Canocapavir Is a Novel Capsid Assembly Modulator Inducing a Conformational Change of the Linker Region of HBV Core Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Antiviral Compounds
2.2. DNA Constructs
2.3. Antibodies
2.4. siRNA and Transfection
2.5. Southern Blot Analysis of HBV Replicative Mediates
2.6. Analysis of Viral Particles
2.7. Western Blot Assay
2.8. Ultracentrifugation of HBV Capsids
2.9. Immunofluorescence Staining
2.10. Immunoprecipitation of Virions in Supernatant
2.11. Co-Immunoprecipitation Assay
2.12. Molecule Docking Analysis
2.13. ELISA
2.14. Real-Time PCR HBV
2.15. Statistical Analysis
3. Results
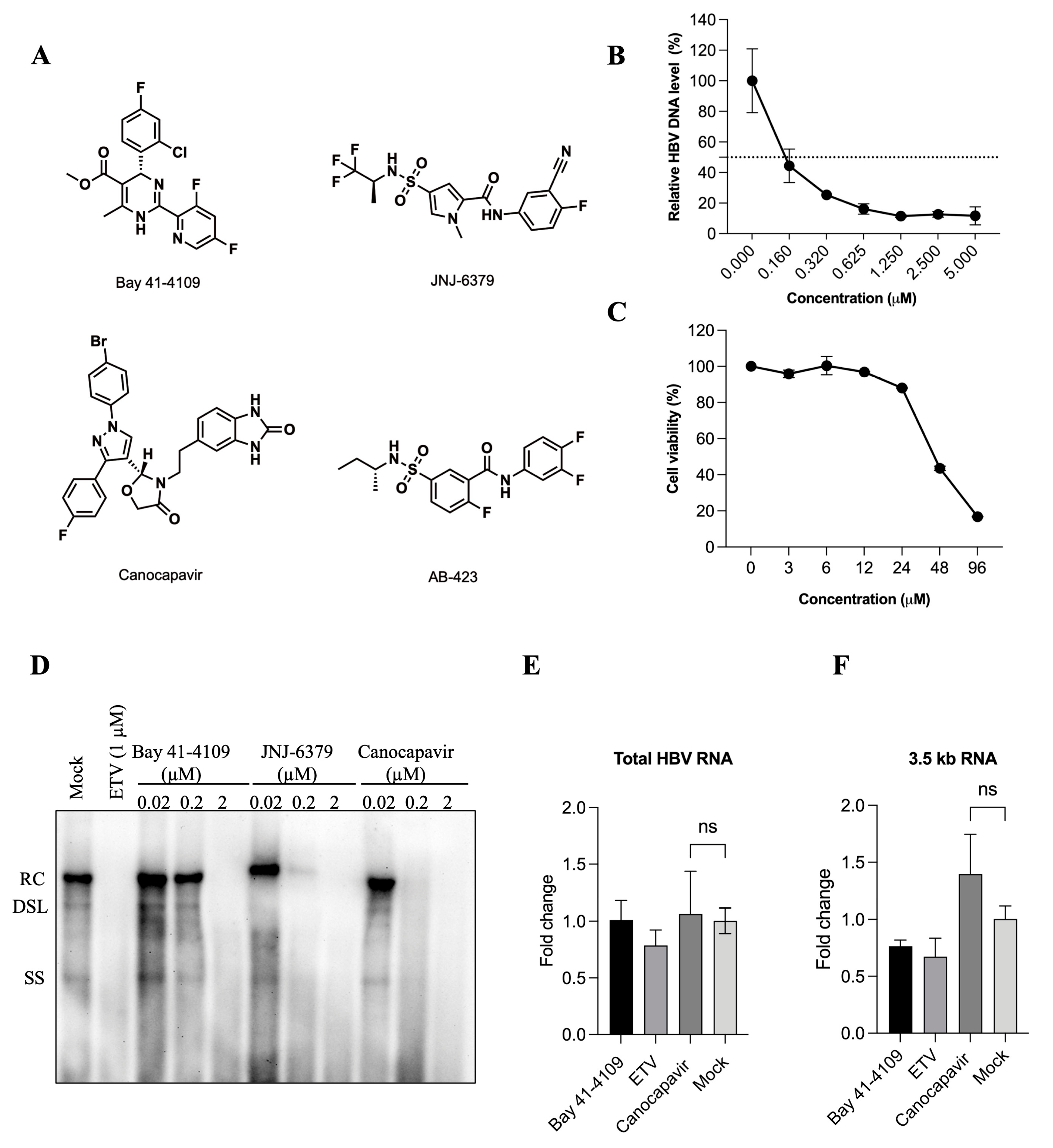
3.1. Characterization of the Antiviral Activity of Canocapavir
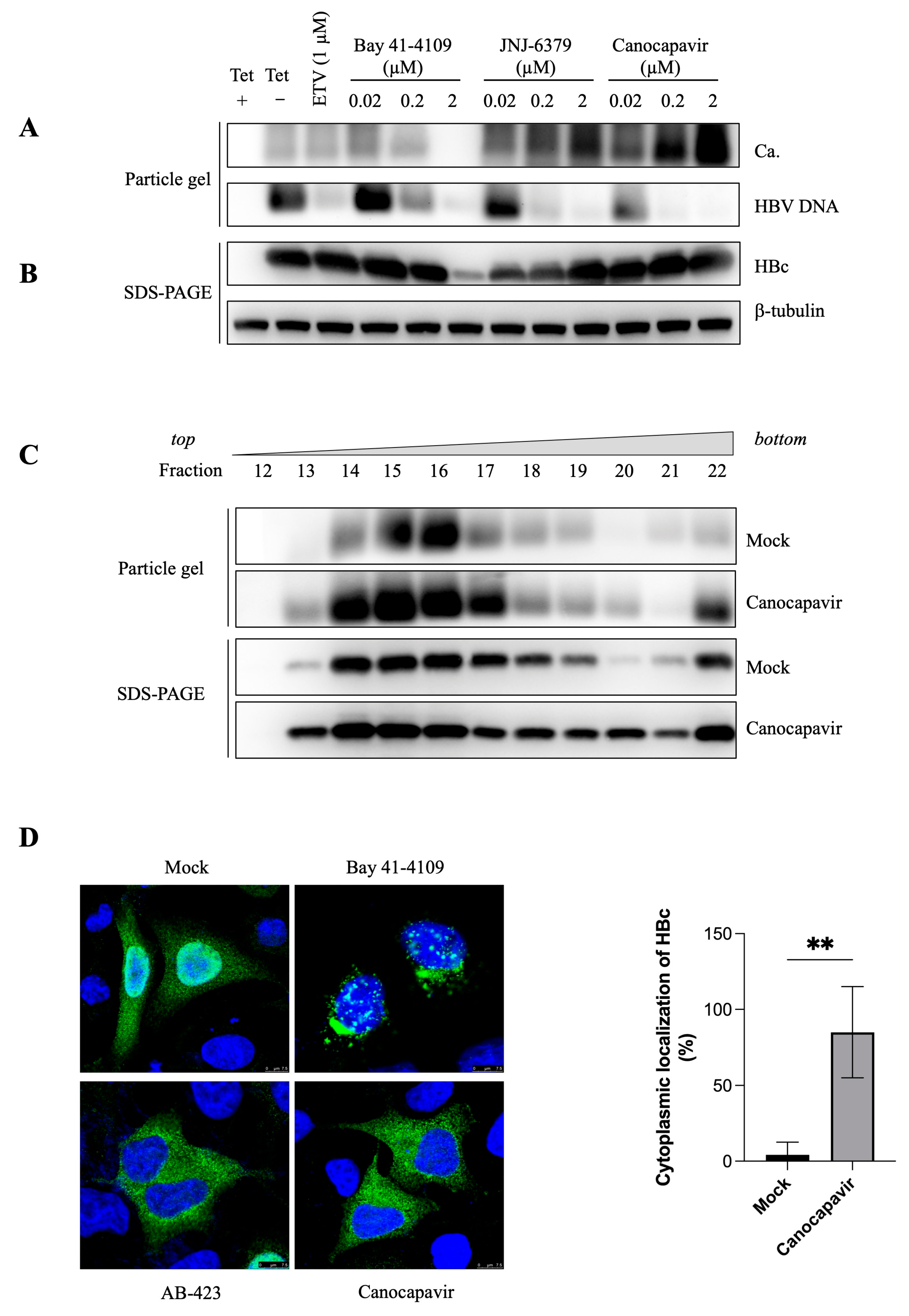
3.2. Canocapavir Promotes Cytoplasmic Accumulation of Empty Nucleocapsids
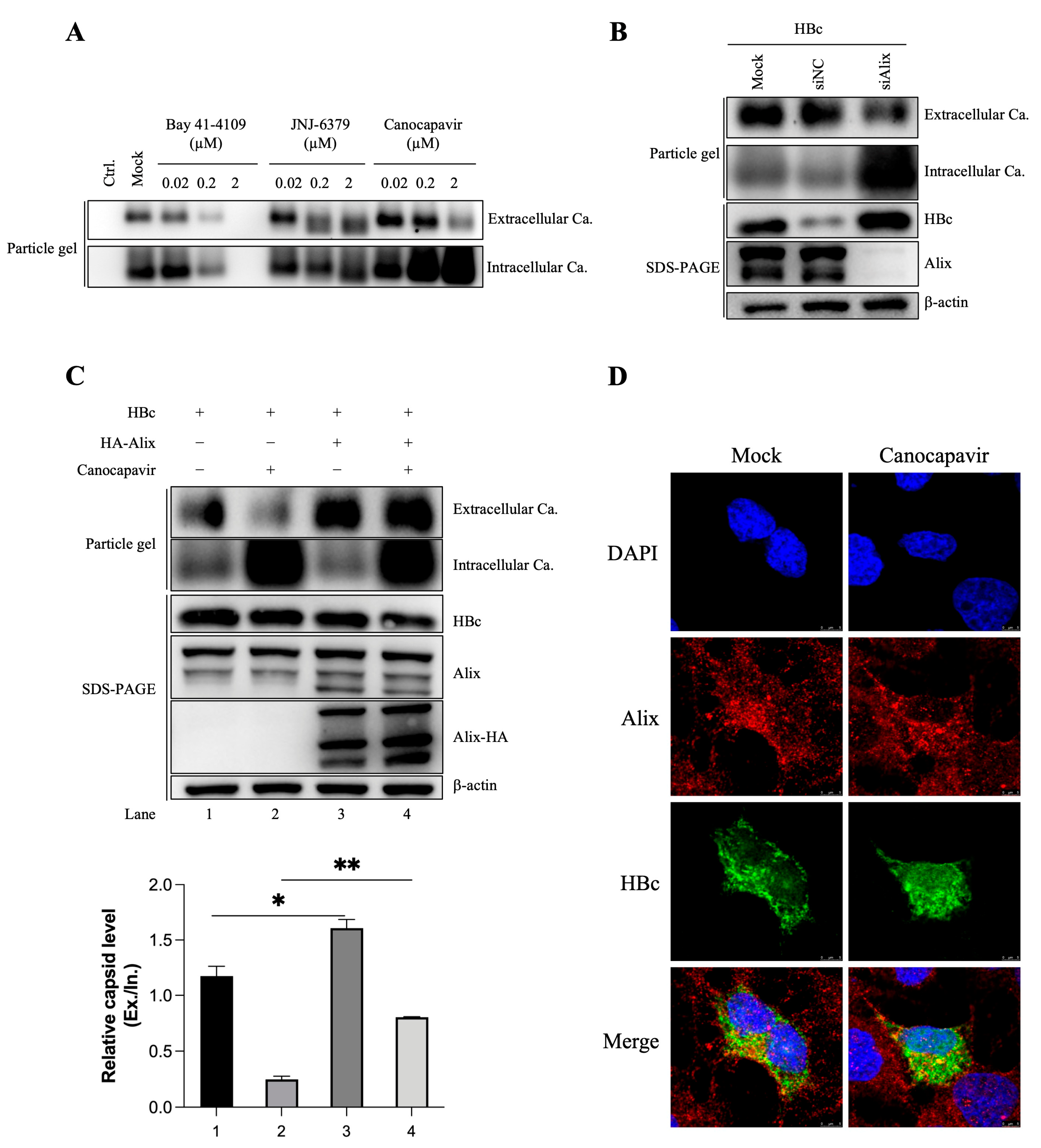
3.3. Overexpressing Alix Abolishes the Inhibitory Effect of Canocapavir on Naked Capsid Egress
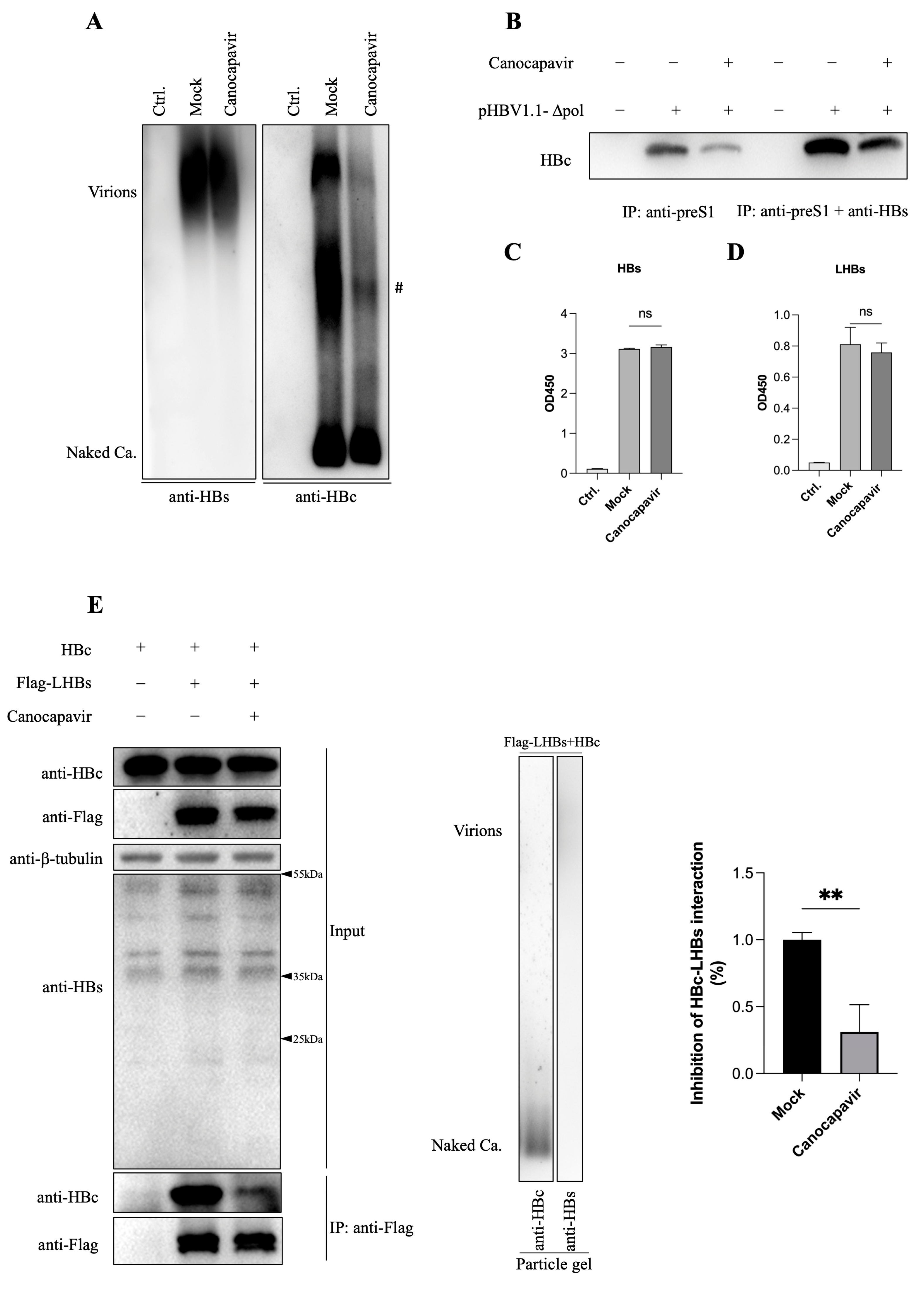
3.4. Canocapavir Inhibits HBV Envelopment by Interrupting the Interaction between HBc and LHBs
3.5. Canocapavir Induces a Conformational Change at the HBc Linker Region
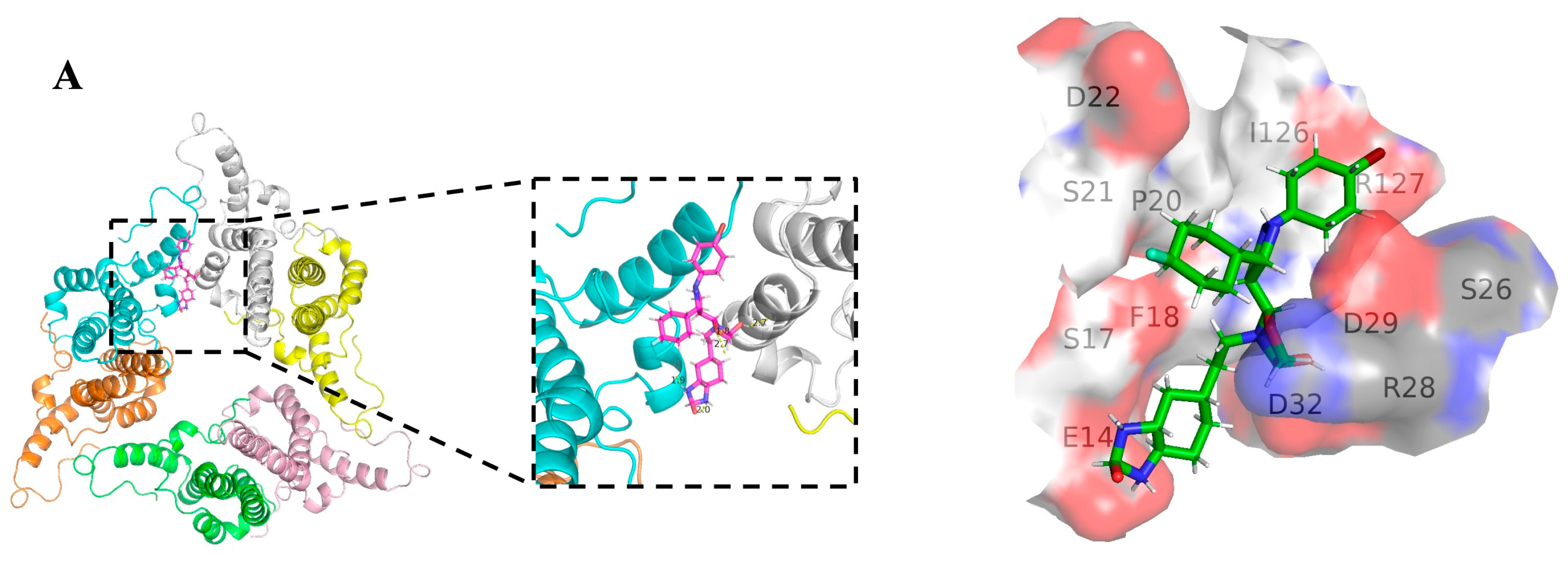
3.6. The Binding Site of Canocapavir Is Located at the Hydrophobic (HAP) Pocket
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: A modelling study. Lancet Gastroenterol. Hepatol. 2018, 3, 383–403. [CrossRef]
