HSV1716 Prevents Myeloma Cell Regrowth When Combined with Bortezomib In Vitro and Significantly Reduces Systemic Tumor Growth in Mouse Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Culture
2.2. HSV1716-GFP Infection Assays
2.3. Oncolysis Assays
2.4. ICP0 and ICP8 Gene Expression Analyses
2.5. Apoptosis Analysis
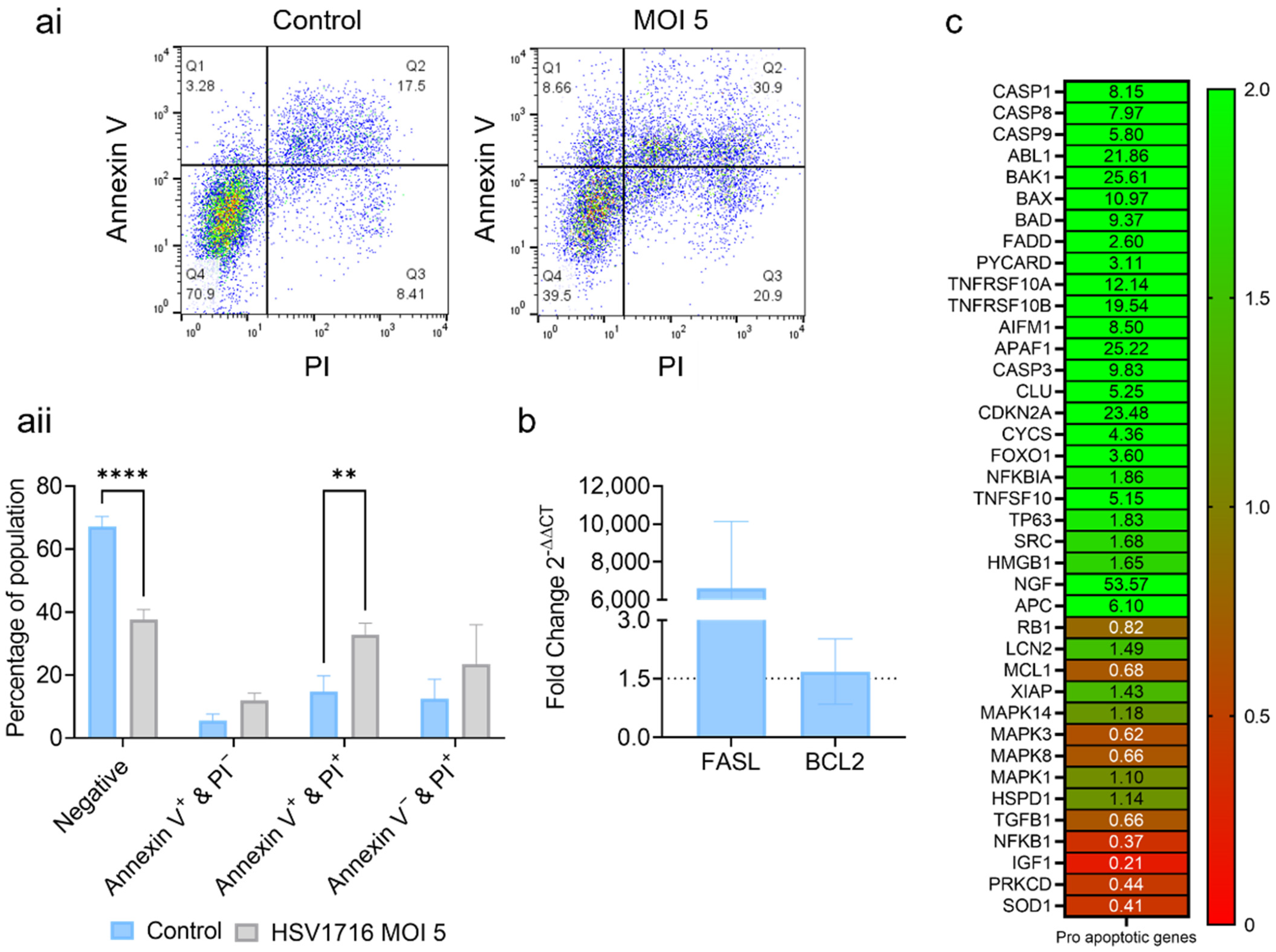
