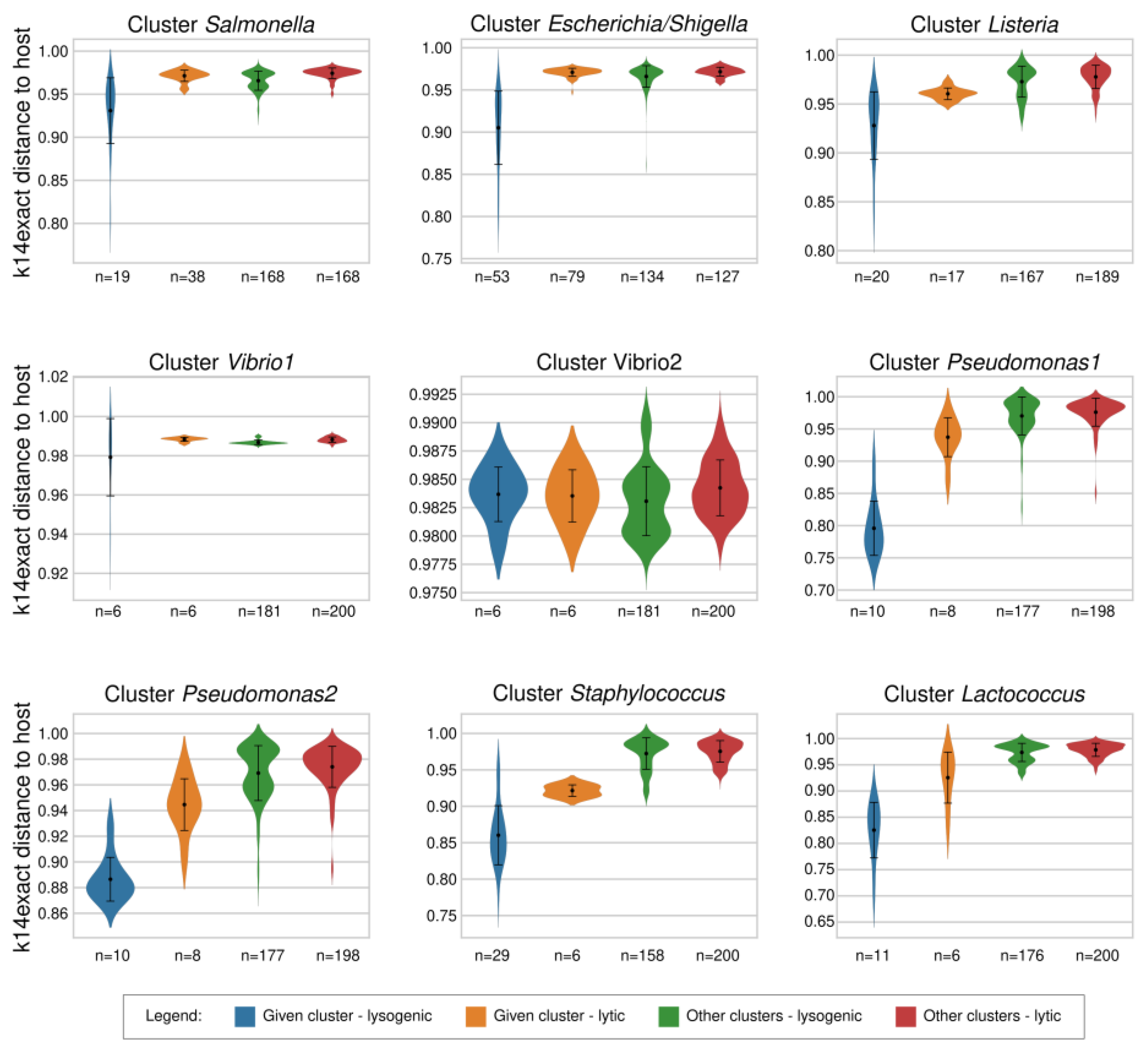
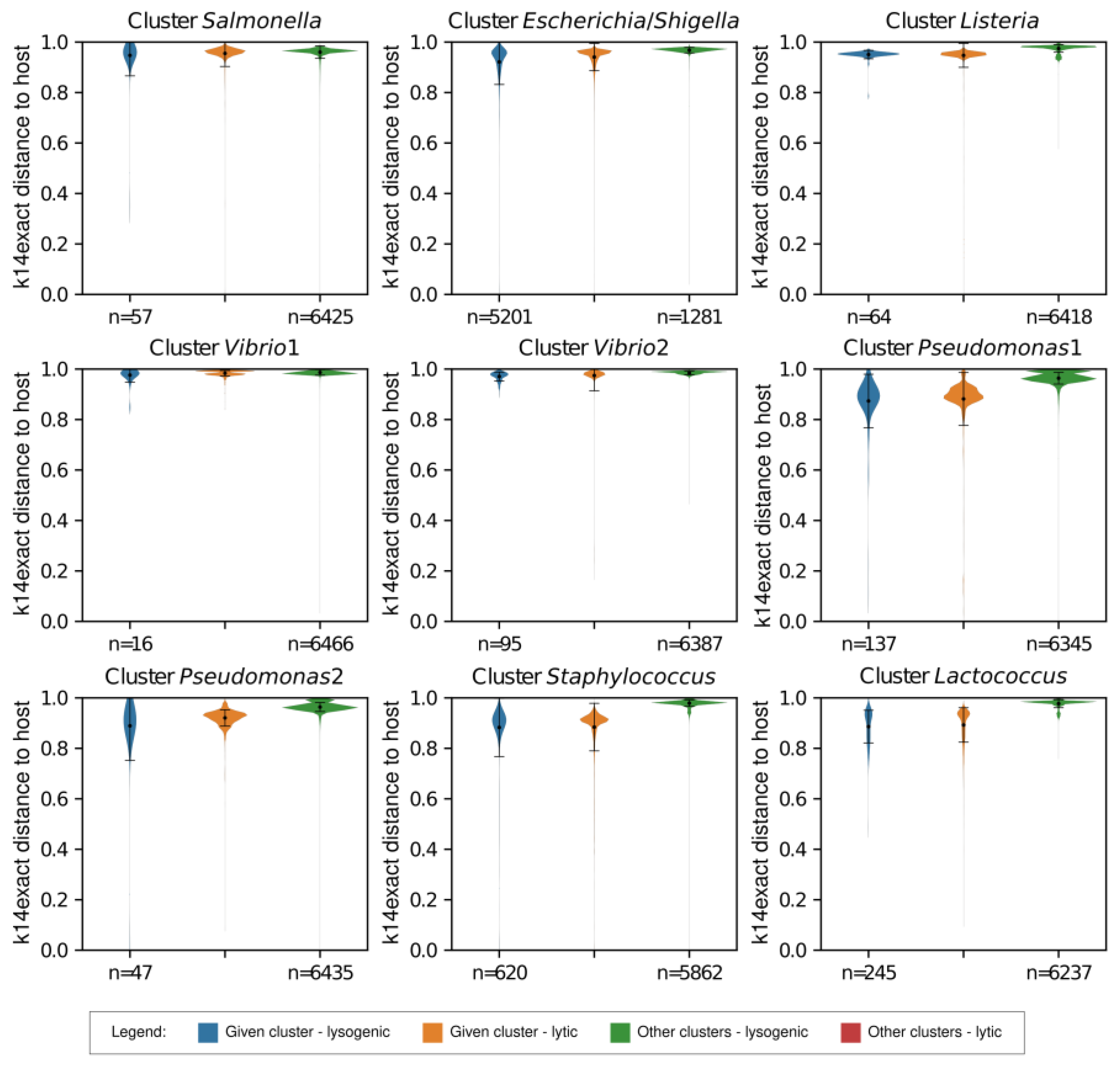
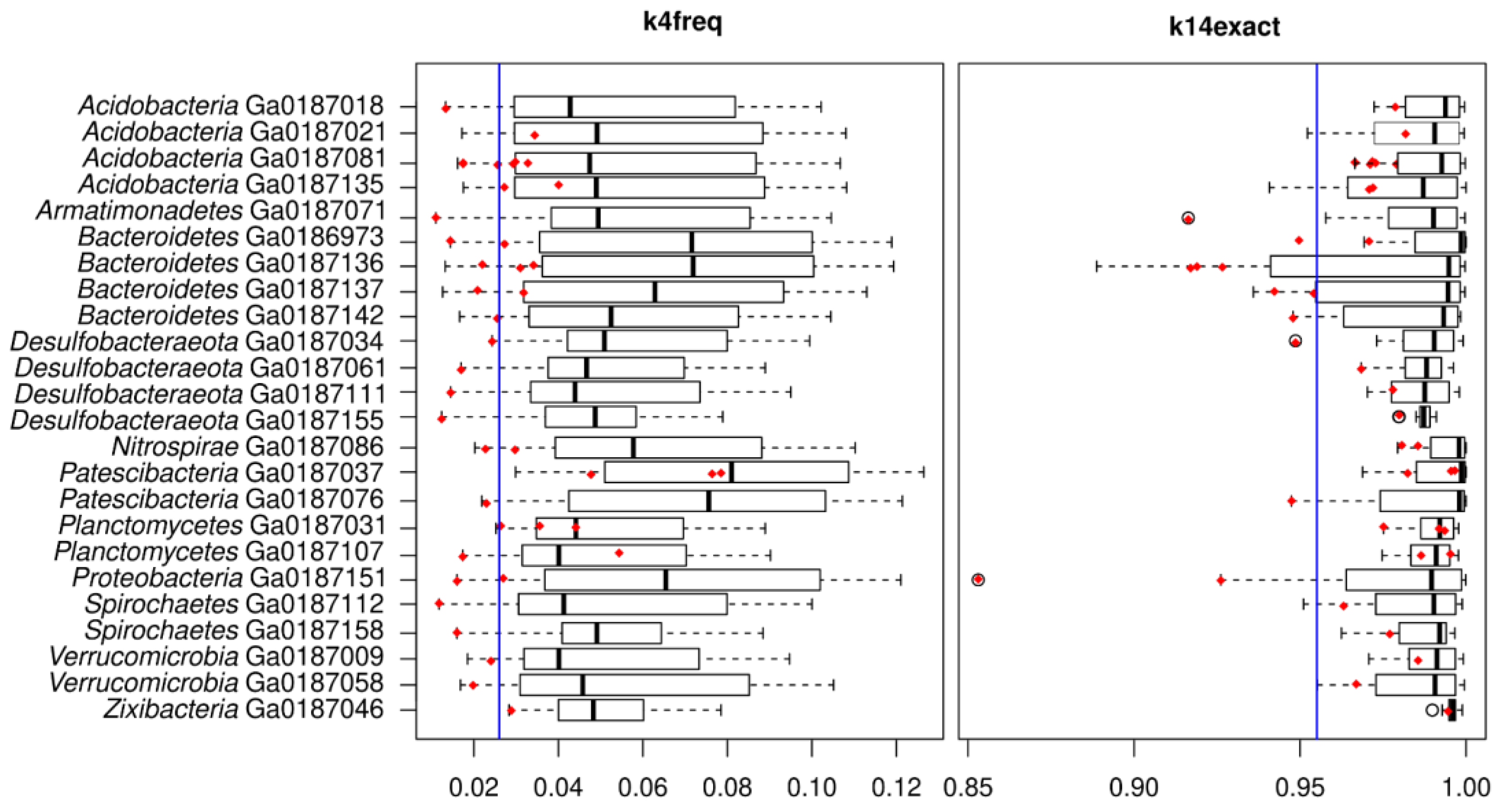
2.3. Comparison of k14exact and k4freq Methods
2.4. Reference Genomes Set Tested for the Strain-Level Association of Plasmids
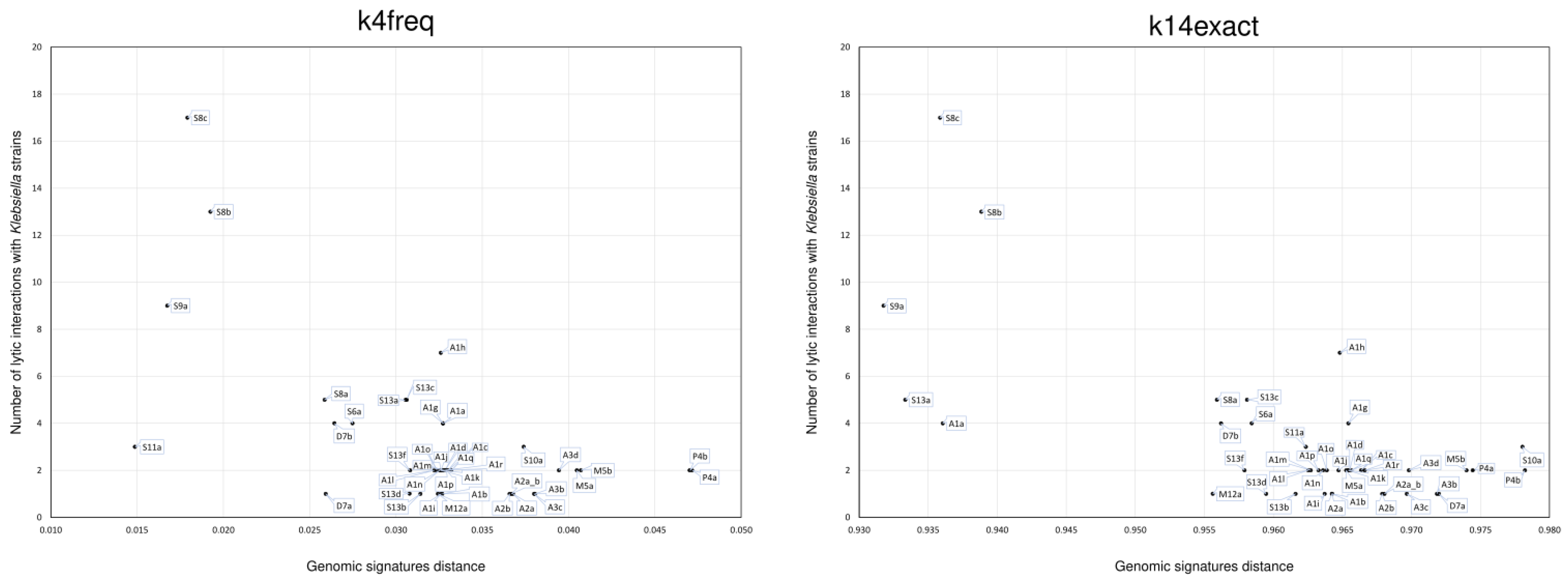
2.5. Interactions of Klebsiella Pneumoniae Strains with Their Lytic Phages
2.6. Host–Phage Interactions Predicted from Bacterial Single Cells
3. Discussion
4. Methods
4.1. Datasets
4.2. Genomic Signatures Based on Oligonucloetide Frequencies
4.3. Alignment-Free Genomic Distances
4.4. Determination of Thresholds for Distinguishing Lytic and Lysogenic Phages
4.5. Determination of Lysogenic Activity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; Ponsero, A.; Thornton, J.; U’Ren, J.M. Phage hunters: Computational strategies for finding phages in large-scale ‘omics datasets. Virus Res. 2018, 244, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Chevallereau, A.; Pons, B.J.; van Houte, S.; Westra, E.R. Interactions between bacterial and phage communities in natural environments. Nat. Rev. Microbiol. 2022, 20, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Batinovic, S.; Wassef, F.; Knowler, S.A.; Rice, D.T.F.; Stanton, C.R.; Rose, J.; Tucci, J.; Nittami, T.; Vinh, A.; Drummond, G.R.; et al. Bacteriophages in natural and artificial environments. Pathogens 2019, 8, 100. [Google Scholar] [CrossRef]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI-bacteriophage life cycle recognition with machine learning and natural language processing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19. [Google Scholar] [CrossRef]
