Validation of HIV-1 MA Shell Structural Arrangements and Env Protein Interactions Predict a Role of the MA Shell in Viral Maturation
Abstract
1. Introduction
2. Materials and Methods
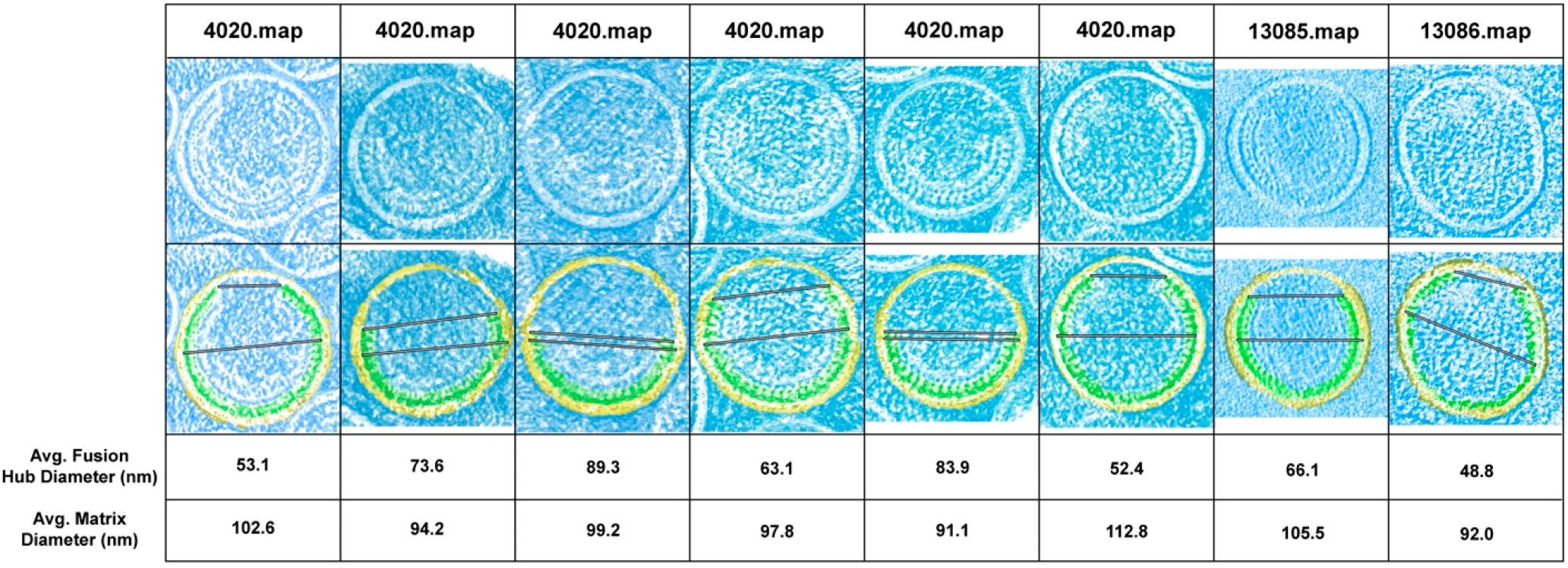
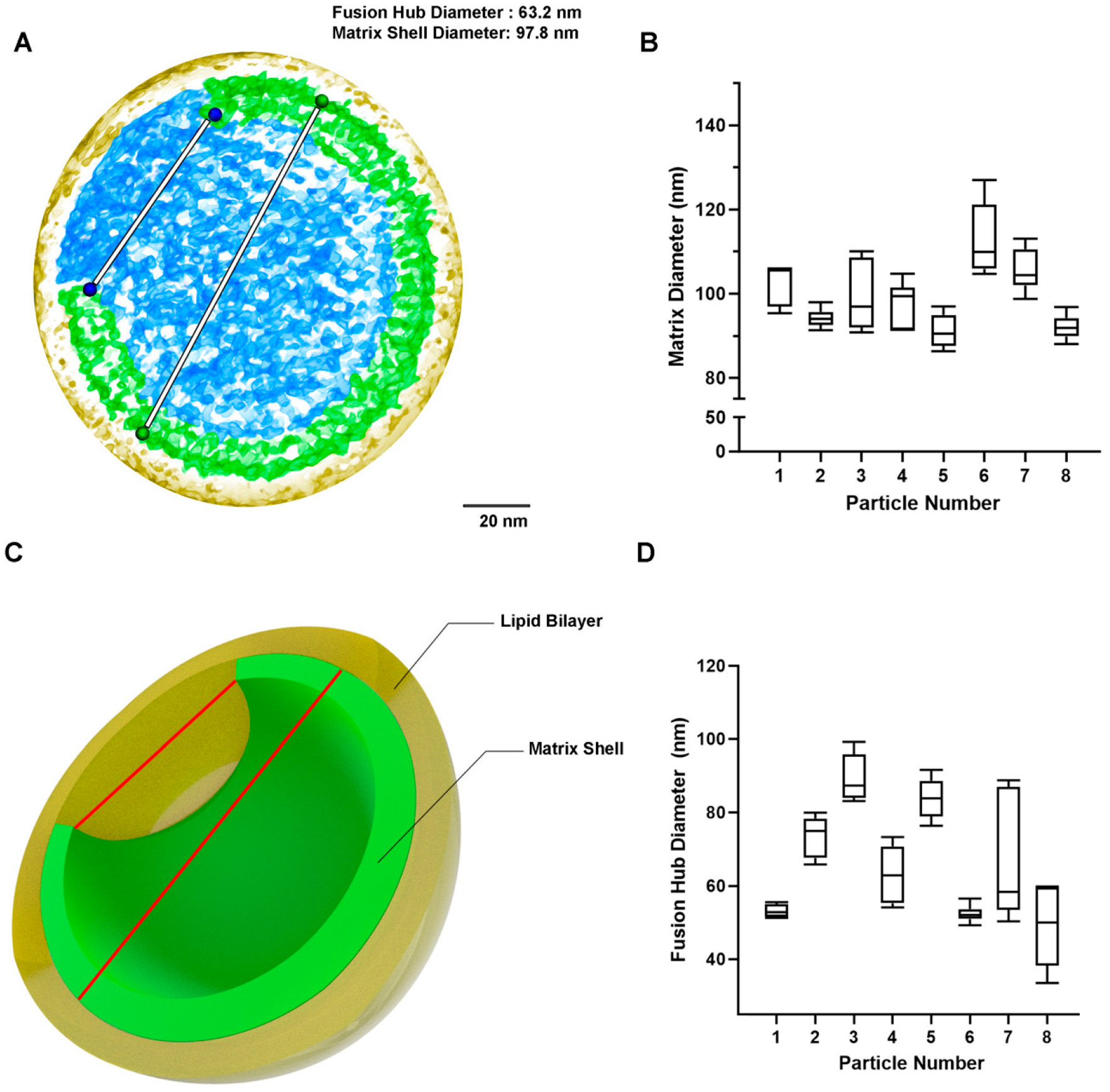
2.1. Quantification of MA Shell Diameter and Gap
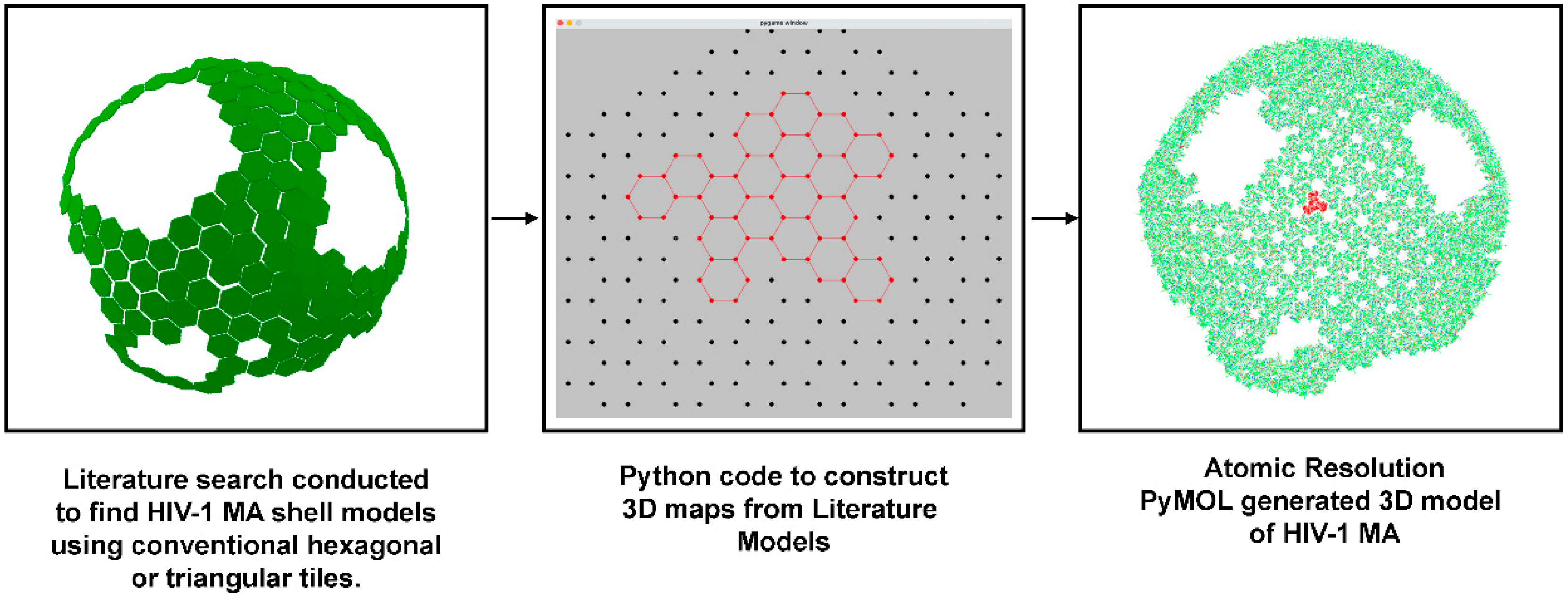
2.2. Visualization of HIV-1 MA Shells from the Literature
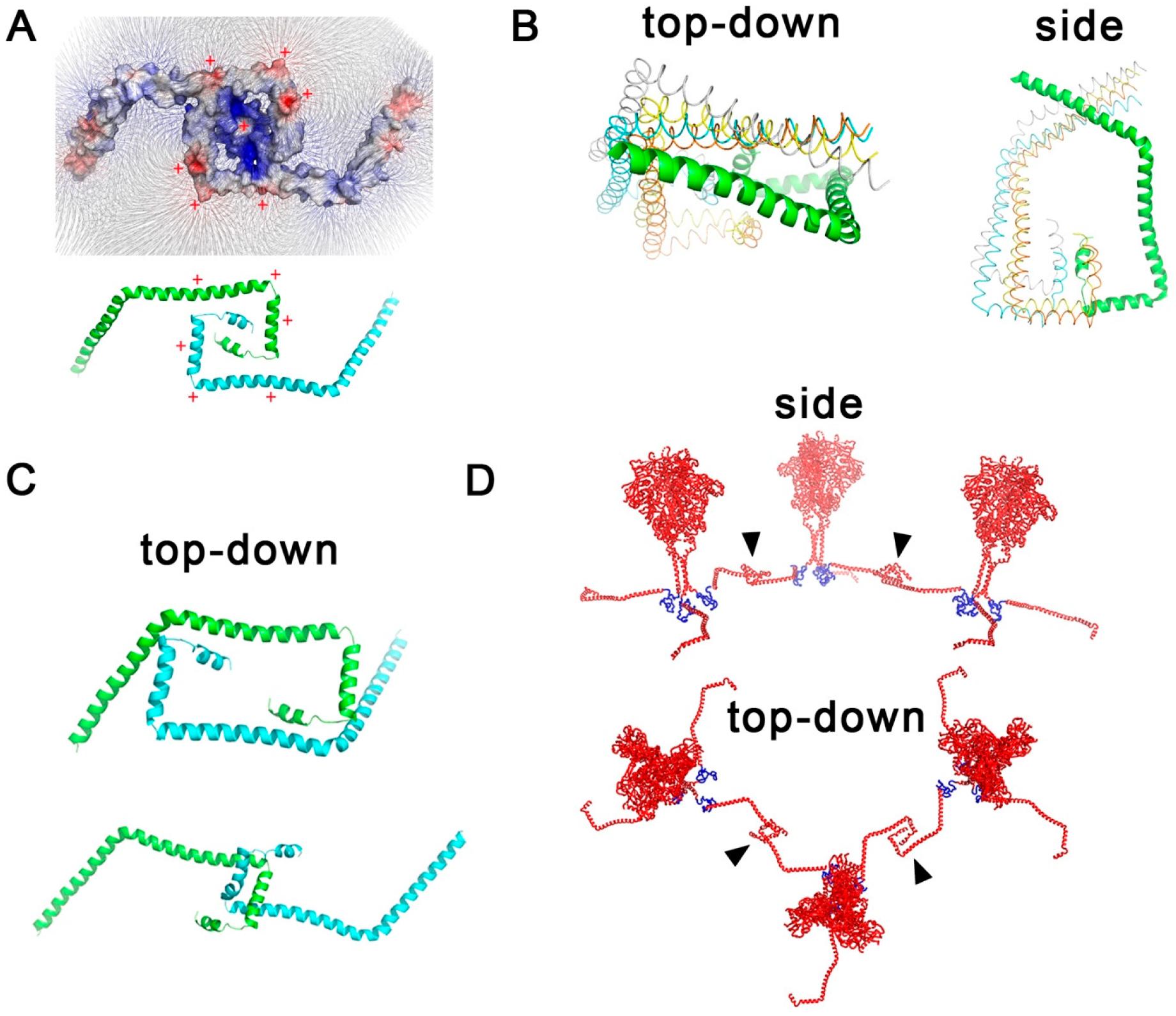
2.3. Interaction Models for Two Env CT Domains
3. Results
3.1. Estimation of MA Shell Diameter and Fusion Hub Gap Dimensions
3.2. Validation of Published MA Shell Models
3.3. Interactions of Two Env CT Domains
3.4. An Updated Model of the HIV-1 MA Shell
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alfadhli, A.; Barklis, R.L.; Barklis, E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology 2009, 387, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Brandenberg, O.F.; Magnus, C.; Regoes, R.R.; Trkola, A. The HIV-1 Entry Process: A Stoichiometric View. Trends Microbiol. 2015, 23, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Brandenberg, O.F.; Magnus, C.; Rusert, P.; Regoes, R.R.; Trkola, A. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog. 2015, 11, e1004595. [Google Scholar] [CrossRef]