2.6. PrimePCRTM Human Apoptosis Microarray Array
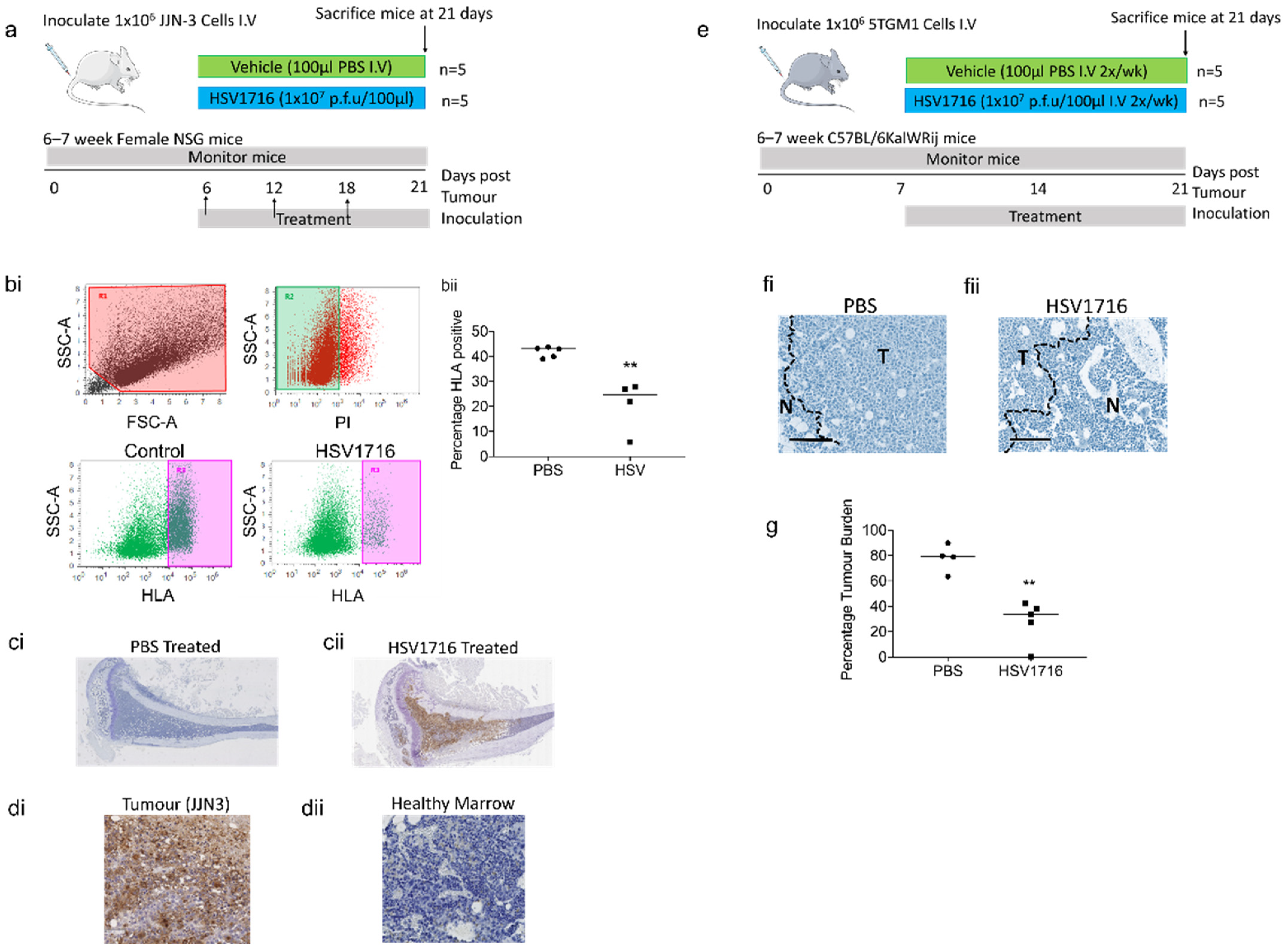
2.7. JJN-3 Xenograft Model of Myeloma
2.8. 5TGM1 Syngeneic Murine Model of Myeloma
2.9. Assessment of Tumor Burden
2.10. Assessment of Bone Disease
2.11. Statistical Analyses
3. Results
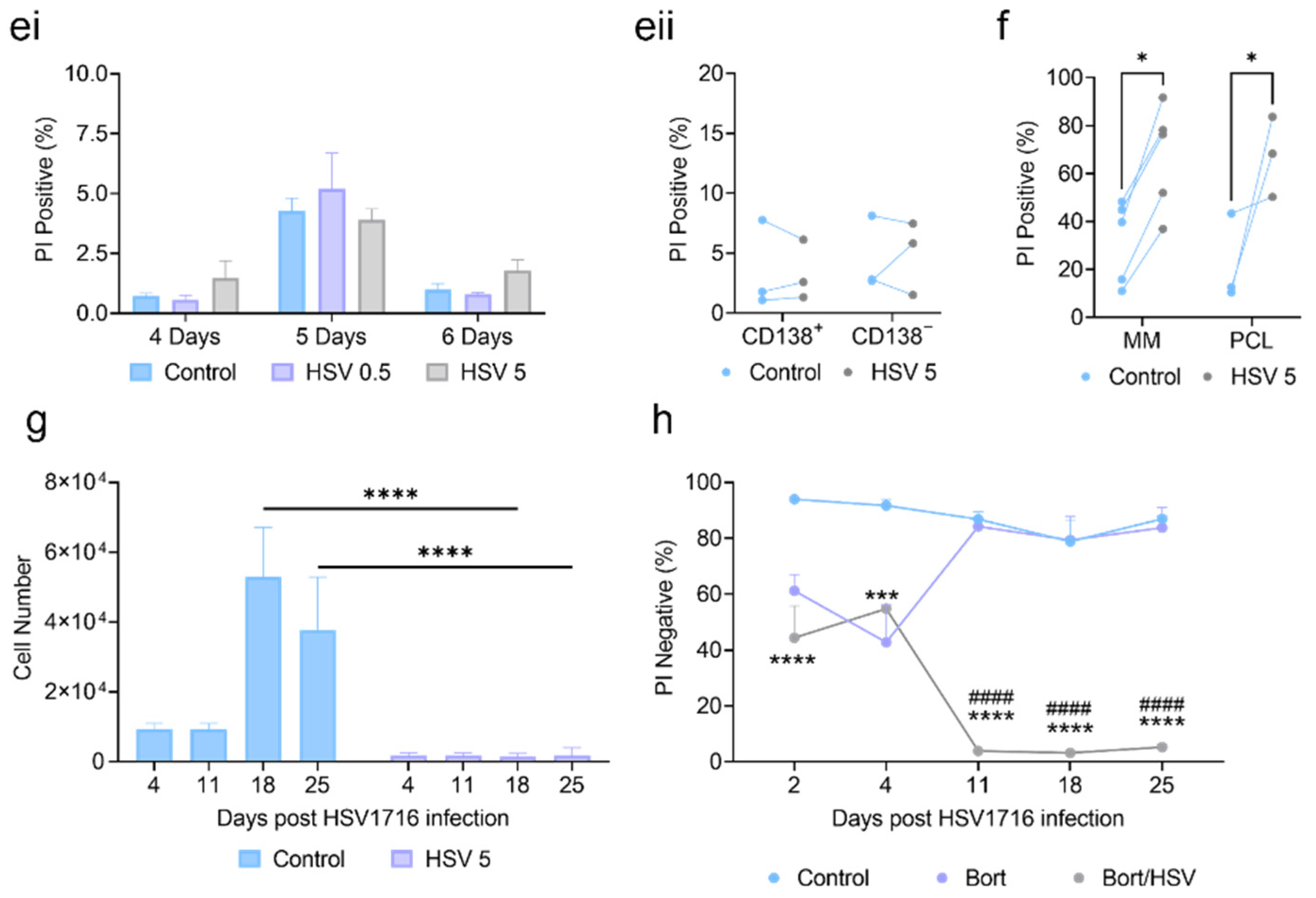
3.1. HSV1716 Induces Potent Cell Death in Human Myeloma Cell Lines and Primary Patient Samples
3.2. HSV1716 Induces Cell Death via Apoptosis
3.3. Oncolytic Virus Therapy Significantly Lowers Tumor Burden in Murine Myeloma Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Research UK. Myeloma Statistics|Cancer Research UK. Myeloma Statistics. 2017. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/myeloma/survival2018 (accessed on 1 October 2022).
