Intact Proviral DNA Analysis of the Brain Viral Reservoir and Relationship to Neuroinflammation in People with HIV on Suppressive Antiretroviral Therapy
Abstract
1. Introduction
2. Methods
2.1. Cohort Selection and Characterization
2.2. Nucleic Acid Extraction and Brain HIV IPDA and Viral Load Measurements
2.3. Gene Expression Profiling
2.4. Data Analysis
3. Results
3.1. Characteristics of the Study Cohort
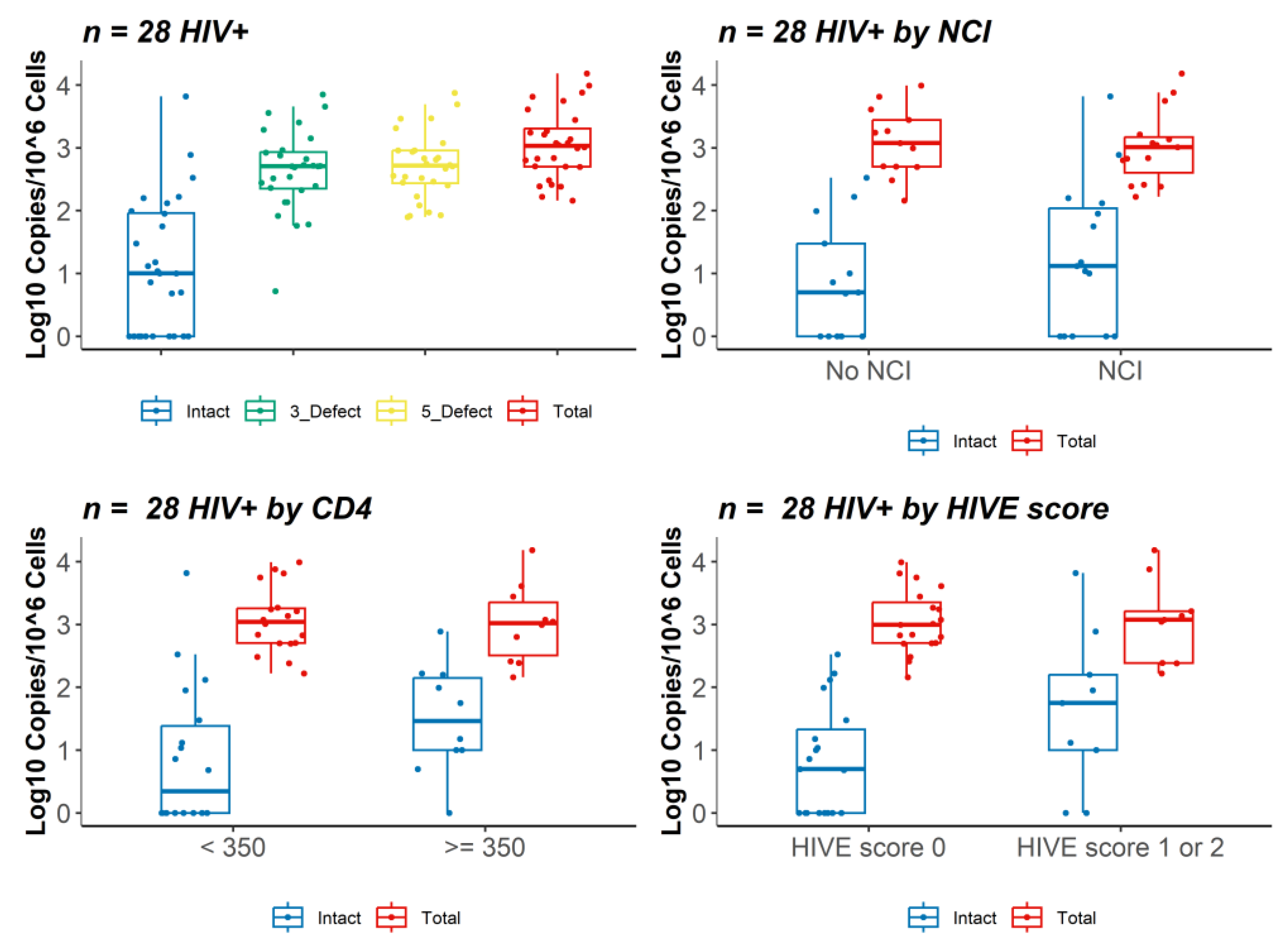
3.2. IPDA Analysis and HIV DNA and RNA Brain Viral Load
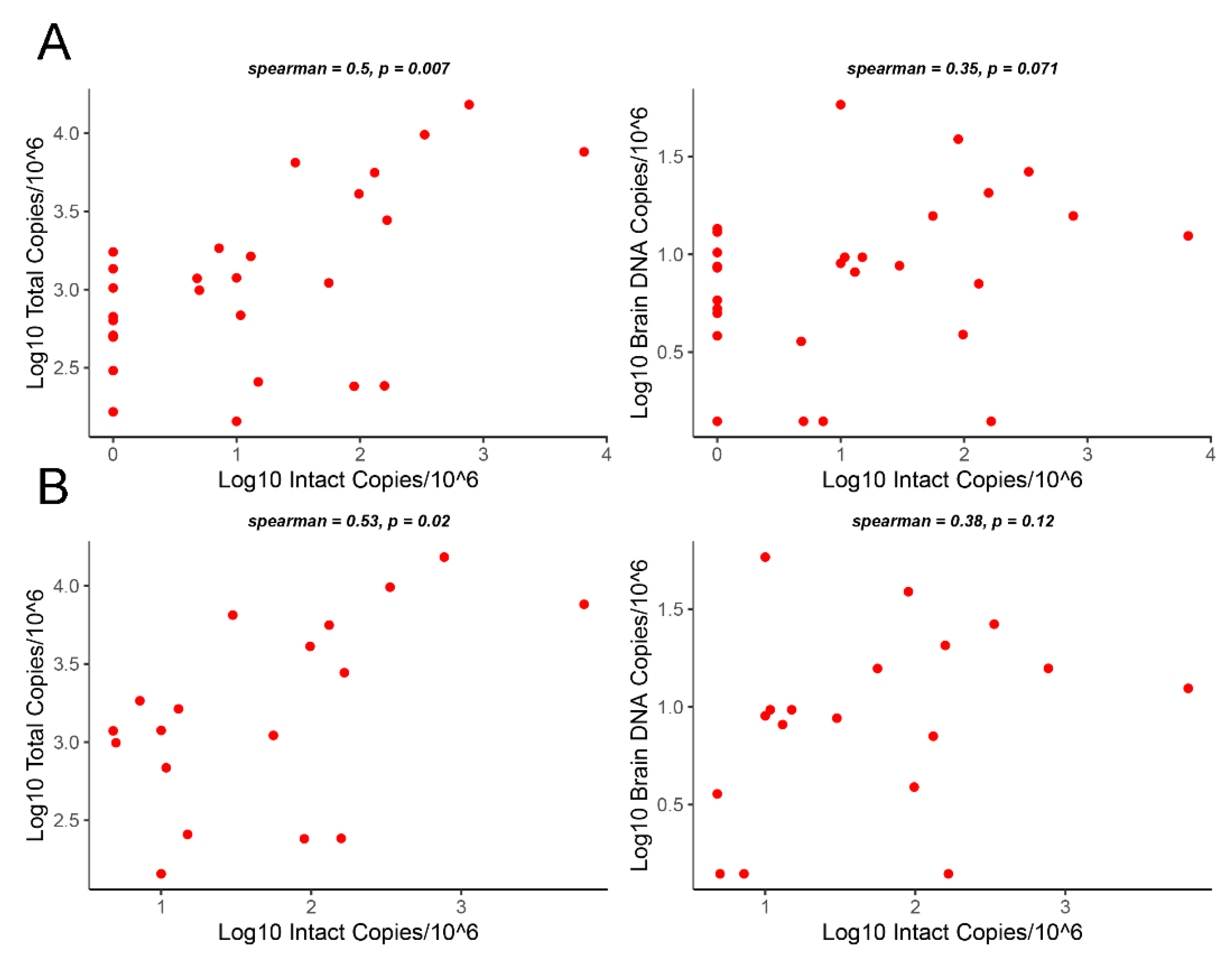
3.3. Correlation Analysis of Intact Proviruses versus Total Proviruses or Gag DNA Levels
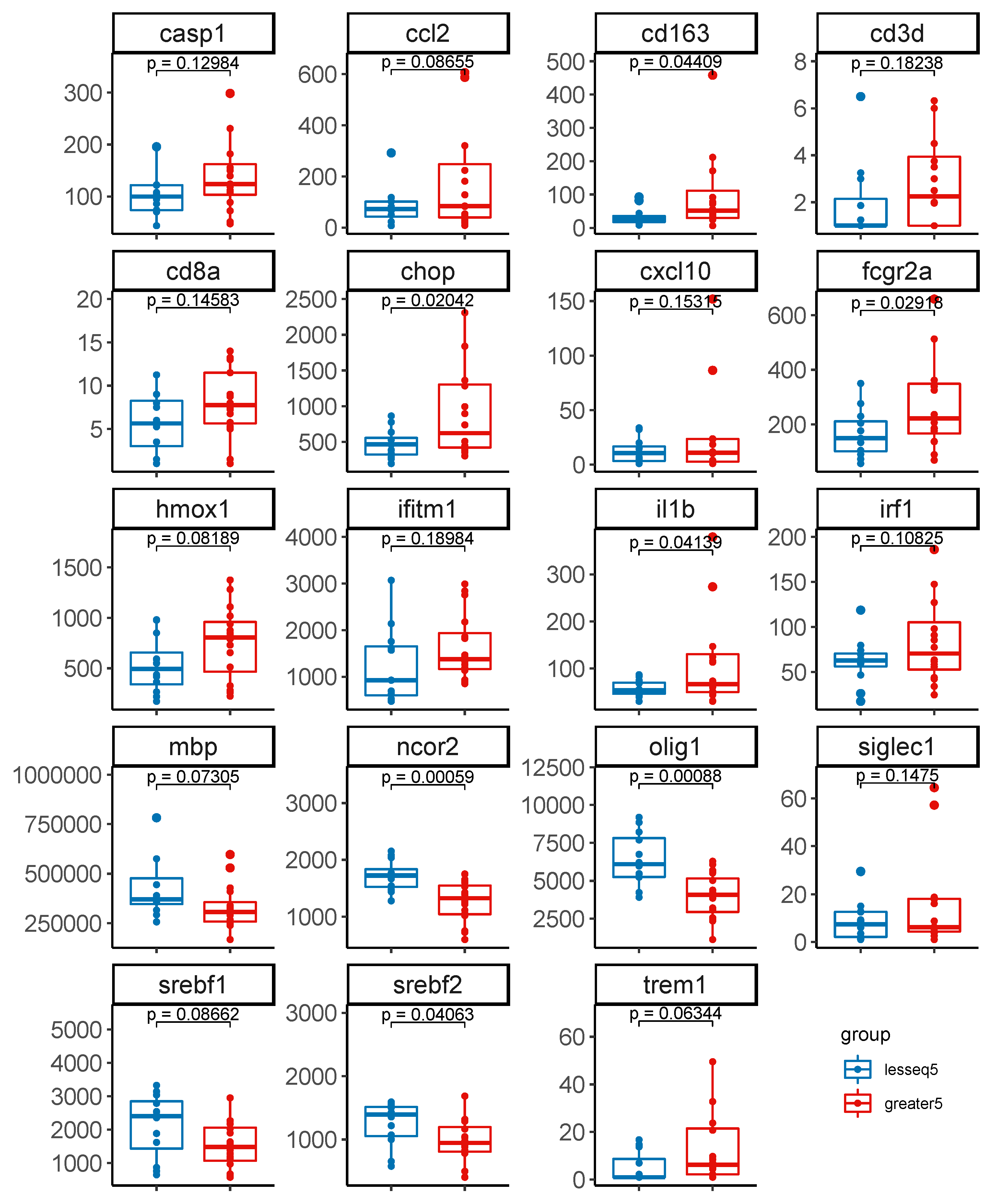
3.4. Gene Expression Profiles Stratified by Level of Intact Proviruses in Brain Tissue
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gelman, B.B.; Lisinicchia, J.G.; Morgello, S.; Masliah, E.; Commins, D.; Achim, C.L.; Fox, H.S.; Kolson, D.L.; Grant, I.; Singer, E.; et al. Neurovirological correlation with HIV-associated neurocognitive disorders and encephalitis in a HAART-era cohort. J. Acquir. Immune Defic. Syndr. 2013, 62, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Brew, B.J.; Barnes, S.L. The impact of HIV central nervous system persistence on pathogenesis. AIDS 2019, 33 (Suppl. 2), S113–S121. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Daley, W.; Lawrence, D.; Lorenzo, E.; Perrin, P.; Rao, V.R.; Tsai, S.Y.; Varthakavi, V. Role of macrophages in HIV pathogenesis and cure: NIH perspectives. J. Leukoc. Biol. 2022, 112, 1233–1243. [Google Scholar] [CrossRef]
- Joseph, S.B.; Kincer, L.P.; Bowman, N.M.; Evans, C.; Vinikoor, M.J.; Lippincott, C.K.; Gisslén, M.; Spudich, S.; Menezes, P.; Robertson, K. Human immunodeficiency virus type 1 RNA detected in the central nervous system (CNS) after years of suppressive antiretroviral therapy can originate from a replicating CNS reservoir or clonally expanded cells. Clin. Infect. Dis. 2019, 69, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Spudich, S.; Peterson, J.; Fuchs, D.; Price, R.W.; Gisslen, M. Potential for early antiretroviral therapy to reduce central nervous system HIV-1 persistence. AIDS 2019, 33 (Suppl. 2), S135–S144. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.; Lamers, S.L.; Nolan, D.J.; Maidji, E.; Faria, N.R.; Pybus, O.G.; Dollar, J.J.; Maruniak, S.A.; McAvoy, A.C.; Salemi, M.; et al. HIV Maintains an Evolving and Dispersed Population in Multiple Tissues during Suppressive Combined Antiretroviral Therapy in Individuals with Cancer. J. Virol. 2016, 90, 8984–8993. [Google Scholar] [CrossRef]
