Molecular Interaction of Nonsense-Mediated mRNA Decay with Viruses
Abstract
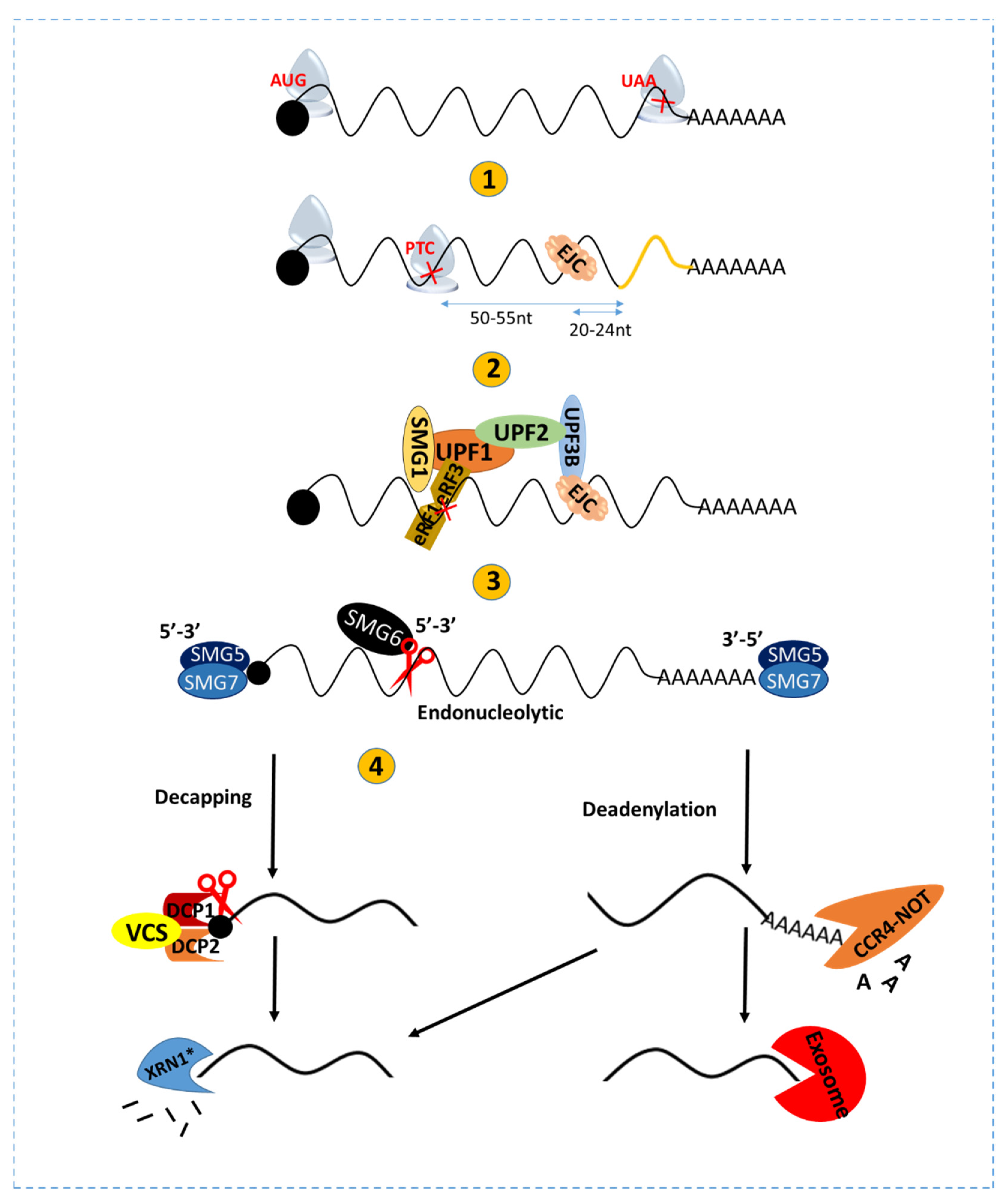
1. Introduction
2. NMD Defense against Viruses
2.1. NMD Targets Positive Sense RNA Viruses
2.2. NMD Targets Double-Stranded RNA Viruses
2.3. NMD Targets Retroviruses
2.4. NMD Targets Viral RNAs of DNA Viruses
3. Diversity of Viral Tactics Counteracts NMD-Mediated Host Defense
3.1. Viral Proteins: Targeting NMD Components and Interfering with their Function
3.1.1. Movement Proteins
3.1.2. Tax and Rex Factors
3.1.3. Capsid Protein (CP)
3.1.4. Core Protein
3.1.5. Trans-Activator Protein (TAV)
3.1.6. N-Protein
3.1.7. Replicase Protein
3.1.8. Non-Structural Protein
3.2. Viral RNA Sequences or Structures
3.2.1. Rous Sarcoma Virus RNA Stability Element (RSE)
3.2.2. Unstructured Element of TCV
3.2.3. Readthrough and Frameshifting Elements
3.3. Others
| Virus Species | Types of Genome | Host | NMD-Eliciting Feature | NMD Resistant Factor | Reference |
|---|---|---|---|---|---|
| PVX | (+) ssRNA | Arabidopsis thaliana | iTC and long 3′ UTR | Unknown | [55] |
| TCV | (+) ssRNA | Nicotiana benthamiana | iTC and long 3′ UTR | Unstructured element | [55] |
| PEMV2 | (+) ssRNA | N. benthamiana | Long 3′ UTR | p26 (MP), frameshifting element | [73] |
| Rotavirus | dsRNA | African green monkey kidney cell line | Unknown | non-structural protein 5 (NSP5) | [59] |
| CaMV | DNA | N. benthamiana | Polycistronic 35S pgRNA | Transactivator Protein (TAV) | [70] |
| HTLV-1 | (+) ssRNA | Hela and HEK cell line | Unknown | TAX and REX factor | [21,64,65] |
| HCV | (+) ssRNA | hepatoma cells | Unknown | Core Protein | [82] |
| SFV | (+) ssRNA | Hela Cell line | Unknown | Replicase /nsp3 protein, capsid protein | [54,80,81,96] |
| MoMLV | (+) ssRNA | Mouse | Long 3′ UTR on gag gene | MoMLU-RT | [67] |
| CTFV | dsRNA | HEK293T cells | Unknown | Hairpin structure (CTFV-HP) | [99,100] |
| Zika Virus | (+) ssRNA | Human NPC cells | Unknown | Capsid Protein | [79] |
| RSV | (+) ssRNA | CEF cell line | uORF and long 3′ UTR | RNA Stability Element | [62] |
| HIV-1 | (+) ssRNA | Hela cell line | REV | [101,103] | |
| MHV | (+) ssRNA | 17Cl-1 Cells, Mouse astrcytoma cell line | uORF and 3′ UTR | N Protein | [57] |
| KSHV | DNA | PEL cell line | Long 3′ UTR of Orf50 mRNA | Unknown | [71,72] |
| EBV | DNA | HEK293T cells | Long 3′ UTR of BRLF1-encoding transcripts | Unknown | [71,72] |
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- May, J.P.; Yuan, X.; Sawicki, E.; Simon, A.E. RNA Virus Evasion of Nonsense-Mediated Decay. PLoS Pathog. 2018, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Kervestin, S.; Jacobson, A. NMD: A Multifaceted Response to Premature Translational Termination. Nat. Rev. Mol. Cell Biol. 2012, 13, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Schweingruber, C.; Rufener, S.C.; Zünd, D.; Yamashita, A.; Mühlemann, O. Nonsense-Mediated MRNA Decay—Mechanisms of Substrate MRNA Recognition and Degradation in Mammalian Cells. Biochim. Biophys. Acta—Gene Regul. Mech. 2013, 1829, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Rebbapragada, I.; Lykke-Andersen, J. A Competition between Stimulators and Antagonists of Upf Complex Recruitment Governs Human Nonsense-Mediated MRNA Decay. PLoS Biol. 2008, 6, e111. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.R.; Goff, S.P. Upf1 Senses 3’UTR Length to Potentiate MRNA Decay. Cell 2010, 143, 379–389. [Google Scholar] [CrossRef]