- Low, S.J.; Džunková, M.; Chaumeil, P.A.; Parks, D.H.; Hugenholtz, P. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat. Microbiol. 2019, 4, 1306–1315. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E.V.; Fitzgerald, C.B.; Stockdale, S.R.; Draper, L.A.; Ross, R.P.; Hill, C. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018, 9, 4781. [Google Scholar] [CrossRef]
- Dang, V.T.; Sullivan, M.B. Emerging methods to study bacteriophage infection at the single-cell level. Front. Microbiol. 2014, 5, 724. [Google Scholar] [CrossRef] [PubMed]
- Marbouty, M.; Thierry, A.; Millot, G.A.; Koszul, R. MetaHiC phage-bacteria infection network reveals active cycling phages of the healthy human gut. eLife 2021, 10, e60608. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.W.; Lance, S.T.; Stedman, K.M.; Abate, A.R. PCR-activated cell sorting as a general, cultivation-free method for high-throughput identification and enrichment of virus hosts. J. Virol. Methods 2017, 242, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Džunková, M.; Low, S.J.; Daly, J.N.; Deng, L.; Rinke, C.; Hugenholtz, P. Defining the human gut host-phage network through single-cell viral tagging. Nat. Microbiol. 2019, 4, 2192–2203. [Google Scholar] [CrossRef] [PubMed]
- Jarett, J.K.; Džunková, M.; Schulz, F.; Roux, S.; Paez-Espino, D.; Eloe-Fadrosh, E.; Jungbluth, S.P.; Ivanova, N.; Spear, J.R.; Carr, S.A.; et al. Insights into the dynamics between viruses and their hosts in a hot spring microbial mat. ISME J. 2020, 14, 2527–2541. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, R.; Díaz-Villanueva, W.; Arnau, V.; Moya, A. Genomic signature in evolutionary biology: A review. Biology 2023, 12, 322. [Google Scholar] [CrossRef]