- Teoh, G.; Chen, L.; Urashima, M.; Tai, Y.T.; Celi, L.A.; Chen, D.; Chauhan, D.; Ogata, A.; Finberg, R.W.; Webb, I.J.; et al. Adenovirus Vector-Based Purging of Multiple Myeloma Cells. Blood 1998, 92, 4591–4601. [Google Scholar] [CrossRef]
- Kelly, K.R.; Espitia, C.M.; Zhao, W.; Wu, K.; Visconte, V.; Anwer, F.; Calton, C.M.; Carew, J.S.; Nawrocki, S.T. Oncolytic reovirus sensitizes multiple myeloma cells to anti-PD-L1 therapy. Leukemia 2018, 32, 230–233. [Google Scholar] [CrossRef]
- Wenthe, J.; Naseri, S.; Hellström, A.-C.; Wiklund, H.J.; Eriksson, E.; Loskog, A. Immunostimulatory oncolytic virotherapy for multiple myeloma targeting 4-1BB and/or CD40. Cancer Gene Ther. 2020, 27, 948–959. [Google Scholar] [CrossRef]
- Lei, W.; Wang, S.; Xu, N.; Chen, Y.; Wu, G.; Zhang, A.; Chen, X.; Tong, Y.; Qian, W. Enhancing therapeutic efficacy of oncolytic vaccinia virus armed with Beclin-1, an autophagic Gene in leukemia and myeloma. Biomed. Pharmacother. 2020, 125, 110030. [Google Scholar] [CrossRef]
- Dispenzieri, A.; Tong, C.; LaPlant, B.; Lacy, M.Q.; Laumann, K.; Dingli, D.; Zhou, Y.; Federspiel, M.J.; Gertz, M.A.; Hayman, S.; et al. Phase I trial of systemic administration of edmonston strain of measles virus genetically engineered to express the sodium iodide symporter in patients with recurrent or refractory multiple myeloma. Leukemia 2017, 31, 2791–2798. [Google Scholar] [CrossRef]
- Masoud, S.J.; Hu, J.B.; Beasley, G.M.; Stewart, J.H.; Mosca, P.J. Efficacy of Talimogene Laherparepvec (T-VEC) Therapy in Patients with In-Transit Melanoma Metastasis Decreases with Increasing Lesion Size. Ann. Surg. Oncol. 2019, 26, 4633–4641. [Google Scholar] [CrossRef]
- Andtbacka, R.H.I.; Collichio, F.; Harrington, K.J.; Middleton, M.R.; Downey, G.; Öhrling, K.; Kaufman, H.L. Final analyses of OPTiM: A randomized phase III trial of talimogene laherparepvec versus granulo-cyte-macrophage colony-stimulating factor in unresectable stage III-IV melanoma. J. ImmunoTherapy Cancer 2019, 7, 145. [Google Scholar] [CrossRef]
- E Müller, L.M.; Migneco, G.; Scott, G.B.; Down, J.; King, S.; Askar, B.; Jennings, V.; Oyajobi, B.; Scott, K.; West, E.; et al. Reovirus-induced cell-mediated immunity for the treatment of multiple myeloma within the resistant bone marrow niche. J. Immunother. Cancer 2021, 9, e001803. [Google Scholar] [CrossRef]
- Cassady, K.A.; Gross, M.; Roizman, B. The Second-Site Mutation in the Herpes Simplex Virus Recombinants Lacking the γ134.5 Genes Precludes Shutoff of Protein Synthesis by Blocking the Phosphorylation of eIF-2α. J. Virol. 1998, 72, 7005–7011. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-S.; Dzojic, H.; Nilsson, B.; Tötterman, T.H.; Essand, M. An oncolytic conditionally replicating adenovirus for hormone-dependent and hormone-independent prostate cancer. Cancer Gene Ther. 2006, 13, 13–20. [Google Scholar] [CrossRef]
- He, B.; Gross, M.; Roizman, B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 843–848. [Google Scholar] [CrossRef]