- Hurt, J.A.; Robertson, A.D.; Burge, C.B. Global Analyses of UPF1 Binding and Function Reveal Expanded Scope of Nonsense-Mediated MRNA Decay. Genome Res. 2013, 23, 1636–1650. [Google Scholar] [CrossRef]
- Colombo, M.; Karousis, E.D.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-Wide Identification of NMD-Targeted Human MRNAs Reveals Extensive Redundancy between SMG6- and SMG7-Mediated Degradation Pathways. RNA 2017, 23, 189–201. [Google Scholar] [CrossRef]
- Wachter, A.; Hartmann, L. NMD: Nonsense-Mediated Defense. Cell Host Microbe 2014, 16, 273–275. [Google Scholar] [CrossRef]
- He, F.; Jacobson, A. Nonsense-Mediated MRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu. Rev. Genet. 2015, 49, 339–366. [Google Scholar] [CrossRef]
- Dai, Y.; Li, W.; An, L. NMD Mechanism and the Functions of Upf Proteins in Plant. Plant Cell Rep. 2016, 35, 5–15. [Google Scholar] [CrossRef]
- Karousis, E.D.; Mühlemann, O. Nonsense-Mediated MRNA Decay Begins Where Translation Ends. Cold Spring Harb. Perspect. Biol. 2019, 11, a032862. [Google Scholar] [CrossRef] [PubMed]
- Peccarelli, M.; Kebaara, B.W. Regulation of Natural MRNAs by the Nonsense-Mediated MRNA Decay Pathway. Eukaryot. Cell 2014, 13, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Muhlrad, D.; Parker, R. Aberrant MRNAs with Extended 3’ UTRs Are Substrates for Rapid Degradation by MRNA Surveillance. RNA 1999, 5, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Crespo, M.; Palacios, I.M. Nonsense-Mediated MRNA Decay and Development: Shoot the Messenger to Survive? Biochem. Soc. Trans. 2011, 38, 1500–1505. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R.; Wilkinson, M.F. Nonsense-Mediated RNA Decay in the Brain: Emerging Modulator of Neural Development and Disease. Nat. Rev. Neurosci. 2018, 19, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Metzstein, M.M.; Krasnow, M.A. Functions of the Nonsense-Mediated MRNA Decay Pathway in Drosophila Development. PLoS Genet. 2006, 2, e180. [Google Scholar] [CrossRef]
- Nickless, A.; Bailis, J.M.; You, Z. Control of Gene Expression through the Nonsense-Mediated RNA Decay Pathway. Cell Biosci. 2017, 7, 26. [Google Scholar] [CrossRef]
- Brogna, S.; Wen, J. Nonsense-Mediated MRNA Decay (NMD) Mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Maquat, L.E. Nonsense-Mediated MRNA Decay: Splicing, Translation and MRNP Dynamics. Nat. Rev. Mol. Cell Biol. 2004, 5, 89–99. [Google Scholar] [CrossRef]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a Novel SMG-1-Upf1-ERF1-ERF3 Complex (SURF) to the Exon Junction Complex Triggers Upf1 Phosphorylation and Nonsense-Mediated MRNA Decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Karasawa, N.; Hashizume, M.; Tanaka, Y.; Ohsugi, T.; Uchimaru, K.; Watanabe, T. Elucidation of the Mechanism of Host NMD Suppression by HTLV-1 Rex: Dissection of Rex to Identify the NMD Inhibitory Domain. Viruses 2022, 14, 344. [Google Scholar] [CrossRef]