- Deschavanne, P.; DuBow, M.S.; Regeard, C. The use of genomic signature distance between bacteriophages and their hosts displays evolutionary relationships and phage growth cycle determination. Virol. J. 2010, 7, 163. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Ochman, H. Amelioration of bacterial genomes: Rates of change and exchange. J. Mol. Evol. 1997, 44, 383–397. [Google Scholar] [CrossRef]
- Pride, D.T.; Wassenaar, T.M.; Ghose, C.; Blaser, M.J. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genom. 2006, 7, 8. [Google Scholar] [CrossRef]
- Zielezinski, A.; Vinga, S.; Almeida, J.; Karlowski, W.M. Alignment-free sequence comparison: Benefits, applications, and tools. Genome Biol. 2017, 18, 186. [Google Scholar] [CrossRef]
- Elois, M.A.; Silva, R.D.; Pilati, G.V.T.; Rodríguez-Lázaro, D.; Fongaro, G. Bacteriophages as biotechnological tools. Viruses 2023, 15, 349. [Google Scholar] [CrossRef] [PubMed]
- Beamud, B.; García-González, N.; Gómez-Ortega, M.; González-Candelas, F.; Domingo-Calap, P.; Sanjuan, R. Genetic determinants of host tropism in Klebsiella phages. Cell. Rep. 2023, 42, 112048. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.P.; Nayfach, S.; Chen, I.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Ritter, S.J.; Reddy, T.B.K.; Mukherjee, S.; Schulz, F.; et al. IMG/VR v4: An expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res. 2023, 51, D733–D743. [Google Scholar] [CrossRef]
- Holmfeldt, K.; Middelboe, M.; Nybroe, O.; Riemann, L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl. Environ. Microbiol. 2007, 73, 6730–6739. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Reeves, P.R.; Beatson, S.A.; Hugenholtz, P. Reclassification of Shigella species as later heterotypic synonyms of Escherichia coli in the Genome Taxonomy Database. bioRxiv 2021. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 2016, 11, e0150183. [Google Scholar] [CrossRef]