- Lamers, S.L.; Rose, R.; Maidji, E.; Agsalda-Garcia, M.; Nolan, D.J.; Fogel, G.B.; Salemi, M.; Garcia, D.L.; Bracci, P.; Yong, W.; et al. HIV DNA Is Frequently Present within Pathologic Tissues Evaluated at Autopsy from Combined Antiretroviral Therapy-Treated Patients with Undetectable Viral Loads. J. Virol. 2016, 90, 8968–8983. [Google Scholar] [CrossRef]
- Abreu, C.; Shirk, E.N.; Queen, S.E.; Mankowski, J.L.; Gama, L.; Clements, J.E. A Quantitative Approach to SIV Functional Latency in Brain Macrophages. J. Neuroimmune Pharmacol. 2019, 14, 23–32. [Google Scholar] [CrossRef]
- Gama, L.; Abreu, C.; Shirk, E.N.; Queen, S.E.; Beck, S.E.; Metcalf Pate, K.A.; Bullock, B.T.; Zink, M.C.; Mankowski, J.L.; Clements, J.E. SIV Latency in Macrophages in the CNS. Curr. Top. Microbiol. Immunol. 2018, 417, 111–130. [Google Scholar] [CrossRef]
- Ko, A.; Kang, G.; Hattler, J.B.; Galadima, H.I.; Zhang, J.; Li, Q.; Kim, W.K. Macrophages but not Astrocytes Harbor HIV DNA in the Brains of HIV-1-Infected Aviremic Individuals on Suppressive Antiretroviral Therapy. J. Neuroimmune Pharmacol. 2019, 14, 110–119. [Google Scholar] [CrossRef]
- Solomon, I.H.; Chettimada, S.; Misra, V.; Lorenz, D.R.; Gorelick, R.J.; Gelman, B.B.; Morgello, S.; Gabuzda, D. White Matter Abnormalities Linked to Interferon, Stress Response, and Energy Metabolism Gene Expression Changes in Older HIV-Positive Patients on Antiretroviral Therapy. Mol. Neurobiol. 2020, 57, 1115–1130. [Google Scholar] [CrossRef] [PubMed]
- Farhadian, S.F.; Lindenbaum, O.; Zhao, J.; Corley, M.J.; Im, Y.; Walsh, H.; Vecchio, A.; Garcia-Milian, R.; Chiarella, J.; Chintanaphol, M.; et al. HIV viral transcription and immune perturbations in the CNS of people with HIV despite ART. JCI Insight 2022, 7, e160267. [Google Scholar] [CrossRef] [PubMed]
- Spudich, S.; Robertson, K.R.; Bosch, R.J.; Gandhi, R.T.; Cyktor, J.C.; Mar, H.; Macatangay, B.J.; Lalama, C.M.; Rinaldo, C.; Collier, A.C.; et al. Persistent HIV-infected cells in cerebrospinal fluid are associated with poorer neurocognitive performance. J. Clin. Investig. 2019, 129, 3339–3346. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Zaunders, J.; Gates, T.M.; Levert, A.; Butterly, S.; Liu, Z.; Ishida, T.; Palmer, S.; Rae, C.D.; Juge, L.; et al. Elevation of cell-associated HIV-1 transcripts in CSF CD4+ T cells, despite effective antiretroviral therapy, is linked to brain injury. Proc. Natl. Acad. Sci. USA 2022, 119, e2210584119. [Google Scholar] [CrossRef]
