Identification and Profiling of a Novel Bombyx mori latent virus Variant Acutely Infecting Helicoverpa armigera and Trichoplusia ni
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics
2.1.1. Virus Discovery
2.1.2. Genome Assembly
2.1.3. vsRNA Analysis and piRNA Fingerprinting
2.1.4. Phylogenetic Analysis
2.2. Isolation of BmLV from Hi5 Cells
2.3. Infection of IOZCAS-Ha-I by BmLV
2.4. Helicoverpa armigera Rearing, Infection with BmLV, and Dissection
2.4.1. H. armigera Rearing
2.4.2. H. armigera Infection with BmLV
2.4.3. H. armigera Dissection
2.5. Total RNA Extraction
2.5.1. From Cells and Tissues
2.5.2. From Whole Insects
2.6. cDNA Synthesis
2.7. Quantitative Real-Time PCR
2.8. IOZCAS-Ha-I Species Identification and BmLV Genome Sequencing
2.9. Confocal Immunofluorescence Microscopy
2.10. Statistical Analysis
2.11. Next Generation Sequencing
3. Results
3.1. Prevalence and Phylogeny of BmLV in Lepidopteran Cell Lines and Insects
3.2. Prior Characterization of the Lepidopteran IOZCAS-Ha-I Cell Line
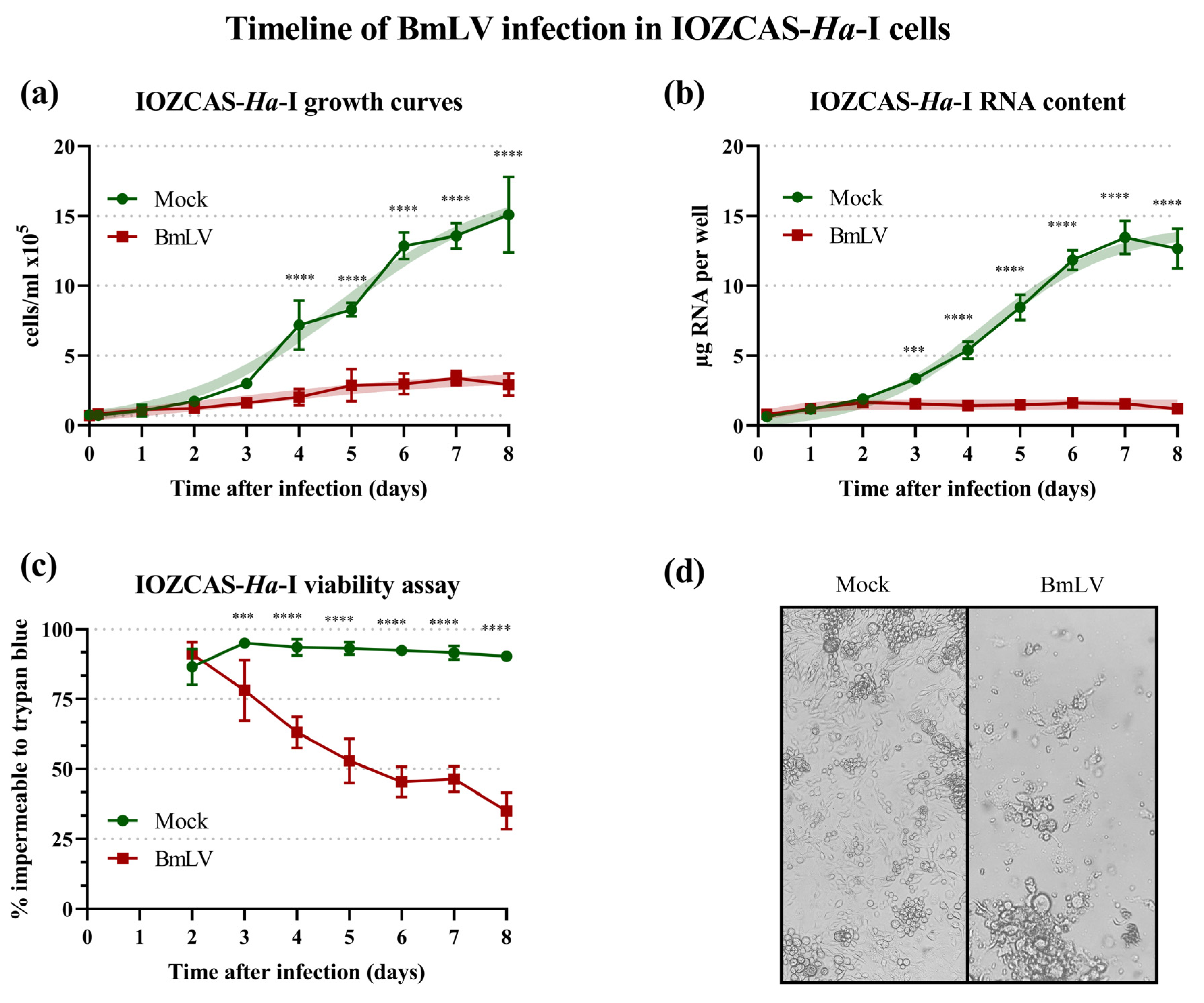
3.3. BmLV Causes Acute and Lethal Infection in the Lepidopteran IOZCAS-Ha-I Cell Line
3.4. BmLV Replication Co-Localizes with Mitochondria
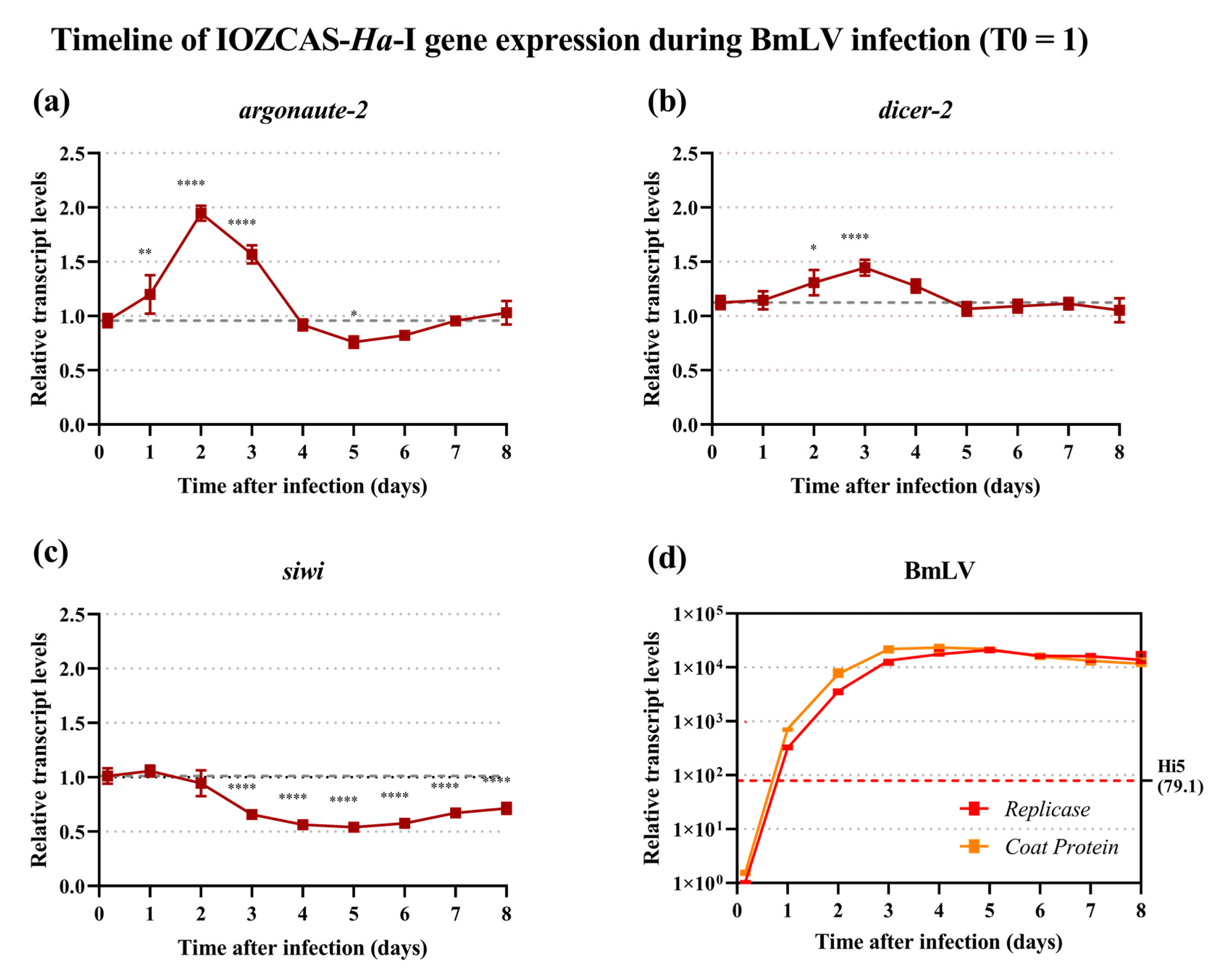
3.5. Regulation of siRNAi and piRNAi Machinery during BmLV Infection in IOZCAS-Ha-I Cells
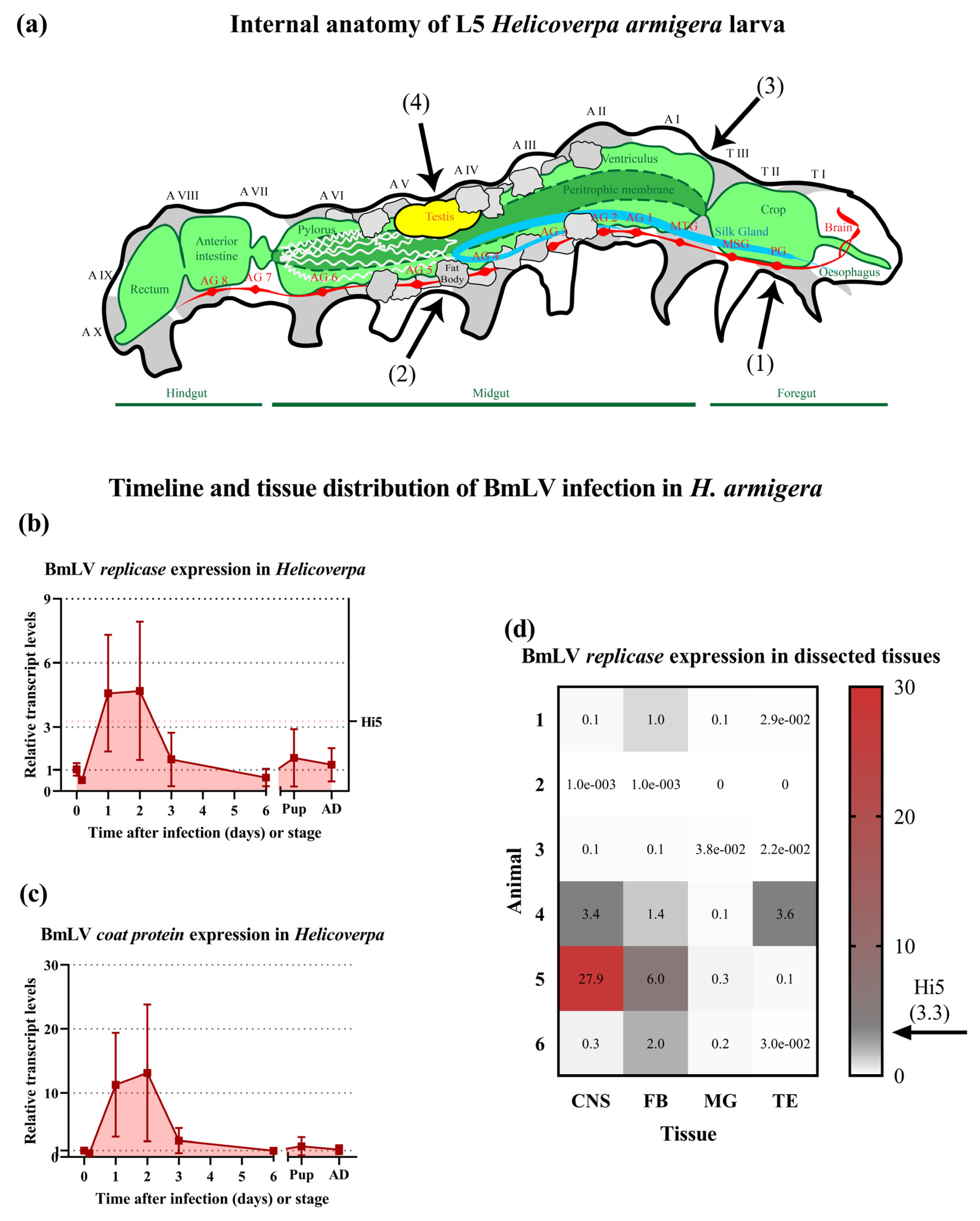
3.6. BmLV Causes Acute Followed by Persistent Infection in H. armigera In Vivo
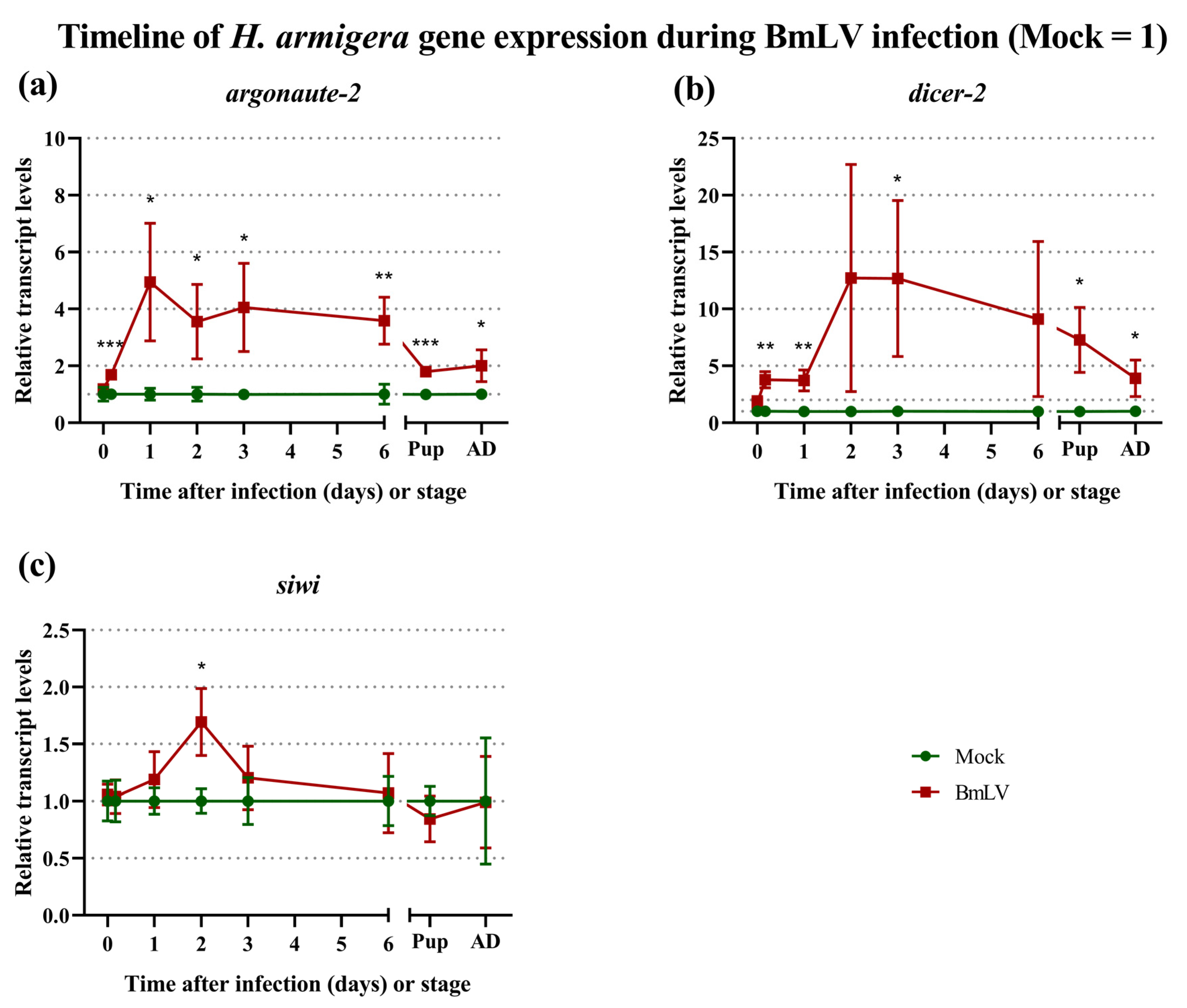
3.7. Regulation of siRNAi and piRNAi Machinery during BmLV Infection in H. armigera In Vivo
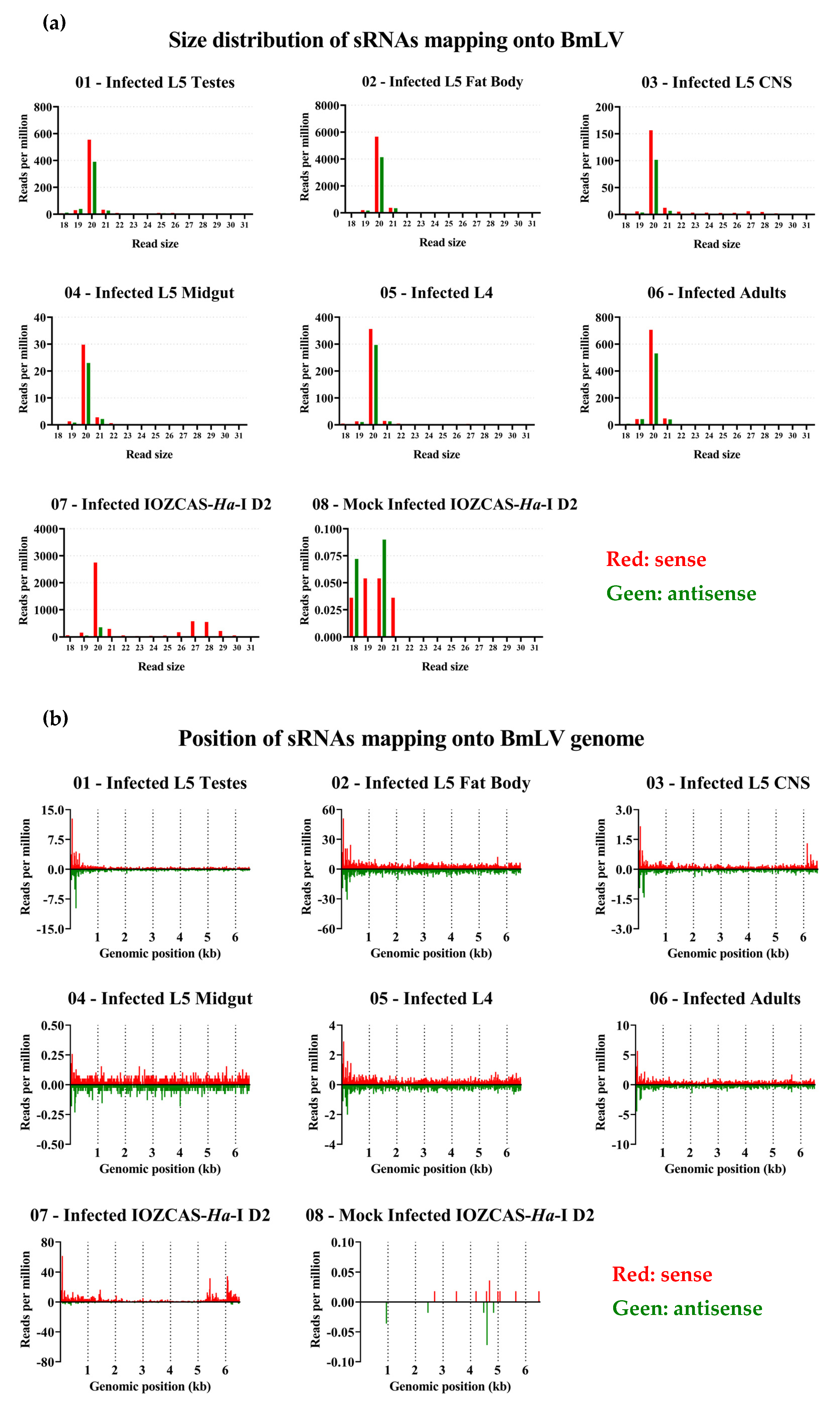
3.8. BmLV Infection in IOZCAS-Ha-I Cells and in H. armigera In Vivo Is Associated Mainly with Viral siRNAs
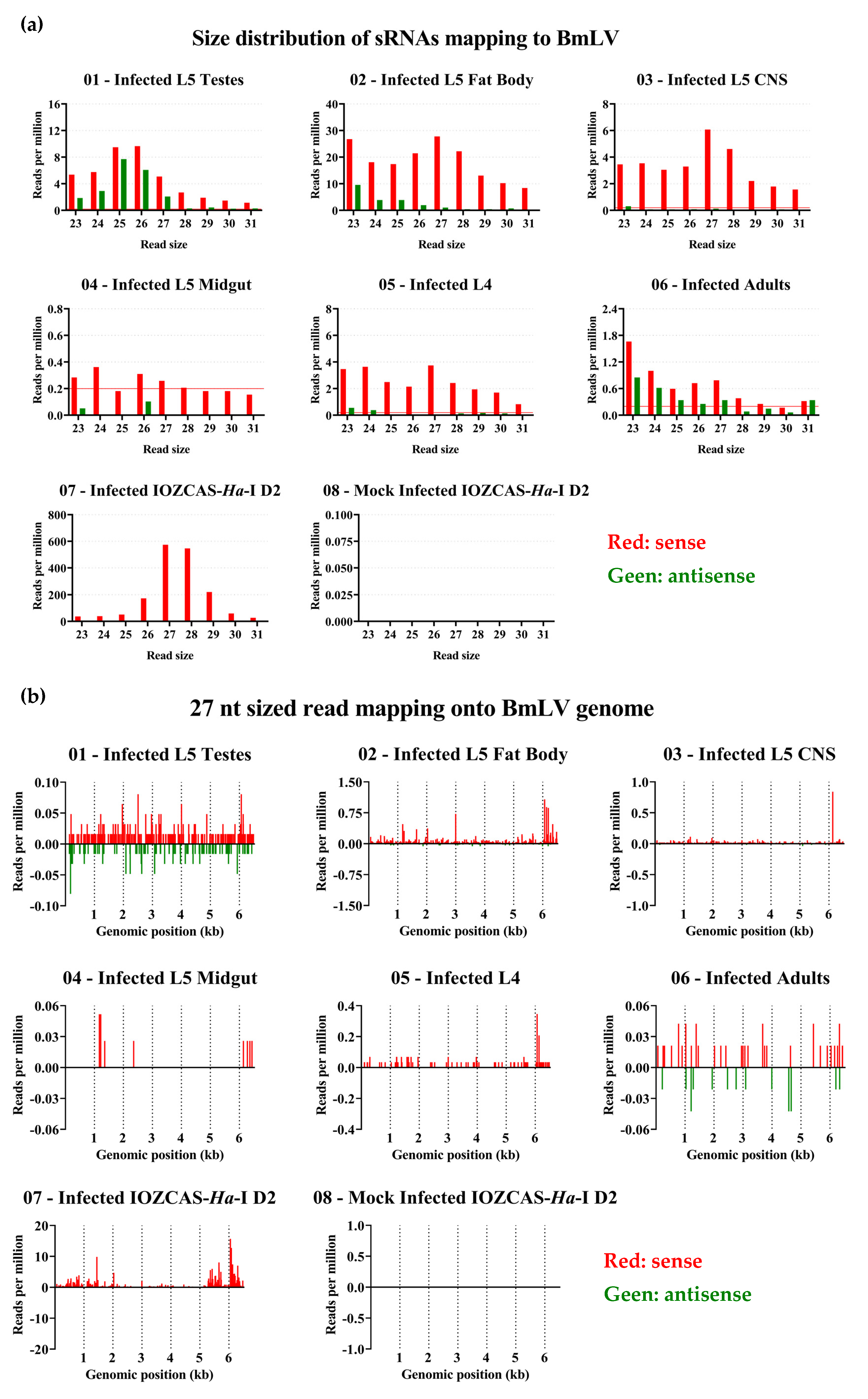
3.9. piRNA Biogenesis Pathways are Active in IOZCAS-Ha-I Cells and in the Studied H. armigera Tissues
4. Discussion
4.1. The Diversity of BmLV Variants and Its Implications
4.2. Persistent Viral Infections in Cell Lines and Their Implications