- Hill, C.P.; Worthylake, D.; Bancroft, D.P.; Christensen, A.M.; Sundquist, W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 1996, 93, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.R.L.; Sun, W.; Mangukia, T.A.; Reyes-Serratos, E.; Marcet-Palacios, M. Challenging the Existing Model of the Hexameric HIV-1 Gag Lattice and MA Shell Superstructure: Implications for Viral Entry. Viruses 2021, 13, 1515. [Google Scholar] [CrossRef]
- Sun, W.; Reyes-Serratos, E.; Barilla, D.; Santos, J.R.L.; Bujold, M.; Graves, S.; Marcet-Palacios, M. Mathematical determination of the HIV-1 matrix shell structure and its impact on the biology of HIV-1. PLoS ONE 2019, 14, e0224965. [Google Scholar] [CrossRef]
- Benjamin, J.; Ganser-Pornillos, B.K.; Tivol, W.F.; Sundquist, W.I.; Jensen, G.J. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J. Mol. Biol. 2005, 346, 577–588. [Google Scholar] [CrossRef]
- Qu, K.; Ke, Z.; Zila, V.; Anders-Osswein, M.; Glass, B.; Mucksch, F.; Muller, R.; Schultz, C.; Muller, B.; Krausslich, H.G.; et al. Maturation of the matrix and viral membrane of HIV-1. Science 2021, 373, 700–704. [Google Scholar] [CrossRef]