- Churchill, M.J.; Gorry, P.R.; Cowley, D.; Lal, L.; Sonza, S.; Purcell, D.F.; Thompson, K.A.; Gabuzda, D.; McArthur, J.C.; Pardo, C.A.; et al. Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J. Neurovirol. 2006, 12, 146–152. [Google Scholar] [CrossRef]
- Gelman, B.B. Neuropathology of HAND With Suppressive Antiretroviral Therapy: Encephalitis and Neurodegeneration Reconsidered. Curr. HIV/AIDS Rep. 2015, 12, 272–279. [Google Scholar] [CrossRef]
- Chung, H.K.; Hattler, J.B.; Narola, J.; Babbar, H.; Cai, Y.; Abdel-Mohsen, M.; Kim, W.K. Development of Droplet Digital PCR-Based Assays to Quantify HIV Proviral and Integrated DNA in Brain Tissues from Viremic Individuals with Encephalitis and Virally Suppressed Aviremic Individuals. Microbiol. Spectr. 2022, 10, e0085321. [Google Scholar] [CrossRef]
- Cochrane, C.R.; Angelovich, T.A.; Byrnes, S.J.; Waring, E.; Guanizo, A.C.; Trollope, G.S.; Zhou, J.; Vue, J.; Senior, L.; Wanicek, E.; et al. Intact HIV Proviruses Persist in the Brain Despite Viral Suppression with ART. Ann. Neurol. 2022, 92, 532–544. [Google Scholar] [CrossRef]
- Mukerji, S.S.; Misra, V.; Lorenz, D.R.; Uno, H.; Morgello, S.; Franklin, D.; Ellis, R.J.; Letendre, S.; Gabuzda, D. Impact of Antiretroviral Regimens on Cerebrospinal Fluid Viral Escape in a Prospective Multicohort Study of Antiretroviral Therapy-Experienced Human Immunodeficiency Virus-1-Infected Adults in the United States. Clin. Infect. Dis. 2018, 67, 1182–1190. [Google Scholar] [CrossRef]
- Winston, A.; Antinori, A.; Cinque, P.; Fox, H.S.; Gisslen, M.; Henrich, T.J.; Letendre, S.; Persaud, D.; Price, R.W.; Spudich, S. Defining cerebrospinal fluid HIV RNA escape: Editorial review AIDS. AIDS 2019, 33 (Suppl. 2), S107–S111. [Google Scholar] [CrossRef]
- Joseph, J.; Cinque, P.; Colosi, D.; Dravid, A.; Ene, L.; Fox, H.; Gabuzda, D.; Gisslen, M.; Beth Joseph, S.; Letendre, S.; et al. Highlights of the Global HIV-1 CSF Escape Consortium Meeting, 9 June 2016, Bethesda, MD, USA. J. Virus Erad. 2016, 2, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Saylor, D.; Dickens, A.M.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-associated neurocognitive disorder--pathogenesis and prospects for treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Winston, A.; Spudich, S. Cognitive disorders in people living with HIV. Lancet HIV 2020, 7, e504–e513. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.M.; Munoz-Moreno, J.A.; McClernon, D.R.; Ellis, R.J.; Cookson, D.; Clifford, D.B.; Collier, A.C.; Gelman, B.B.; Marra, C.M.; McArthur, J.C.; et al. Prevalence and Correlates of Persistent HIV-1 RNA in Cerebrospinal Fluid During Antiretroviral Therapy. J. Infect. Dis. 2017, 215, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Mukerji, S.S.; Petersen, K.J.; Pohl, K.M.; Dastgheyb, R.M.; Fox, H.S.; Bilder, R.M.; Brouillette, M.J.; Gross, A.L.; Scott-Sheldon, L.A.J.; Paul, R.H.; et al. Machine learning approaches to understand cognitive phenotypes in people with HIV. J. Infect. Dis. 2022; in press. [Google Scholar] [CrossRef]
- Solomon, I.H.; De Girolami, U.; Chettimada, S.; Misra, V.; Singer, E.J.; Gabuzda, D. Brain and liver pathology, amyloid deposition, and interferon responses among older HIV-positive patients in the late HAART era. BMC Infect. Dis. 2017, 17, 151. [Google Scholar] [CrossRef]
- Tavazzi, E.; Morrison, D.; Sullivan, P.; Morgello, S.; Fischer, T. Brain inflammation is a common feature of HIV-infected patients without HIV encephalitis or productive brain infection. Curr. HIV Res. 2014, 12, 97–110. [Google Scholar] [CrossRef]
- Gelman, B.B.; Chen, T.; Lisinicchia, J.G.; Soukup, V.M.; Carmical, J.R.; Starkey, J.M.; Masliah, E.; Commins, D.L.; Brandt, D.; Grant, I.; et al. The National NeuroAIDS Tissue Consortium brain gene array: Two types of HIV-associated neurocognitive impairment. PLoS ONE 2012, 7, e46178. [Google Scholar] [CrossRef]
- Sanna, P.P.; Fu, Y.; Masliah, E.; Lefebvre, C.; Repunte-Canonigo, V. Central nervous system (CNS) transcriptomic correlates of human immunodeficiency virus (HIV) brain RNA load in HIV-infected individuals. Sci. Rep. 2021, 11, 12176. [Google Scholar] [CrossRef]
- Borjabad, A.; Morgello, S.; Chao, W.; Kim, S.Y.; Brooks, A.I.; Murray, J.; Potash, M.J.; Volsky, D.J. Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders. PLoS Pathog. 2011, 7, e1002213. [Google Scholar] [CrossRef]
- Plaza-Jennings, A.L.; Valada, A.; O’Shea, C.; Iskhakova, M.; Hu, B.; Javidfar, B.; Ben Hutta, G.; Lambert, T.Y.; Murray, J.; Kassim, B.; et al. HIV integration in the human brain is linked to microglial activation and 3D genome remodeling. Mol. Cell 2022, 82, 4647–4663.e4648. [Google Scholar] [CrossRef]