- Lejeune, F.; Ishigaki, Y.; Li, X.; Maquat, L.E. The Exon Junction Complex Is Detected on CBP80-Bound but Not EIF4E-Bound MRNA in Mammalian Cells: Dynamics of MRNP Remodeling. EMBO J. 2002, 21, 3536–3545. [Google Scholar] [CrossRef]
- Hwang, J.; Maquat, L.E. Nonsense-Mediated MRNA Decay (NMD) in Animal Embryogenesis: To Die or Not to Die, That Is the Question. Curr. Opin. Genet. Dev. 2011, 21, 422–430. [Google Scholar] [CrossRef]
- Rigby, R.E.; Rehwinkel, J. RNA Degradation in Antiviral Immunity and Autoimmunity. Trends Immunol. 2015, 36, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The Exon-Exon Junction Complex Provides a Binding Platform for Factors Involved in MRNA Export and Nonsense-Mediated MRNA Decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a Novel Phosphatidylinositol 3-Kinase-Related Protein Kinase, Associates with Components of the MRNA Surveillance Complex and Is Involved in the Regulation of Nonsense-Mediated MRNA Decay. Genes Dev. 2001, 15, 2215–2228. [Google Scholar] [CrossRef] [PubMed]
- Okada-Katsuhata, Y.; Yamashita, A.; Kutsuzawa, K.; Izumi, N.; Hirahara, F.; Ohno, S. N-and C-Terminal Upf1 Phosphorylations Create Binding Platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012, 40, 1251–1266. [Google Scholar] [CrossRef]
- Chamieh, H.; Ballut, L.; Bonneau, F.; Le Hir, H. NMD Factors UPF2 and UPF3 Bridge UPF1 to the Exon Junction Complex and Stimulate Its RNA Helicase Activity. Nat. Struct. Mol. Biol. 2008, 15, 85–93. [Google Scholar] [CrossRef]
- Yepiskoposyan, H.; Aeschimann, F.; Nilsson, D.; Okoniewski, M.; Mühlemann, O. Autoregulation of the Nonsense-Mediated MRNA Decay Pathway in Human Cells. RNA 2011, 17, 2108–2118. [Google Scholar] [CrossRef]
- Gatfield, D.; Unterholzner, L.; Ciccarelli, F.D.; Bork, P.; Izaurralde, E. Nonsense-Mediated MRNA Decay in Drosophila: At the Intersection of the Yeast and Mammalian Pathways. EMBO J. 2003, 22, 3960–3970. [Google Scholar] [CrossRef]
- Wen, J.; Brogna, S. Splicing-Dependent NMD Does Not Require the EJC in Schizosaccharomyces Pombe. EMBO J. 2010, 29, 1537–1551. [Google Scholar] [CrossRef]
- Tian, M.; Yang, W.; Zhang, J.; Dang, H.; Lu, X.; Fu, C.; Miao, W. Nonsense-Mediated MRNA Decay in Tetrahymena Is EJC Independent and Requires a Protozoa-Specific Nuclease. Nucleic Acids Res. 2017, 45, 6848–6863. [Google Scholar] [CrossRef]
- Metze, S.; Herzog, V.A.; Ruepp, M.D.; Mühlemann, O. Comparison of EJC-Enhanced and EJC-Independent NMD in Human Cells Reveals Two Partially Redundant Degradation Pathways. RNA 2013, 19, 1432–1448. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, A. RNA-Targeted Antiviral Immunity: More Than Just RNA Silencing. Trends Microbiol. 