- Block, T.M.; Guo, H.; Guo, J.-T. Molecular Virology of Hepatitis B Virus for Clinicians. Clin. Liver Dis. 2007, 11, 685–706. [Google Scholar] [CrossRef]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, K. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses 2017, 9, 56. [Google Scholar] [CrossRef]
- Bai, L.; Zhang, X.; Kozlowski, M.; Li, W.; Wu, M.; Liu, J.; Chen, L.; Zhang, J.; Huang, Y.; Yuan, Z. Extracellular Hepatitis B Virus RNAs Are Heterogeneous in Length and Circulate as Capsid-Antibody Complexes in Addition to Virions in Chronic Hepatitis B Patients. J. Virol. 2018, 92, e00798-18. [Google Scholar] [CrossRef]
- Ning, X.; Nguyen, D.; Mentzer, L.; Adams, C.; Lee, H.; Ashley, R.; Hafenstein, S.; Hu, J. Secretion of Genome-Free Hepatitis B Virus—Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus. PLoS Pathog. 2011, 7, e1002255. [Google Scholar] [CrossRef] [PubMed]
- Zlotnick, A.; Venkatakrishnan, B.; Tan, Z.; Lewellyn, E.; Turner, W.; Francis, S. Core protein: A pleiotropic keystone in the HBV lifecycle. Antivir. Res. 2015, 121, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Wynne, S.; Crowther, R.; Leslie, A. The Crystal Structure of the Human Hepatitis B Virus Capsid. Mol. Cell 1999, 3, 771–780. [Google Scholar] [CrossRef]
- Bruss, V. Hepatitis B virus morphogenesis. World J. Gastroenterol. 2007, 13, 65–73. [Google Scholar] [CrossRef]