- Bruner, K.M.; Wang, Z.; Simonetti, F.R.; Bender, A.M.; Kwon, K.J.; Sengupta, S.; Fray, E.J.; Beg, S.A.; Antar, A.A.R.; Jenike, K.M.; et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019, 566, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.N.; Hughes, S.M.; Roychoudhury, P.; Reeves, D.B.; Amstuz, C.; Zhu, H.; Huang, M.L.; Wei, Y.; Bull, M.E.; Cassidy, N.A.J.; et al. A highly multiplexed droplet digital PCR assay to measure the intact HIV-1 proviral reservoir. Cell Rep. Med. 2021, 2, 100243. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Catalano, A.A.; Ho, Y.C. Measuring the size and decay dynamics of the HIV-1 latent reservoir. Cell Rep. Med. 2021, 2, 100249. [Google Scholar] [CrossRef]
- Massanella, M.; Richman, D.D. Measuring the latent reservoir in vivo. J. Clin. Investig. 2016, 126, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Peluso, M.J.; Bacchetti, P.; Ritter, K.D.; Beg, S.; Lai, J.; Martin, J.N.; Hunt, P.W.; Henrich, T.J.; Siliciano, J.D.; Siliciano, R.F.; et al. Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight 2020, 5, e132997. [Google Scholar] [CrossRef]
- Simonetti, F.R.; White, J.A.; Tumiotto, C.; Ritter, K.D.; Cai, M.; Gandhi, R.T.; Deeks, S.G.; Howell, B.J.; Montaner, L.J.; Blankson, J.N.; et al. Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc. Natl. Acad. Sci. USA 2020, 117, 18692–18700. [Google Scholar] [CrossRef]
- Gandhi, R.T.; Cyktor, J.C.; Bosch, R.J.; Mar, H.; Laird, G.M.; Martin, A.; Collier, A.C.; Riddler, S.A.; Macatangay, B.J.; Rinaldo, C.R.; et al. Selective Decay of Intact HIV-1 Proviral DNA on Antiretroviral Therapy. J. Infect. Dis. 2021, 223, 225–233. [Google Scholar] [CrossRef]
- Martin, A.R.; Bender, A.M.; Hackman, J.; Kwon, K.J.; Lynch, B.A.; Bruno, D.; Martens, C.; Beg, S.; Florman, S.S.; Desai, N.; et al. Similar Frequency and Inducibility of Intact Human Immunodeficiency Virus-1 Proviruses in Blood and Lymph Nodes. J. Infect. Dis. 2021, 224, 258–268. [Google Scholar] [CrossRef]
- Morgello, S.; Gelman, B.; Kozlowski, P.; Vinters, H.; Masliah, E.; Cornford, M.; Cavert, W.; Marra, C.; Grant, I.; Singer, E. The National NeuroAIDS Tissue Consortium: A new paradigm in brain banking with an emphasis on infectious disease. Neuropathol. Appl. Neurobiol. 2001, 27, 326–335. [Google Scholar] [CrossRef]
- Antinori, A.; Arendt, G.; Becker, J.T.; Brew, B.J.; Byrd, D.A.; Cherner, M.; Clifford, D.B.; Cinque, P.; Epstein, L.G.; Goodkin, K.; et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology 2007, 69, 1789–1799. [Google Scholar] [CrossRef] [PubMed]