2019, 27, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Jonas, S.; Izaurralde, E. The SMG5-SMG7 Heterodimer Directly Recruits the CCR4-NOT Deadenylase Complex to MRNAs Containing Nonsense Codons via Interaction with POP2. Genes Dev. 2013, 27, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Eberle, A.B.; Lykke-Andersen, S.; Mühlemann, O.; Jensen, T.H. SMG6 Promotes Endonucleolytic Cleavage of Nonsense MRNA in Human Cells. Nat. Struct. Mol. Biol. 2009, 16, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jones, C.I.; Newbury, S.F.; Green, P.J. XRN 5′→3′ Exoribonucleases: Structure, Mechanisms and Functions. Biochim. Biophys. Acta-Gene Regul. Mech. 2013, 1829, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Kerényi, Z.; Mérai, Z.; Hiripi, L.; Benkovics, A.; Gyula, P.; Lacomme, C.; Barta, E.; Nagy, F.; Silhavy, D. Inter-Kingdom Conservation of Mechanism of Nonsense-Mediated MRNA Decay. EMBO J. 2008, 27, 1585–1595. [Google Scholar] [CrossRef]
- Causier, B.; Li, Z.; De Smet, R.; Lloyd, J.P.B.; Van De Peer, Y.; Davies, B. Conservation of Nonsense-Mediated MRNA Decay Complex Components Throughout Eukaryotic Evolution. Sci. Rep. 2017, 7, 16692. [Google Scholar] [CrossRef]
- Lloyd, J.P.B. The Evolution and Diversity of the Nonsense-Mediated MRNA Decay Pathway [Version 1; Peer Review: 4 Approved]. F1000Research 2018, 7, 1299. [Google Scholar] [CrossRef]
- Miras, M.; Allen Miller, W.; Truniger, V.; Aranda, M.A. Non-Canonical Translation in Plant RNA Viruses. Front. Plant Sci. 2017, 8, 1385–1409. [Google Scholar] [CrossRef]
- Ho, J.S.Y.; Zhu, Z.; Marazzi, I. Unconventional Viral Gene Expression Mechanisms as Therapeutic Targets. Nature 2021, 593, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Skuzeski, J.M.; Nichols, L.M.; Gesteland, R.F.; Atkins, J.F. The Signal for a Leaky UAG Stop Codon in Several Plant Viruses Includes the Two Downstream Codons. J. Mol. Biol. 1991, 218, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D. Mechanisms and Implications of Programmed Translational Frameshifting. Wiley Interdiscip. Rev. RNA 2012, 3, 661–673. [Google Scholar] [CrossRef]
- Jaafar, Z.A.; Kieft, J.S. Viral RNA Structure-Based Strategies to Manipulate Translation. Nat. Rev. Microbiol. 2019, 17, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.A.; Marcotrigiano, J. Viral Precursor Polyproteins: Keys of Regulation from Replication to Maturation. Curr. Opin. Virol. 2013, 3, 137–142. [Google Scholar] [CrossRef]
- Withers, J.B.; Beemon, K.L. Structural Features in the Rous Sarcoma Virus RNA Stability Element Are Necessary for Sensing the Correct Termination Codon. Retrovirology 2010, 7, 65. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921. [Google Scholar] [CrossRef]
- Bühler, M.; Steiner, S.; Mohn, F.; Paillusson, A.; Mühlemann, O. EJC-Independent Degradation of Nonsense Immunoglobulin-μ MRNA Depends on 3′ UTR Length. Nat. Struct. Mol. Biol. 2006, 13, 462–464. [Google Scholar] [CrossRef]
- Kertész, S.; Kerényi, Z.; Mérai, Z.; Bartos, I.; Pálfy, T.; Barta, E.; Silhavy, D. Both Introns and Long 3′-UTRs Operate as Cis-Acting Elements to Trigger Nonsense-Mediated Decay in Plants. Nucleic Acids Res. 2006, 34, 6147–6157. [Google Scholar] [CrossRef]
- Karousis, E.D.; Nasif, S.; Mühlemann, O. Nonsense-Mediated MRNA Decay: Novel Mechanistic Insights and Biological Impact. Wiley Interdiscip. Rev. RNA 2016, 7, 661–682. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The Alphaviruses: Gene Expression, Replication, and Evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Wernet, M.F.; Klovstad, M.; Clandinin, T.R. Generation of Infectious Virus Particles from Inducible Transgenic Genomes. Curr. Biol. 2014, 24, 107–108. [Google Scholar] [CrossRef]