4.3. BmLV Acutely Infects Two Noctuidae Species In Vivo and In Vitro
4.4. sRNA-Based Immune Response to BmLV Infection
4.5. Secondary piRNA Biogenesis Pathway in Somatic vs. Germline Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Katsuma, S.; Tanaka, S.; Omuro, N.; Daimon, T.; Imanishi, S.; Iwanaga, M.; Mita, K.; Maeda, S.; Takabuchi, L.; Yamashita, S.; et al. Novel Macula-Like Virus Identified in Bombyx Mori Cultured Cells. Society 2005, 79, 5577–5584. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Kawamoto, M.; Shoji, K.; Aizawa, T.; Kiuchi, T.; Izumi, N.; Ogawa, M.; Mashiko, T.; Kawasaki, H.; Sugano, S.; et al. Transcriptome Profiling Reveals Infection Strategy of an Insect Maculavirus. DNA Res. 2018, 25, 277–286. [Google Scholar] [CrossRef]
- Dreher, T.W.; Edwards, M.C.; Gibbs, A.J.; Haenni, A.-L.; Hammond, R.W.; Jupin, I.; Koenig, R.; Sabanadzovic, S.; Martelli, G.P. Tymoviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; pp. 944–952. ISBN 9780123846846. [Google Scholar]
- Iwanaga, M. Persistent Virus in the Silkworm Cell Lines: Bombyx Mori Latent Virus. Uirusu 2019, 68, 137–146. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Scott Cornman, R.; Evans, J.D.; Semberg, E.; Haddad, N.; Neumann, P.; Gauthier, L. Genome Characterization, Prevalence and Distribution of a Macula-like Virus from Apis Mellifera and Varroa Destructor. Viruses 2015, 7, 3586–3602. [Google Scholar] [CrossRef]
- Wang, L.; Lv, X.; Zhai, Y.; Fu, S.; Wang, D.; Rayner, S.; Tang, Q.; Liang, G. Genomic Characterization of a Novel Virus of the Family Tymoviridae Isolated from Mosquitoes. PLoS ONE 2012, 7, e39845. [Google Scholar] [CrossRef]
- Charles, J.; Tangudu, C.S.; Hurt, S.L.; Tumescheit, C.; Firth, A.E.; Garcia-Rejon, J.E.; Machain-Williams, C.; Blitvich, B.J. Discovery of a Novel Tymoviridae-like Virus in Mosquitoes from Mexico. Arch. Virol. 2019, 164, 649–652. [Google Scholar] [CrossRef]