- Heaton, R.K.; Clifford, D.B.; Franklin, D.R., Jr.; Woods, S.P.; Ake, C.; Vaida, F.; Ellis, R.J.; Letendre, S.L.; Marcotte, T.D.; Atkinson, J.H.; et al. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 2010, 75, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Woods, S.P.; Rippeth, J.D.; Frol, A.B.; Levy, J.K.; Ryan, E.; Soukup, V.M.; Hinkin, C.H.; Lazzaretto, D.; Cherner, M.; Marcotte, T.D.; et al. Interrater reliability of clinical ratings and neurocognitive diagnoses in HIV. J. Clin. Exp. Neuropsychol. 2004, 26, 759–778. [Google Scholar] [CrossRef] [PubMed]
- Underwood, J.; Cole, J.H.; Caan, M.; De Francesco, D.; Leech, R.; Van Zoest, R.A.; Su, T.; Geurtsen, G.J.; Schmand, B.A.; Portegies, P.; et al. Gray and White Matter Abnormalities in Treated Human Immunodeficiency Virus Disease and Their Relationship to Cognitive Function. Clin. Infect. Dis. 2017, 65, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Cassol, E.; Misra, V.; Morgello, S.; Gabuzda, D. Applications and limitations of inflammatory biomarkers for studies on neurocognitive impairment in HIV infection. J. Neuroimmune Pharmacol. 2013, 8, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- McArthur, J.C.; Johnson, T.P. Chronic inflammation mediates brain injury in HIV infection: Relevance for cure strategies. Curr. Opin. Neurol. 2020, 33, 397–404. [Google Scholar] [CrossRef]
- Guha, D.; Lorenz, D.R.; Misra, V.; Chettimada, S.; Morgello, S.; Gabuzda, D. Proteomic analysis of cerebrospinal fluid extracellular vesicles reveals synaptic injury, inflammation, and stress response markers in HIV patients with cognitive impairment. J. Neuroinflamm. 2019, 16, 254. [Google Scholar] [CrossRef]
- Gill, A.J.; Kovacsics, C.E.; Cross, S.A.; Vance, P.J.; Kolson, L.L.; Jordan-Sciutto, K.L.; Gelman, B.B.; Kolson, D.L. Heme oxygenase-1 deficiency accompanies neuropathogenesis of HIV-associated neurocognitive disorders. J. Clin. Investig. 2014, 124, 4459–4472. [Google Scholar] [CrossRef]
- Voskuhl, R.R.; Itoh, N.; Tassoni, A.; Matsukawa, M.A.; Ren, E.; Tse, V.; Jang, E.; Suen, T.T.; Itoh, Y. Gene expression in oligodendrocytes during remyelination reveals cholesterol homeostasis as a therapeutic target in multiple sclerosis. Proc. Natl. Acad. Sci. USA 2019, 116, 10130–10139. [Google Scholar] [CrossRef]
- Camargo, N.; Goudriaan, A.; van Deijk, A.F.; Otte, W.M.; Brouwers, J.F.; Lodder, H.; Gutmann, D.H.; Nave, K.A.; Dijkhuizen, R.M.; Mansvelder, H.D.; et al. Oligodendroglial myelination requires astrocyte-derived lipids. PLoS Biol. 2017, 15, e1002605. [Google Scholar] [CrossRef]