- Brown, S.M.; MacLean, A.R.; A McKie, E.; Harland, J. The herpes simplex virus virulence factor ICP34.5 and the cellular protein MyD116 complex with proliferating cell nuclear antigen through the 63-amino-acid domain conserved in ICP34.5, MyD116, and GADD34. J. Virol. 1997, 71, 9442–9449. [Google Scholar] [CrossRef]
- Brown, S.M.; Harland, J.; MacLean, A.R.; Podlech, J.; Clements, J.B. Cell type and cell state determine differential in vitro growth of non-neurovirulent ICP34.5-negative herpes simplex virus types 1 and 2. J. Gen. Virol. 1994, 75, 2367–2377. [Google Scholar] [CrossRef]
- Detta, A.; Harland, J.; Hanif, I.; Brown, S.M.; Cruickshank, G. Proliferative activity and in vivo replication of HSV1716 in human metastatic brain tumours. J. Gene Med. 2003, 5, 681–689. [Google Scholar] [CrossRef]
- Farassati, F.; Yang, A.-D.; Lee, P.W.K. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nature 2001, 3, 745–750. [Google Scholar] [CrossRef]
- Rampling, R.; Cruickshank, G.; Papanastassiou, V.; Nicoll, J.; Hadley, D.; Brennan, D.; Petty, R.; MacLean, A.; Harland, J.; McKie, E.; et al. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther. 2000, 7, 859–866. [Google Scholar] [CrossRef]
- Papanastassiou, V.; Rampling, R.; Fraser, M.; Petty, R.; Hadley, D.; Nicoll, J.; Harland, J.; Mabbs, R.; Brown, M. The potential for efficacy of the modified (ICP 34.5-) herpes simplex virus HSV1716 following intra-tumoural injection into malignant glioma: A proof of principle study. Gene Ther. 2002, 9, 398–406. [Google Scholar] [CrossRef]
- Harland, J.; Papanastassiou, V.; Brown, S.M. HSV1716 persistence in primary human glioma cells in vitro. Gene Ther. 2002, 9, 1194–1198. [Google Scholar] [CrossRef]
- Lasner, T.M.; Kesari, S.; Brown, S.M.; Lee, V.M.-Y.; Fraser, N.W.; Trojanowski, J.Q. Therapy of a murine model of pediatric brain tumors using a herpes simplex virus type-1 ICP34.5 mutant and demonstration of viral replication within the CNS. J. Neuropathol. Exp. Neurol. 1996, 55, 1259–1269. [Google Scholar] [CrossRef]
- Randazzo, B.P.; Bhat, M.G.; Kesari, S.; Fraser, N.W.; Brown, S.M. Treatment of experimental subcutaneous human mel-anoma with a replication-restricted herpes simplex virus mutant. J. Investig. Dermatol. 1997, 108, 933–937. [Google Scholar] [CrossRef]
- MacKie, R.M.; Stewart, B.; Brown, S.M. Intralesional injection of herpes simplex virus 1716 in metastatic melanoma. Lancet 2001, 357, 525–526. [Google Scholar] [CrossRef]
- Oku, M.; Ishino, R.; Uchida, S.; Imataki, O.; Sugimoto, N.; Todo, T.; Kadowaki, N. Oncolytic herpes simplex virus type 1 (HSV-1) in combination with lenalidomide for plasma cell neoplasms. Br. J. Haematol. 2021, 192, 343–353. [Google Scholar] [CrossRef]
- Ghose, J.; Dona, A.; Murtadha, M.; Gunes, E.G.; Caserta, E.; Yoo, J.Y.; Russell, L.; Jaime-Ramirez, A.C.; Barwick, B.G.; Gupta, V.A.; et al. Oncolytic herpes simplex virus infects myeloma cells in vitro and in vivo. Mol. Ther.-Oncolytics 2021, 20, 519–531. [Google Scholar] [CrossRef]