- Balistreri, G.; Horvath, P.; Schweingruber, C.; Zünd, D.; McInerney, G.; Merits, A.; Mühlemann, O.; Azzalin, C.; Helenius, A. The Host Nonsense-Mediated MRNA Decay Pathway Restricts Mammalian RNA Virus Replication. Cell Host Microbe 2014, 16, 403–411. [Google Scholar] [CrossRef]
- Garcia, D.; Garcia, S.; Voinnet, O. Nonsense-Mediated Decay Serves as a General Viral Restriction Mechanism in Plants. Cell Host Microbe 2014, 16, 391–402. [Google Scholar] [CrossRef]
- Imamachi, N.; Salam, K.A.; Suzuki, Y.; Akimitsu, N. A GC-Rich Sequence Feature in the 3′ UTR Directs UPF1-Dependent MRNA Decay in Mammalian Cells. Genome Res. 2017, 27, 407–418. [Google Scholar] [CrossRef]
- Wada, M.; Lokugamage, K.G.; Nakagawa, K.; Narayanan, K.; Makino, S. Interplay between Coronavirus, a Cytoplasmic RNA Virus, and Nonsense-Mediated MRNA Decay Pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E10157–E10166. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Sarkar, R.; Banerjee, S.; Mukherjee, A.; Chawla-Sarkar, M. Rotaviral Nonstructural Protein 5 (NSP5) Promotes Proteasomal Degradation of up-Frameshift Protein 1 (UPF1), a Principal Mediator of Nonsense-Mediated MRNA Decay (NMD) Pathway, to Facilitate Infection. Cell Signal. 2022, 89, 110180. [Google Scholar] [CrossRef] [PubMed]
- Bolinger, C.; Boris-Lawrie, K. Mechanisms Employed by Retroviruses to Exploit Host Factors for Translational Control of a Complicated Proteome. Retrovirology 2009, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.J.; Beemon, K.L. Unspliced Rous Sarcoma Virus Genomic RNAs Are Translated and Subjected to Nonsense-Mediated MRNA Decay before Packaging. J. Virol. 2004, 78, 5139–5146. [Google Scholar] [CrossRef]
- Quek, B.L.; Beemon, K. Retroviral Strategy to Stabilize Viral RNA. Curr. Opin. Microbiol. 2014, 18, 78–82. [Google Scholar] [CrossRef]
- Barker, G.F.; Beemon, K. Rous Sarcoma Virus RNA Stability Requires an Open Reading Frame in the Gag Gene and Sequences Downstream of the Gag-Pol Junction. Mol. Cell. Biol. 1994, 14, 1986–1996. [Google Scholar] [CrossRef]
- Fiorini, F.; Robin, J.P.; Kanaan, J.; Borowiak, M.; Croquette, V.; Le Hir, H.; Jalinot, P.; Mocquet, V. HTLV-1 Tax Plugs and Freezes UPF1 Helicase Leading to Nonsense-Mediated MRNA Decay Inhibition. Nat. Commun. 2018, 9, 431. [Google Scholar] [CrossRef]
- Mocquet, V.; Neusiedler, J.; Rende, F.; Cluet, D.; Robin, J.-P.; Terme, J.-M.; Duc Dodon, M.; Wittmann, J.; Morris, C.; Le Hir, H.; et al. The Human T-Lymphotropic Virus Type 1 Tax Protein Inhibits Nonsense-Mediated MRNA Decay by Interacting with INT6/EIF3E and UPF1. J. Virol. 2012, 86, 7530–7543. [Google Scholar] [CrossRef] [PubMed]
- Ajamian, L.; Abrahamyan, L.; Milev, M.; Ivanov, P.V.; Kulozik, A.E.; Gehring, N.H.; Mouland, A.J. Unexpected Roles for UPF1 in HIV-1 RNA Metabolism and Translation. RNA 2008, 14, 914–927. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhu, Y.; Baker, S.L.; Bowler, M.W.; Chen, B.J.; Chen, C.; Hogg, J.R.; Goff, S.P.; Song, H. Structural Basis of Suppression of Host Translation Termination by Moloney Murine Leukemia Virus. Nat. Commun. 2016, 7, 12070. [Google Scholar] [CrossRef]