- Basagoudanavar, S.H.; Perlman, D.H.; Hu, J. Regulation of Hepadnavirus Reverse Transcription by Dynamic Nucleocapsid Phosphorylation. J. Virol. 2007, 81, 1641–1649. [Google Scholar] [CrossRef]
- Ludgate, L.; Liu, K.; Luckenbaugh, L.; Streck, N.; Eng, S.; Voitenleitner, C.; Delaney, W.E.; Hu, J. Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation. J. Virol. 2016, 90, 5830–5844. [Google Scholar] [CrossRef]
- Zhao, Q.; Hu, Z.; Cheng, J.; Wu, S.; Luo, Y.; Chang, J.; Hu, J.; Guo, J.-T. Hepatitis B Virus Core Protein Dephosphorylation Occurs during Pregenomic RNA Encapsidation. J. Virol. 2018, 92, e02139-17. [Google Scholar] [CrossRef]
- Hu, Z.; Ban, H.; Zheng, H.; Liu, M.; Chang, J.; Guo, J.-T. Protein phosphatase 1 catalyzes HBV core protein dephosphorylation and is co-packaged with viral pregenomic RNA into nucleocapsids. PLoS Pathog. 2020, 16, e1008669. [Google Scholar] [CrossRef]
- Liu, K.; Luckenbaugh, L.; Ning, X.; Xi, J.; Hu, J. Multiple roles of core protein linker in hepatitis B virus replication. PLoS Pathog. 2018, 14, e1007085. [Google Scholar] [CrossRef]
- Xi, J.; Luckenbaugh, L.; Hu, J. Multiple roles of PP2A binding motif in hepatitis B virus core linker and PP2A in regulating core phosphorylation state and viral replication. PLoS Pathog. 2021, 17, e1009230. [Google Scholar] [CrossRef]
- Xi, J. Region-Specific Hepatitis B Virus Genome Exposure from Nucleocapsid Modulated by Capsid Linker Sequence and In-hibitor: Implications for Uncoating. J. Virol. 2022, 96, e0039922. [Google Scholar] [CrossRef]