- Briggs, J.A.; Riches, J.D.; Glass, B.; Bartonova, V.; Zanetti, G.; Krausslich, H.G. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. USA 2009, 106, 11090–11095. [Google Scholar] [CrossRef]
- Tan, A.; Pak, A.J.; Morado, D.R.; Voth, G.A.; Briggs, J.A.G. Immature HIV-1 assembles from Gag dimers leaving partial hexamers at lattice edges as potential substrates for proteolytic maturation. Proc. Natl. Acad. Sci. USA 2021, 118, e2020054118. [Google Scholar] [CrossRef]
- Mattei, S.; Tan, A.; Glass, B.; Muller, B.; Krausslich, H.G.; Briggs, J.A.G. High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc. Natl. Acad. Sci. USA 2018, 115, E9401–E9410. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Samal, A.B.; Vlach, J.; Saad, J.S. Solution Structure and Membrane Interaction of the Cytoplasmic Tail of HIV-1 gp41 Protein. Structure 2017, 25, 1708–1718. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins 2017, 85, 435–444. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Huang, S.Y.; Zou, X. A knowledge-based scoring function for protein-RNA interactions derived from a statistical mechanics-based iterative method. Nucleic Acids Res. 2014, 42, e55. [Google Scholar] [CrossRef]
- Yan, Y.; Wen, Z.; Wang, X.; Huang, S.Y. Addressing recent docking challenges: A hybrid strategy to integrate template-based and free protein-protein docking. Proteins 2017, 85, 497–512. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.Y. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Bharat, T.A.; Castillo Menendez, L.R.; Hagen, W.J.; Lux, V.; Igonet, S.; Schorb, M.; Schur, F.K.; Krausslich, H.G.; Briggs, J.A. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 8233–8238. [Google Scholar] [CrossRef]
- Mangala Prasad, V.; Leaman, D.P.; Lovendahl, K.N.; Croft, J.T.; Benhaim, M.A.; Hodge, E.A.; Zwick, M.B.; Lee, K.K. Cryo-ET of Env on intact HIV virions reveals structural variation and positioning on the Gag lattice. Cell 2022, 185, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Piai, A.; Fu, Q.; Cai, Y.; Ghantous, F.; Xiao, T.; Shaik, M.M.; Peng, H.; Rits-Volloch, S.; Chen, W.; Seaman, M.S.; et al. Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein. Nat. Commun. 2020, 11, 2317. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangukia, T.A.; Santos, J.R.L.; Sun, W.; Cesarz, D.; Ortíz Hidalgo, C.D.; Marcet-Palacios, M. Validation of HIV-1 MA Shell Structural Arrangements and Env Protein Interactions Predict a Role of the MA Shell in Viral Maturation. Viruses 2023, 15, 893. https://doi.org/10.3390/v15040893
Mangukia TA, Santos JRL, Sun W, Cesarz D, Ortíz Hidalgo CD, Marcet-Palacios M. Validation of HIV-1 MA Shell Structural Arrangements and Env Protein Interactions Predict a Role of the MA Shell in Viral Maturation. Viruses. 2023; 15(4):893. https://doi.org/10.3390/v15040893
Chicago/Turabian StyleMangukia, Tarana A., Joy Ramielle L. Santos, Weijie Sun, Dominik Cesarz, Carlos D. Ortíz Hidalgo, and Marcelo Marcet-Palacios. 2023. "Validation of HIV-1 MA Shell Structural Arrangements and Env Protein Interactions Predict a Role of the MA Shell in Viral Maturation" Viruses 15, no. 4: 893. https://doi.org/10.3390/v15040893
APA StyleMangukia, T. A., Santos, J. R. L., Sun, W., Cesarz, D., Ortíz Hidalgo, C. D., & Marcet-Palacios, M. (2023). Validation of HIV-1 MA Shell Structural Arrangements and Env Protein Interactions Predict a Role of the MA Shell in Viral Maturation. Viruses, 15(4), 893. https://doi.org/10.3390/v15040893