- Parmentier, L.; Smagghe, G.; de Graaf, D.C.; Meeus, I. Varroa Destructor Macula-like Virus, Lake Sinai Virus and Other New RNA Viruses in Wild Bumblebee Hosts (Bombus pascuorum, Bombus lapidarius and Bombus pratorum). J. Invertebr. Pathol. 2016, 134, 6–11. [Google Scholar] [CrossRef]
- Swevers, L.; Ioannidis, K.; Kolovou, M.; Zografidis, A.; Labropoulou, V.; Santos, D.; Wynant, N.; Vanden Broeck, J.; Wang, L.; Cappelle, K.; et al. Persistent RNA Virus Infection of Lepidopteran Cell Lines: Interactions with the RNAi Machinery. J. Insect Physiol. 2016, 93–94, 81–93. [Google Scholar] [CrossRef]
- Innami, K.; Aizawa, T.; Tsukui, T.; Katsuma, S.; Imanishi, S.; Kawasaki, H.; Iwanaga, M. Infection Studies of Nontarget Mammalian Cell Lines with Bombyx Mori Macula-like Virus. J. Virol. Methods 2016, 229, 24–26. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, X.; Kumar, D.; Kuang, S.; Liu, B.; Hu, X.; Zhu, M.; Liang, Z.; Cao, G.; Xue, R.; et al. Transient Propagation of BmLV and Dysregulation of Gene Expression in Nontarget Cells Following BmLV Infection. J. Asian Pac. Entomol. 2021, 24, 893–902. [Google Scholar] [CrossRef]
- Santos, D.; Wynant, N.; Van den Brande, S.; Verdonckt, T.-W.; Mingels, L.; Peeters, P.; Kolliopoulou, A.; Swevers, L.; Vanden Broeck, J. Insights into RNAi-Based Antiviral Immunity in Lepidoptera: Acute and Persistent Infections in Bombyx Mori and Trichoplusia Ni Cell Lines. Sci. Rep. 2018, 8, 2423. [Google Scholar] [CrossRef]
- Katsuma, S.; Shoji, K.; Suzuki, Y.; Kiuchi, T. CRISPR/Cas9-Mediated Mutagenesis of Ago2 and Siwi in Silkworm Cultured Cells. Gene 2021, 768, 145314. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, M.; Hitotsuyama, T.; Katsuma, S.; Ishihara, G.; Daimon, T.; Shimada, T.; Imanishi, S.; Kawasaki, H. Infection Study of Bombyx Mori Macula-like Virus (BmMLV) Using a BmMLV-Negative Cell Line and an Infectious CDNA Clone. J. Virol. Methods 2012, 179, 316–324. [Google Scholar] [CrossRef]