- Takemura, A.F.; Chien, D.M.; Polz, M.F. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 2014, 5, 38. [Google Scholar] [CrossRef]
- Swain, M.T.; Vickers, M. Interpreting alignment-free sequence comparison: What makes a score a good score? NAR Genom. Bioinform. 2022, 4, lqac062. [Google Scholar] [CrossRef]
- Song, K. Classifying the lifestyle of metagenomically-derived phages sequences using alignment-free methods. Front. Microbiol. 2020, 11, 567769. [Google Scholar] [CrossRef]
- Wu, S.; Fang, Z.; Tan, J.; Li, M.; Wang, C.; Guo, Q.; Xu, C.; Jiang, X.; Zhu, H. DeePhage: Distinguishing virulent and temperate phage-derived sequences in metavirome data with a deep learning approach. Gigascience 2021, 10, giab056. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, L.; Bowler, L.; Caplin, J.; Dedi, C.; Diston, D.; Cheek, E.; Taylor, H.; Ebdon, J.E.; Jones, B.V. Genome signature-based dissection of human gut metagenomes to extract subliminal viral sequences. Nat. Commun. 2013, 4, 2420. [Google Scholar] [CrossRef] [PubMed]
- Sinha, V.; Goyal, A.; Svenningsen, S.L.; Semsey, S.; Krishna, S. In silico Evolution of lysis-lysogeny strategies reproduces observed lysogeny propensities in temperate bacteriophages. Front. Microbiol. 2017, 8, 1386. [Google Scholar] [CrossRef] [PubMed]
- Luo, E.; Eppley, J.M.; Romano, A.E.; Mende, D.R.; DeLong, E.F. Double-stranded DNA virioplankton dynamics and reproductive strategies in the oligotrophic open ocean water column. ISME J. 2020, 14, 1304–1315. [Google Scholar] [CrossRef]
- Roux, S.; Hallam, S.J.; Woyke, T.; Sullivan, M.B. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. eLife 2015, 4, e08490. [Google Scholar] [CrossRef]