- Liu, H.; Xi, J.; Hu, J. Regulation of Hepatitis B Virus Replication by Cyclin Docking Motifs in Core Protein. J. Virol. 2021, 95, e00230-21. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, U.; Mani, N.; Hu, Z.; Ban, H.; Du, Y.; Hu, J.; Chang, J.; Guo, J.-T. Targeting the multifunctional HBV core protein as a potential cure for chronic hepatitis B. Antivir. Res. 2020, 182, 104917. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Mai, J.; Wu, M.; Chen, H.; Li, X.; Li, C.; Liu, J.; Liu, C.; Hu, Y.; Zhu, X.; et al. Safety, tolerability, pharmacokinetics, and antiviral activity of the novel core protein allosteric modulator ZM-H1505R (Canocapavir) in chronic hepatitis B patients: A randomized multiple-dose escalation trial. BMC Med. 2023, 21, 98. [Google Scholar] [CrossRef]
- Qi, Z. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J. Virol. 2014, 88, 8045–8056. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Guo, H.; Pan, X.-B.; Mao, R.; Yu, W.; Xu, X.; Wei, L.; Chang, J.; Block, T.M.; Guo, J.-T. Interferons Accelerate Decay of Replication-Competent Nucleocapsids of Hepatitis B Virus. J. Virol. 2010, 84, 9332–9340. [Google Scholar] [CrossRef] [PubMed]