- Katsuma, S.; Shoji, K.; Suzuki, Y.; Iwanaga, M. Potential for Small RNA Production against Bombyx Mori Latent Virus in Bombyx Mori Ovaries. Arch. Insect Biochem. Physiol. 2020, 106, e21761. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral Immunity Directed by Small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.-W.; Vanden Broeck, J. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef]
- Santos, D.; Verdonckt, T.-W.; Mingels, L.; Van den Brande, S.; Kolliopoulou, A.; Swevers, L.; Vanden Broeck, J.; Wynant, N.; Geens, B.; Van Nieuwerburgh, F.; et al. PIWI Proteins Play an Antiviral Role in Lepidopteran Cell Lines. Viruses 2022, 14, 1442. [Google Scholar] [CrossRef]
- Santos, D.; Feng, M.; Kolliopoulou, A.; Taning, C.N.T.; Sun, J.; Swevers, L. What Are the Functional Roles of Piwi Proteins and PiRNAs in Insects? Insects 2023, 14, 187. [Google Scholar] [CrossRef]
- Miesen, P.; Joosten, J.; van Rij, R.P. PIWIs Go Viral: Arbovirus-Derived PiRNAs in Vector Mosquitoes. PLoS Pathog. 2016, 12, e1006017. [Google Scholar] [CrossRef]
- Varjak, M.; Leggewie, M.; Schnettler, E. The Antiviral PiRNA Response in Mosquitoes? J. Gen. Virol. 2018, 99, 1551–1562. [Google Scholar] [CrossRef]
- Petit, M.; Mongelli, V.; Frangeul, L.; Blanc, H.; Jiggins, F.; Saleh, M.-C. PiRNA Pathway Is Not Required for Antiviral Defense in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 2016, 113, E4218–E4227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.A.; Qin, Q.; Wang, Y.; Li, X.; Miao, L.; Yin, Z.; Zhang, A.; Qu, L.; Ding, C. A New Cell Line from Larval Fat Bodies of the Bollworm, Helicoverpa Armigera (Lepidoptera: Noctuidae). In Vitro Cell. Dev. Biol. Anim. 2006, 42, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M. The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinform. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/ (accessed on 13 April 2023).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.-X.; Ding, S.-W. Virus Discovery by Deep Sequencing and Assembly of Virus-Derived Small Silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Chilton, J.M.; Grüning, B.; Johnson, J.E.; Soranzo, N. NCBI BLAST+ Integrated into Galaxy. Gigascience 2015, 4, 39. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Varlamov, A.; Vaskin, Y.; Efremov, I.; German Grehov, O.G.; Kandrov, D.; Rasputin, K.; Syabro, M.; et al. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Watson, M.; Schnettler, E.; Kohl, A. ViRome: An R Package for the Visualization and Analysis of Viral Small RNA Sequence Datasets. Bioinformatics 2013, 29, 1902–1903. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A Web-Compatible Database Search or Multiple Alignment Viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef]
- Nguyen, L.-T.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Verdonckt, T.-W.; Vanden Broeck, J. Methods for the Cost-Effective Production of Bacteria-Derived Double-Stranded RNA for in Vitro Knockdown Studies. Front. Physiol. 2022, 13, 836106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-Ddct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.R.; Dawes, J.M.; McMahon, S.B.; Bennett, D.L.H.; Orengo, C.; Kohl, M. ReadqPCR and NormqPCR: R Packages for the Reading, Quality Checking and Normalisation of RT-QPCR Quantification Cycle (Cq) Data. BMC Genom. 2012, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Jiggins, F.M.; Halligan, D.L.; Little, T.J. Natural Selection Drives Extremely Rapid Evolution in Antiviral RNAi Genes. Curr. Biol. 2006, 16, 580–585. [Google Scholar] [CrossRef]
- Wynant, N.; Santos, D.; Vanden Broeck, J. The Evolution of Animal Argonautes: Evidence for the Absence of Antiviral AGO Argonautes in Vertebrates. Sci. Rep. 2017, 7, 9230. [Google Scholar] [CrossRef]
- Weber, F.; Wagner, V.; Simon, B.; Hartmann, R.; Paludan, S.R.; Rasmussen, S.B. Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef]
- Schonborn, J.; Oberstraß, J.; Breyel, E.; Tittgen, J.; Schumacher, J.; Lukacs, N. Monoclonal Antibodies to Double-Stranded RNA as Probes of RNA Structure in Crude Nucleic Acid Extracts. Nucleic Acids Res. 1991, 19, 2993–3000. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A. A PIWI-Interacting Small RNAs: The Vanguard of Genome Defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Kawaoka, S.; Arai, Y.; Kadota, K.; Suzuki, Y.; Hara, K.; Sugano, S.; Shimizu, K.; Tomari, Y.; Shimada, T.; Katsuma, S. Zygotic Amplification of Secondary PiRNAs during Silkworm Embryogenesis. RNA 2011, 17, 1401–1407. [Google Scholar] [CrossRef]