- Hohn, T.; Rothnie, H. Plant Pararetroviruses: Replication and Expression. Curr. Opin. Virol. 2013, 3, 621–628. [Google Scholar] [CrossRef]
- Bouton, C.; Geldreich, A.; Ramel, L.; Ryabova, L.A.; Dimitrova, M.; Keller, M. Cauliflower Mosaic Virus Transcriptome Reveals a Complex Alternative Splicing Pattern. PLoS ONE 2015, 10, e0132665. [Google Scholar] [CrossRef]
- Lukhovitskaya, N.; Ryabova, L.A. Cauliflower Mosaic Virus Transactivator Protein (TAV) Can Suppress Nonsense-Mediated Decay by Targeting VARICOSE, a Scaffold Protein of the Decapping Complex. Sci. Rep. 2019, 9, 7042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ye, X.; Shehata, M.; Dunker, W.; Xie, Z.; Karijolich, J. The RNA Quality Control Pathway Nonsense-Mediated MRNA Decay Targets Cellular and Viral RNAs to Restrict KSHV. Nat. Commun. 2020, 11, 3345. [Google Scholar] [CrossRef]
- Van Gent, M.; Reich, A.; Velu, S.E.; Gack, M.U. Nonsense-Mediated Decay Controls the Reactivation of the Oncogenic Herpesviruses Ebv and Kshv. PLoS Biol. 2021, 19, e3001097. [Google Scholar] [CrossRef] [PubMed]
- May, J.P.; Johnson, P.Z.; Ilyas, M.; Gao, F.; Simon, A.E. The Multifunctional Long-Distance Movement Protein of Pea Enation Mosaic Virus 2 Protects Viral and Host Transcripts from Nonsense-Mediated Decay. MBio 2020, 11, e00204–e00220. [Google Scholar] [CrossRef]
- Franks, T.M.; Singh, G.; Lykke-Andersen, J. Upf1 ATPase-Dependent MRNP Disassembly Is Required for Completion of Nonsense-Mediated MRNA Decay. Cell 2010, 143, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Lemasson, I.; Lewis, M.R.; Polakowski, N.; Hivin, P.; Cavanagh, M.-H.; Thébault, S.; Barbeau, B.; Nyborg, J.K.; Mesnard, J.-M. Human T-Cell Leukemia Virus Type 1 (HTLV-1) BZIP Protein Interacts with the Cellular Transcription Factor CREB To Inhibit HTLV-1 Transcription. J. Virol. 2007, 81, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Wittmann, J.; Jäck, H.M.; Jalinot, P. Human INT6/EIF3e Is Required for Nonsense-Mediated MRNA Decay. EMBO Rep. 2007, 8, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Ando, T.; Yamagishi, M.; Yokoyama, K.; Ishida, T.; Ohsugi, T.; Tanaka, Y.; Brighty, D.W.; Watanabe, T. Viral Interference with Host MRNA Surveillance, the Nonsense-Mediated MRNA Decay (NMD) Pathway, through a New Function of HTLV-1 Rex: Implications for Retroviral Replication. Microbes Infect. 2013, 15, 491–505. [Google Scholar] [CrossRef]
- Li, M.; Johnson, J.R.; Truong, B.; Kim, G.; Weinbren, N.; Dittmar, M.; Shah, P.S.; Von Dollen, J.; Newton, B.W.; Jang, G.M.; et al. Identification of Antiviral Roles for the Exon–Junction Complex and Nonsense-Mediated Decay in Flaviviral Infection. Nat. Microbiol. 2019, 4, 985–995. [Google Scholar] [CrossRef]
- Fontaine, K.A.; Leon, K.E.; Khalid, M.M.; Tomar, S.; Jimenez-Morales, D.; Dunlap, M.; Kaye, J.A.; Shah, P.S.; Finkbeiner, S.; Krogan, N.J.; et al. The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. MBio 2018, 9, e02126-18. [Google Scholar] [CrossRef]
- Wei-lin Popp, M.; Cho, H.; Maquat, L.E. Viral Subversion of Nonsense-Mediated MRNA Decay nonsense-mediated mrna decay in plants and animals: An overview. RNA 2020, 26, 1509–1518. [Google Scholar] [CrossRef]