- Ferry, T.; Kolenda, C.; Laurent, F.; Leboucher, G.; Merabischvilli, M.; Djebara, S.; Gustave, C.A.; Perpoint, T.; Barrey, C.; Pirnay, J.P.; et al. Personalized bacteriophage therapy to treat pandrug-resistant spinal Pseudomonas aeruginosa infection. Nat. Commun. 2022, 13, 4239. [Google Scholar] [CrossRef]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef]
- Sánchez-Osuna, M.; Barbé, J.; Erill, I. Systematic in silico assessment of antimicrobial resistance dissemination across the global plasmidome. Antibiotics 2023, 12, 281. [Google Scholar] [CrossRef]
- Aytan-Aktug, D.; Clausen, P.T.L.C.; Szarvas, J.; Munk, P.; Otani, S.; Nguyen, M.; Davis, J.J.; Lund, O.; Aarestrup, F.M. PlasmidHostFinder: Prediction of plasmid hosts using random forest. mSystems 2022, 7, e0118021. [Google Scholar] [CrossRef]
- Quince, C.; Nurk, S.; Raguideau, S.; James, R.; Soyer, O.S.; Summers, J.K.; Limasset, A.; Eren, A.M.; Chikhi, R.; Darling, A.E. STRONG: Metagenomics strain resolution on assembly graphs. Genome Biol. 2021, 22, 214. [Google Scholar] [CrossRef]
- Suzuki, H.; Brown, C.J.; Top, E.M. Genomic signature analysis to predict plasmid host range. In Molecular Life Sciences; Wells, R.D., Bond, J.S., Klinman, J., Masters, B.S.S., Eds.; Springer: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Meng, M.; Li, Y.; Yao, H. Plasmid-mediated transfer of antibiotic resistance genes in soil. Antibiotics 2022, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Mavrich, T.N.; Hatfull, G.F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2017, 2, 17112. [Google Scholar] [CrossRef] [PubMed]
- Vinga, S.; Almeida, J. Alignment-free sequence comparison—A review. Bioinformatics 2003, 19, 513–523. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Screening for lysogen activity in therapeutically relevant bacteriophages. Bio. Protoc. 2021, 11, e3997. [Google Scholar] [CrossRef]