- Geisler, C.; Jarvis, D.L. Adventitious Viruses in Insect Cell Lines Used for Recombinant Protein Expression. Protein Expr. Purif. 2018, 144, 25–32. [Google Scholar] [CrossRef]
- Grace, T.D.C. Establishment of a Line of Cells from the Silkworm Bombyx Mori. Nature 1967, 216, 613. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, M.; Adachi, Y.; Uchiyama, K.; Tsukui, K.; Katsuma, S.; Kawasaki, H. Long-Term Adaptation of the Bombyx Mori BmN4 Cell Line to Grow in Serum-Free Culture. In Vitro Cell. Dev. Biol.—Anim. 2014, 50, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yang, Y.; Zhang, H.; Farley, G.; Wang, J.; Quarles, K.A.; Weng, Z.; Zamore, P.D. The Genome of the Hi5 Germ Cell Line from Trichoplusia Ni, an Agricultural Pest and Novel Model for Small RNA Biology. eLife 2018, 7, e31628. [Google Scholar] [CrossRef]
- Li, T.-C.; Scotti, P.D.; Miyamura, T.; Takeda, N. Latent Infection of a New Alphanodavirus in an Insect Cell Line. J. Virol. 2007, 81, 10890–10896. [Google Scholar] [CrossRef] [PubMed]
- Tsukui, K.; Yagisawa, C.; Fujimoto, S.; Ogawa, M.; Kokusho, R.; Nozawa, M.; Kawasaki, H.; Katsuma, S.; Iwanaga, M. Infectious Virions of Bombyx Mori Latent Virus Are Incorporated into Bombyx Mori Nucleopolyhedrovirus Occlusion Bodies. Viruses 2019, 11, 316. [Google Scholar] [CrossRef]
- Wang, L.; Cappelle, K.; Santos, D.; Vanden Broeck, J.; Smagghe, G.; Swevers, L. Short-Term Persistence Precedes Pathogenic Infection: Infection Kinetics of Cricket Paralysis Virus in Silkworm-Derived Bm5 Cells. J. Insect Physiol. 2019, 115, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.E.; Summers, M.D.; Fraser, M.J. Production of Human Beta Interferon in Insect Cells Infected with a Baculovirus Expression Vector. Mol. Cell Biol. 1983, 3, 2156–2165. [Google Scholar] [CrossRef]