- Deres, K.; Schroder, C.H.; Paessens, A.; Goldmann, S.; Hacker, H.J.; Weber, O.; Rubsamen-Waigmann, H. Faculty Opinions recommendation of Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 2003, 299, 184414. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-C.; Huang, E.-Y.; Su, P.-Y.; Wu, S.-Y.; Yang, C.-C.; Lin, Y.-S.; Chang, W.-C.; Shih, C. Nuclear Export and Import of Human Hepatitis B Virus Capsid Protein and Particles. PLoS Pathog. 2010, 6, e1001162. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yin, L.; Xu, X.-Z.; Sun, H.-C.; Huang, Z.-H.; Ni, X.-Y.; Chen, Y.; Lin, X. Bay41-4109-induced aberrant polymers of hepatitis b capsid proteins are removed via STUB1-promoted p62-mediated macroautophagy. PLoS Pathog. 2022, 18, e1010204. [Google Scholar] [CrossRef]
- Watanabe, T.; Sorensen, E.M.; Naito, A.; Schott, M.; Kim, S.; Ahlquist, P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. USA 2007, 104, 10205–10210. [Google Scholar] [CrossRef]
- Lambert, C.; Döring, T.; Prange, R. Hepatitis B Virus Maturation Is Sensitive to Functional Inhibition of ESCRT-III, Vps4, and γ2-Adaptin. J. Virol. 2007, 81, 9050–9060. [Google Scholar] [CrossRef]
- Bardens, A.; Döring, T.; Stieler, J.; Prange, R. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell Microbiol. 2010, 13, 602–619. [Google Scholar] [CrossRef]
- Wang, S.-J.; Chen, Z.-M.; Wei, M.; Liu, J.-Q.; Li, Z.-L.; Shi, T.-S.; Nian, S.; Fu, R.; Wu, Y.-T.; Zhang, Y.-L.; et al. Specific determination of hepatitis B e antigen by antibodies targeting precore unique epitope facilitates clinical diagnosis and drug evaluation against hepatitis B virus infection. Emerg. Microbes Infect. 2021, 10, 37–50. [Google Scholar] [CrossRef]