- Contu, L.; Balistreri, G.; Domanski, M.; Uldry, A.C.; Mühlemann, O. Characterisation of the Semliki Forest Virus-Host Cell Interactome Reveals the Viral Capsid Protein as an Inhibitor of Nonsense-Mediated MRNA Decay. PLoS Pathog. 2021, 17, 1–29. [Google Scholar] [CrossRef]
- Ramage, H.R.; Kumar, G.R.; Verschueren, E.; Johnson, J.R.; VonDollen, J.; Johnson, T.; Newton, B.; Shah, P.; Horner, J.; Krogan, N.J.; et al. A Combined Proteomics/Genomics Approach Links Hepatitis C Virus Infection with Nonsense-Mediated MRNA Decay. Mol. Cell 2015, 57, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Thiébeauld, O.; Schepetilnikov, M.; Park, H.S.; Geldreich, A.; Kobayashi, K.; Keller, M.; Hohn, T.; Ryabova, L.A. A New Plant Protein Interacts with EIF3 and 60S to Enhance Virus-Activated Translation Re-Initiation. EMBO J. 2009, 28, 3171–3184. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Himmelbach, A.; Browning, K.S.; Hohn, T.; Ryabova, L.A. A Plant Viral “Reinitiation” Factor Interacts with the Host Translational Machinery. Cell 2001, 106, 723–733. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Kobayashi, K.; Geldreich, A.; Caranta, C.; Robaglia, C.; Keller, M.; Ryabova, L.A. Viral Factor TAV Recruits TOR/S6K1 Signalling to Activate Reinitiation after Long ORF Translation. EMBO J. 2011, 30, 1343–1356. [Google Scholar] [CrossRef]
- Ryabova, L.A.; Pooggin, M.M.; Hohn, T. Translation Reinitiation and Leaky Scanning in Plant Viruses. Virus Res. 2006, 119, 52–62. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martínez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 Promote Translation Reinitiation of UORF-Containing MRNAs via Phosphorylation of EIF3h. EMBO J. 2013, 32, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, R.S.; Deshotel, M.J.; Johnson, K.; Adler, F.R.; Sieburth, L.E. Arabidopsis MRNA Decay Landscape Arises from Specialized RNA Decay Substrates, Decapping-Mediated Feedback, and Redundancy. Proc. Natl. Acad. Sci. USA 2018, 115, E1485–E1494. [Google Scholar] [CrossRef]
- Masters, P.S. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 65, 193–292. [Google Scholar] [CrossRef]
- Hsin, W.C.; Chang, C.H.; Chang, C.Y.; Peng, W.H.; Chien, C.L.; Chang, M.F.; Chang, S.C. Nucleocapsid Protein-Dependent Assembly of the RNA Packaging Signal of Middle East Respiratory Syndrome Coronavirus. J. Biomed. Sci. 2018, 25, 47. [Google Scholar] [CrossRef]
- Chang, C.K.; Lo, S.C.; Wang, Y.S.; Hou, M.H. Recent Insights into the Development of Therapeutics against Coronavirus Diseases by Targeting N Protein. Drug Discov. Today 2016, 21, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.; Koetzner, C.A.; Hurst, K.R.; Masters, P.S. Recognition of the Murine Coronavirus Genomic RNA Packaging Signal Depends on the Second RNA-Binding Domain of the Nucleocapsid Protein. J. Virol. 2014, 88, 4451–4465. [Google Scholar] [CrossRef] [PubMed]
- Emmott, E.; Munday, D.; Bickerton, E.; Britton, P.; Rodgers, M.A.; Whitehouse, A.; Zhou, E.-M.; Hiscox, J.A. The Cellular Interactome of the Coronavirus Infectious Bronchitis Virus Nucleocapsid Protein and Functional Implications for Virus Biology. J. Virol. 2013, 87, 9486–9500. [Google Scholar] [CrossRef]