- Felberbaum, R.S. The Baculovirus Expression Vector System: A Commercial Manufacturing Platform for Viral Vaccines and Gene Therapy Vectors. Biotechnol. J. 2015, 10, 702–714. [Google Scholar] [CrossRef]
- Guebre-Xabier, M.; Patel, N.; Tian, J.H.; Zhou, B.; Maciejewski, S.; Lam, K.; Portnoff, A.D.; Massare, M.J.; Frieman, M.B.; Piedra, P.A.; et al. NVX-CoV2373 Vaccine Protects Cynomolgus Macaque Upper and Lower Airways against SARS-CoV-2 Challenge. Vaccine 2020, 38, 7892–7896. [Google Scholar] [CrossRef]
- Geisler, C. A New Approach for Detecting Adventitious Viruses Shows Sf-Rhabdovirus-Negative Sf-RVN Cells Are Suitable for Safe Biologicals Production. BMC Biotechnol. 2018, 18, 8. [Google Scholar] [CrossRef]
- Castellano, M.A.; Martelli, G.P. Ultrastructure and Nature of Vesiculated Bodies Associated with Isometric Virus-like Particles in Diseased Grapevines. J. Ultrasruct. Res. 1984, 89, 56–64. [Google Scholar] [CrossRef]
- Santos, D.; Mingels, L.; Vogel, E.; Wang, L.; Christiaens, O.; Cappelle, K.; Wynant, N.; Gansemans, Y.; Van Nieuwerburgh, F.; Smagghe, G.; et al. Generation of Virus-and DsRNA-Derived SiRNAs with Species-Dependent Length in Insects. Viruses 2019, 11, 738. [Google Scholar] [CrossRef]
- Feng, M.; Kolliopoulou, A.; Zhou, Y.H.; Fei, S.G.; Xia, J.M.; Swevers, L.; Sun, J.C. The PiRNA Response to BmNPV Infection in the Silkworm Fat Body and Midgut. Insect Sci. 2021, 28, 662–679. [Google Scholar] [CrossRef] [PubMed]
- Zografidis, A.; Van Nieuwerburgh, F.; Kolliopoulou, A.; Apostolou-Karampelis, K.; Head, S.R.; Deforce, D.; Smagghe, G.; Swevers, L. Viral Small RNA Analysis of Bombyx Mori Larval Midgut during Persistent and Pathogenic Cytoplasmic Polyhedrosis Virus Infection. J. Virol. 2015, 89, 11473–11486. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.H.; Jiang, L.; Zhu, L.; Cheng, T.C.; Niu, W.H.; Yan, Y.F.; Xia, Q.Y. Characterization of Argonaute Family Members in the Silkworm, Bombyx Mori. Insect Sci. 2013, 20, 78–91. [Google Scholar] [CrossRef]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-Arthropod Analysis Reveals Somatic PiRNAs as an Ancestral Defence against Transposable Elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef]