- Shimano, H.; Sato, R. SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat. Rev. Endocrinol. 2017, 13, 710–730. [Google Scholar] [CrossRef] [PubMed]
- Collora, J.A.; Liu, R.; Pinto-Santini, D.; Ravindra, N.; Ganoza, C.; Lama, J.R.; Alfaro, R.; Chiarella, J.; Spudich, S.; Mounzer, K.; et al. Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity 2022, 55, 1013–1031.e1017. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.J.; Farhadian, S.F. Emerging Single-cell Approaches to Understand HIV in the Central Nervous System. Curr. HIV/AIDS Rep. 2022, 19, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Riggs, P.K.; Chaillon, A.; Jiang, G.; Letendre, S.L.; Tang, Y.; Taylor, J.; Kaytes, A.; Smith, D.M.; Dube, K.; Gianella, S. Lessons for Understanding Central Nervous System HIV Reservoirs from the Last Gift Program. Curr. HIV/AIDS Rep. 2022, 19, 566–579. [Google Scholar] [CrossRef] [PubMed]
| Variable | Total (n = 28) | No NCI (n = 13) | NCI (n = 15) | p-Value |
|---|---|---|---|---|
| Age (years) | 55.50 (50.00, 59.50) | 52.00 (50.00, 59.00) | 56.00 (52.00, 60.00) | 0.5182 |
| Male gender, n (%) | 22 (78.6) | 12 (92.3) | 10 (66.7) | 0.2351 |
| Race/ethnicity, n (%) | 0.5685 | |||
| White | 12 (42.9) | 7 (53.8) | 5 (33.3) | |
| Black | 11 (39.3) | 4 (30.8) | 7 (46.7) | |
| Hispanic | 4 (14.3) | 2 (15.4) | 2 (13.3) | |
| Other | 1 (3.6) | 0 (0.0) | 1 (6.7) | |
| PMI (hours) | 12.00 (5.88, 16.25) | 12.00 (6.00, 15.00) | 11.00 (6.75, 18.25) | 0.9082 |
| Duration of HIV infection (years) | 17.00 (13.00, 21.50) | 18.50 (10.00, 21.25) | 17.00 (14.00, 20.75) | 0.7322 |
| CD4 count (cells/μL) | 287.50 (126.25, 436.50) | 309.00 (128.00, 427.00) | 283.00 (126.50, 454.00) | 0.8719 |
| CD4 nadir (cells/μL) | 98.00 (53.50, 147.50) | 78.00 (17.00, 128.00) | 98.00 (65.00, 185.00) | 0.4201 |
| Plasma VL (copies/mL) * | 40.00 (40.00, 40.00) | 40.00 (40.00, 40.00) | 40.00 (40.00, 77.00) | 0.0123 |
| Time between last plasma VL and death (years) | 0.40 (0.20, 0.62) | 0.40 (0.20, 0.50) | 0.40 (0.15, 0.65) | 0.7801 |
| CSF VL (copies/mL) * | 40.00 (40.00, 40.00) | 40.00 (40.00, 40.00) | 40.00 (40.00, 137.50) | 0.1119 |
| Time between last CSF VL and death (years) | 0.30 (0.00, 1.00) | 0.20 (0.00, 0.75) | 0.35 (0.07, 1.23) | 0.6065 |
| Maximum CSF VL in study (copies/mL) | 40.00 (40.00, 170.00) | 40.00 (40.00, 77,174.00) | 40.00 (40.00, 40.00) | 0.3102 |