- Narayanan, K.; Makino, S. Interplay between Viruses and Host MRNA Degradation. Biochim. Biophys. Acta-Gene Regul. Mech. 2013, 1829, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Weil, J.E.; Hadjithomas, M.; Beemon, K.L. Structural Characterization of the Rous Sarcoma Virus RNA Stability Element. J. Virol. 2009, 83, 2119–2129. [Google Scholar] [CrossRef]
- Weil, J.E.; Beemon, K.L. A 3′ UTR Sequence Stabilizes Termination Codons in the Unspliced RNA of Rous Sarcoma Virus. RNA 2006, 12, 102–110. [Google Scholar] [CrossRef]
- Liu, S.-W.; Wyatt, L.S.; Orandle, M.S.; Minai, M.; Moss, B. The D10 Decapping Enzyme of Vaccinia Virus Contributes to Decay of Cellular and Viral MRNAs and to Virulence in Mice. J. Virol. 2014, 88, 202–211. [Google Scholar] [CrossRef]
- Ge, Z.; Quek, B.L.; Beemon, K.L.; Hogg, J.R. Polypyrimidine Tract Binding Protein 1 Protects MRNAs from Recognition by the Nonsense-Mediated MRNA Decay Pathway. Elife 2016, 5, e11155. [Google Scholar] [CrossRef]
- Baker, S.L.; Hogg, J.R. A System for Coordinated Analysis of Translational Readthrough and Nonsensemediated MRNA Decay. PLoS ONE 2017, 12, e0173980. [Google Scholar] [CrossRef]
- Napthine, S.; Yek, C.; Powell, M.L.; Brown, D.T.K.; Brierley, I. Characterization of the Stop Codon Readthrough Signal of Colorado Tick Fever Virus Segment 9 RNA. RNA 2012, 18, 241–252. [Google Scholar] [CrossRef]
- Ajamian, L.; Abel, K.; Rao, S.; Vyboh, K.; García-de-Gracia, F.; Soto-Rifo, R.; Kulozik, A.E.; Gehring, N.H.; Mouland, A.J. HIV-1 Recruits UPF1 but Excludes UPF2 to Promote Nucleocytoplasmic Export of the Genomic RNA. Biomolecules 2015, 5, 2808–2839. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Amorim, R.; Niu, M.; Breton, Y.; Tremblay, M.J.; Mouland, A.J. Host MRNA Decay Proteins Influence HIV-1 Replication and Viral Gene Expression in Primary Monocyte-Derived Macrophages. Retrovirology 2019, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Toro-Ascuy, D.; Rojas-Araya, B.; Valiente-Echeverría, F.; Soto-Rifo, R. Interactions between the HIV-1 Unspliced MRNA and Host MRNA Decay Machineries. Viruses 2016, 8, 320. [Google Scholar] [CrossRef]
- Leon, K.; Ott, M. An ‘Arms Race’ between the Nonsense-Mediated MRNA Decay Pathway and Viral Infections. Semin. Cell Dev. Biol. 2021, 111, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Speiseder, T.; Dobner, T.; Gonzalez, R.A. DNA Virus Replication Compartments. J. Virol. 2014, 88, 1404–1420. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, M.R.; Du, Z. Molecular Interaction of Nonsense-Mediated mRNA Decay with Viruses. Viruses 2023, 15, 816. https://doi.org/10.3390/v15040816
Ahmed MR, Du Z. Molecular Interaction of Nonsense-Mediated mRNA Decay with Viruses. Viruses. 2023; 15(4):816. https://doi.org/10.3390/v15040816
Chicago/Turabian StyleAhmed, Md Robel, and Zhiyou Du. 2023. "Molecular Interaction of Nonsense-Mediated mRNA Decay with Viruses" Viruses 15, no. 4: 816. https://doi.org/10.3390/v15040816
APA StyleAhmed, M. R., & Du, Z. (2023). Molecular Interaction of Nonsense-Mediated mRNA Decay with Viruses. Viruses, 15(4), 816. https://doi.org/10.3390/v15040816