- Sakakibara, K.; Siomi, M.C. The PIWI-Interacting RNA Molecular Pathway: Insights from Cultured Silkworm Germline Cells. BioEssays 2018, 40, 1700068. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, L.; Yue, D.; Ye, B.; Li, P.; Zhang, B.; Fan, Q. Evaluation of reference genes for normalization of RT-qPCR gene expression data for Trichoplusia ni cells during Antheraea pernyi (Lepidoptera: Saturniidae) Multicapsid Nucleopolyhedrovirus (AnpeNPV) infection. J. Insect Sci. 2019, 19, 4. [Google Scholar] [CrossRef]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verdonckt, T.-W.; Bilsen, A.; Van Nieuwerburgh, F.; De Troij, L.; Santos, D.; Vanden Broeck, J. Identification and Profiling of a Novel Bombyx mori latent virus Variant Acutely Infecting Helicoverpa armigera and Trichoplusia ni. Viruses 2023, 15, 1183. https://doi.org/10.3390/v15051183
Verdonckt T-W, Bilsen A, Van Nieuwerburgh F, De Troij L, Santos D, Vanden Broeck J. Identification and Profiling of a Novel Bombyx mori latent virus Variant Acutely Infecting Helicoverpa armigera and Trichoplusia ni. Viruses. 2023; 15(5):1183. https://doi.org/10.3390/v15051183
Chicago/Turabian StyleVerdonckt, Thomas-Wolf, Anton Bilsen, Filip Van Nieuwerburgh, Loes De Troij, Dulce Santos, and Jozef Vanden Broeck. 2023. "Identification and Profiling of a Novel Bombyx mori latent virus Variant Acutely Infecting Helicoverpa armigera and Trichoplusia ni" Viruses 15, no. 5: 1183. https://doi.org/10.3390/v15051183
APA StyleVerdonckt, T.-W., Bilsen, A., Van Nieuwerburgh, F., De Troij, L., Santos, D., & Vanden Broeck, J. (2023). Identification and Profiling of a Novel Bombyx mori latent virus Variant Acutely Infecting Helicoverpa armigera and Trichoplusia ni. Viruses, 15(5), 1183. https://doi.org/10.3390/v15051183