- Ning, X. Common and Distinct Capsid and Surface Protein Requirements for Secretion of Complete and Genome-Free Hepatitis B Virions. J. Virol. 2018, 92, e00272-18. [Google Scholar] [CrossRef]
- Wu, S.; Luo, Y.; Viswanathan, U.; Kulp, J.; Cheng, J.; Hu, Z.; Xu, Q.; Zhou, Y.; Gong, G.-Z.; Chang, J.; et al. CpAMs induce assembly of HBV capsids with altered electrophoresis mobility: Implications for mechanism of inhibiting pgRNA packaging. Antivir. Res. 2018, 159, 1–12. [Google Scholar] [CrossRef]
- Ruan, L.; Hadden, J.A.; Zlotnick, A. Assembly Properties of Hepatitis B Virus Core Protein Mutants Correlate with Their Resistance to Assembly-Directed Antivirals. J. Virol. 2018, 92, e01082-18. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Cheng, J.; Hu, Z.; Ban, H.; Wu, S.; Hwang, N.; Kulp, J.; Li, Y.; Du, Y.; Chang, J.; et al. Identification of hepatitis B virus core protein residues critical for capsid assembly, pgRNA encapsidation and resistance to capsid assembly modulators. Antivir. Res. 2021, 191, 105080. [Google Scholar] [CrossRef]
- Tan, Z.; Pionek, K.; Unchwaniwala, N.; Maguire, M.L.; Loeb, D.D.; Zlotnick, A. The Interface between Hepatitis B Virus Capsid Proteins Affects Self-Assembly, Pregenomic RNA Packaging, and Reverse Transcription. J. Virol. 2015, 89, 3275–3284. [Google Scholar] [CrossRef] [PubMed]