| Neurocognitive T-score | 43.80 (37.27, 48.69) | 49.14 (47.32, 53.24) | 38.08 (36.16, 43.30) | 0.0002 |
| HAND diagnosis, n (%) | 0.0005 | |||
| MND | 2 (7.4) | 0 (0.0) | 2 (13.3) | |
| HAD | 4 (14.8) | 0 (0.0) | 4 (26.7) | |
| NPI-O | 12 (44.4) | 3 (25.0) | 9 (60.0) | |
| Normal | 9 (33.3) | 9 (75.0) | 0 (0.0) | |
| HIVE score, n (%) | 0.0032 | |||
| 0 | 19 (67.9) | 13 (100.0) | 6 (40.0) | |
| 1 | 7 (25.0) | 0 (0.0) | 7 (46.7) | |
| 2 | 2 (7.1) | 0 (0.0) | 2 (13.3) | |
| Intact proviruses (cps/106 cells) | 10.00 (1.00, 91.96) | 5.00 (1.00, 30.00) | 13.05 (1.00, 110.74) | 0.3831 |
| 3′ defective proviruses (cps/106 cells) | 508.71 (224.80, 857.78) | 530.92 (229.15, 1422.37) | 488.60 (205.88, 590.31) | 0.3001 |
| 5′ defective proviruses (cps/106 cells) | 519.09 (273.18, 905.71) | 645.03 (280.45, 1191.33) | 508.49 (226.47, 686.78) | 0.4472 |
| Total proviruses (cps/106 cells) | 1063.93 (501.28, 2074.33) | 1180.74 (502.72, 2779.76) | 1024.07 (445.79, 1495.22) | 0.6617 |
| HIV gag DNA (cps/106 cells) | 8.71 (4.72, 13.13) | 5.00 (3.59, 8.75) | 9.67 (8.40, 14.36) | 0.0527 |
| HIV gag RNA (cps/106 cells) | 7.45 (1.51, 27.92) | 7.92 (1.12, 25.28) | 6.98 (2.32, 31.08) | 0.8755 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabuzda, D.; Yin, J.; Misra, V.; Chettimada, S.; Gelman, B.B. Intact Proviral DNA Analysis of the Brain Viral Reservoir and Relationship to Neuroinflammation in People with HIV on Suppressive Antiretroviral Therapy. Viruses 2023, 15, 1009. https://doi.org/10.3390/v15041009
Gabuzda D, Yin J, Misra V, Chettimada S, Gelman BB. Intact Proviral DNA Analysis of the Brain Viral Reservoir and Relationship to Neuroinflammation in People with HIV on Suppressive Antiretroviral Therapy. Viruses. 2023; 15(4):1009. https://doi.org/10.3390/v15041009
Chicago/Turabian StyleGabuzda, Dana, Jun Yin, Vikas Misra, Sukrutha Chettimada, and Benjamin B. Gelman. 2023. "Intact Proviral DNA Analysis of the Brain Viral Reservoir and Relationship to Neuroinflammation in People with HIV on Suppressive Antiretroviral Therapy" Viruses 15, no. 4: 1009. https://doi.org/10.3390/v15041009
APA StyleGabuzda, D., Yin, J., Misra, V., Chettimada, S., & Gelman, B. B. (2023). Intact Proviral DNA Analysis of the Brain Viral Reservoir and Relationship to Neuroinflammation in People with HIV on Suppressive Antiretroviral Therapy. Viruses, 15(4), 1009. https://doi.org/10.3390/v15041009