- Hockenberry, A.J.; Wilke, C.O. BACPHLIP: Predicting bacteriophage lifestyle from conserved protein domains. PeerJ 2021, 9, e11396. [Google Scholar] [CrossRef] [PubMed]
- Terzian, P.; Olo Ndela, E.; Galiez, C.; Lossouarn, J.; Pérez Bucio, R.E.; Mom, R.; Toussaint, A.; Petit, M.A.; Enault, F. PHROG: Families of prokaryotic virus proteins clustered using remote homology. NAR Genom. Bioinform. 2021, 3, lqab067. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnau, V.; Díaz-Villanueva, W.; Mifsut Benet, J.; Villasante, P.; Beamud, B.; Mompó, P.; Sanjuan, R.; González-Candelas, F.; Domingo-Calap, P.; Džunková, M. Inference of the Life Cycle of Environmental Phages from Genomic Signature Distances to Their Hosts. Viruses 2023, 15, 1196. https://doi.org/10.3390/v15051196
Arnau V, Díaz-Villanueva W, Mifsut Benet J, Villasante P, Beamud B, Mompó P, Sanjuan R, González-Candelas F, Domingo-Calap P, Džunková M. Inference of the Life Cycle of Environmental Phages from Genomic Signature Distances to Their Hosts. Viruses. 2023; 15(5):1196. https://doi.org/10.3390/v15051196
Chicago/Turabian StyleArnau, Vicente, Wladimiro Díaz-Villanueva, Jorge Mifsut Benet, Paula Villasante, Beatriz Beamud, Paula Mompó, Rafael Sanjuan, Fernando González-Candelas, Pilar Domingo-Calap, and Mária Džunková. 2023. "Inference of the Life Cycle of Environmental Phages from Genomic Signature Distances to Their Hosts" Viruses 15, no. 5: 1196. https://doi.org/10.3390/v15051196
APA StyleArnau, V., Díaz-Villanueva, W., Mifsut Benet, J., Villasante, P., Beamud, B., Mompó, P., Sanjuan, R., González-Candelas, F., Domingo-Calap, P., & Džunková, M. (2023). Inference of the Life Cycle of Environmental Phages from Genomic Signature Distances to Their Hosts. Viruses, 15(5), 1196. https://doi.org/10.3390/v15051196