- Lok, A.S.; Zoulim, F.; Dusheiko, G.; Ghany, M.G. Hepatitis B cure: From discovery to regulatory approval. Hepatology 2017, 66, 1296–1313. [Google Scholar] [CrossRef]
- Perrillo, R. Benefits and risks of interferon therapy for hepatitis B. Hepatology 2009, 49 (Suppl. 5), S103–S111. [Google Scholar] [CrossRef]
- Dienstag, J.L. Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology 2009, 49, S112–S121. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Zlotnick, A. HBV Core Protein Is in Flux between Cytoplasmic, Nuclear, and Nucleolar Compartments. mBio 2021, 12, e03514-20. [Google Scholar] [CrossRef]
- Tan, Z.; Maguire, M.L.; Loeb, D.D.; Zlotnick, A. Genetically Altering the Thermodynamics and Kinetics of Hepatitis B Virus Capsid Assembly Has Profound Effects on Virus Replication in Cell Culture. J. Virol. 2013, 87, 3208–3216. [Google Scholar] [CrossRef]
- Orabi, A.; Bieringer, M.; Geerlof, A.; Bruss, V. An Aptamer against the Matrix Binding Domain on the Hepatitis B Virus Capsid Impairs Virion Formation. J. Virol. 2015, 89, 9281–9287. [Google Scholar] [CrossRef]
- Lauber, C.; Seitz, S.; Mattei, S.; Suh, A.; Beck, J.; Herstein, J.; Börold, J.; Salzburger, W.; Kaderali, L.; Briggs, J.A.; et al. Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses. Cell Host Microbe 2017, 22, 387–399.e6. [Google Scholar] [CrossRef]
- Döring, T.; Prange, R. Rab33B and its autophagic Atg5/12/16L1 effector assist in hepatitis B virus naked capsid formation and release. Cell Microbiol. 2015, 17, 747–764. [Google Scholar] [CrossRef] [PubMed]
- Döring, T. Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does Not Require Atg8/LC3 Lipidation for Viral Maturation. J. Virol. 2018, 92, e01513-17. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Yang, L.; Yu, L.; Zhu, Y.; Wu, Y.; Zhang, Z.; Xia, T.; Deng, Q. Canocapavir Is a Novel Capsid Assembly Modulator Inducing a Conformational Change of the Linker Region of HBV Core Protein. Viruses 2023, 15, 1195. https://doi.org/10.3390/v15051195
Zheng Y, Yang L, Yu L, Zhu Y, Wu Y, Zhang Z, Xia T, Deng Q. Canocapavir Is a Novel Capsid Assembly Modulator Inducing a Conformational Change of the Linker Region of HBV Core Protein. Viruses. 2023; 15(5):1195. https://doi.org/10.3390/v15051195
Chicago/Turabian StyleZheng, Yuan, Le Yang, Lin Yu, Yuanfei Zhu, Yang Wu, Zhijun Zhang, Tian Xia, and Qiang Deng. 2023. "Canocapavir Is a Novel Capsid Assembly Modulator Inducing a Conformational Change of the Linker Region of HBV Core Protein" Viruses 15, no. 5: 1195. https://doi.org/10.3390/v15051195
APA StyleZheng, Y., Yang, L., Yu, L., Zhu, Y., Wu, Y., Zhang, Z., Xia, T., & Deng, Q. (2023). Canocapavir Is a Novel Capsid Assembly Modulator Inducing a Conformational Change of the Linker Region of HBV Core Protein. Viruses, 15(5), 1195. https://doi.org/10.3390/v15051195