- Lawson, M.A.; Paton-Hough, J.M.; Evans, H.R.; Walker, R.E.; Harris, W.; Ratnabalan, D.; Snowden, J.A.; Chantry, A. NOD/SCID-GAMMA mice are an ideal strain to assess the efficacy of therapeutic agents used in the treatment of myeloma bone disease. PLoS ONE 2015, 10, e0119546. [Google Scholar] [CrossRef]
- Muthana, M.; Giannoudis, A.; Scott, S.D.; Fang, H.-Y.; Coffelt, S.B.; Morrow, F.J.; Murdoch, C.; Burton, J.; Cross, N.; Burke, B.; et al. Use of macrophages to target therapeutic adenovirus to human prostate tumors. Cancer Res. 2011, 71, 1805–1815. [Google Scholar] [CrossRef]
- Muthana, M.; Rodrigues, S.; Chen, Y.-Y.; Welford, A.; Hughes, R.; Tazzyman, S.; Essand, M.; Morrow, F.; Lewis, C.E. Macrophage delivery of an oncolytic virus abolishes tumor regrowth and metastasis after chemotherapy or irradiation. Cancer Res 2013, 73, 490–495. [Google Scholar] [CrossRef]
- Green, A.C.; Lath, D.; Hudson, K.; Walkley, B.; Down, J.M.; Owen, R.; Evans, H.; Paton-Hough, J.; Reilly, G.; Lawson, M.; et al. TGFβ Inhibition Stimulates Collagen Maturation to Enhance Bone Repair and Fracture Resistance in a Murine Myeloma Model. J. Bone Miner. Res. 2019, 34, 2311–2326. [Google Scholar] [CrossRef]
- Bouxsein, M.L.; Boyd, S.K.; Christiansen, B.A.; Guldberg, R.E.; Jepsen, K.J.; Müller, R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010, 25, 1468–1486. [Google Scholar] [CrossRef]
- Dempster, D.W.; Compston, J.E.; Drezner, M.K.; Glorieux, F.H.; Kanis, J.A.; Malluche, H.; Meunier, P.J.; Ott, S.M.; Recker, R.R.; Parfitt, A.M. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2013, 28, 2–17. [Google Scholar] [CrossRef]
- Paton-Hough, J.; Chantry, A.; Lawson, M. A review of current murine models of multiple myeloma used to assess the efficacy of therapeutic agents on tumour growth and bone disease. Bone 2015, 77, 57–68. [Google Scholar] [CrossRef]
- Mace, A.T.M.; Ganly, I.; Soutar, D.S.; Brown, S.M. Potential for efficacy of the oncolytic Herpes simplex virus 1716 in patients with oral squamous cell carcinoma. Head Neck 2008, 30, 1045–1051. [Google Scholar] [CrossRef]
- Mace, A.; Harrow, S.; Ganly, I.; Brown, S. Cytotoxic effects of the oncolytic herpes simplex virus HSV1716 alone and in combination with cisplatin in head and neck squamous cell carcinoma. Acta Oto-Laryngologica 2007, 127, 880–887. [Google Scholar] [CrossRef]
- Conner, J.; Braidwood, L.; Brown, S.M. A strategy for systemic delivery of the oncolytic herpes virus HSV1716: Redirected tropism by antibody-binding sites incorporated on the virion surface as a glycoprotein D fusion protein. Gene Ther. 2008, 15, 1579–1592. [Google Scholar] [CrossRef]
- Benencia, F.; Courreges, M.C.; Conejo-García, J.R.; Buckanovich, R.J.; Zhang, L.; Carroll, R.H.; Morgan, M.A.; Coukos, G. Oncolytic HSV exerts direct antiangiogenic activity in ovarian carcinoma. Hum. Gene Ther. 2005, 16, 765–778. [Google Scholar] [CrossRef]
- Braidwood, L.; Learmonth, K.; Graham, A.; Conner, J. Potent efficacy signals from systemically administered oncolytic herpes simplex virus (HSV1716) in hepatocellular carcinoma xenograft models. J. Hepatocell. Carcinoma 2014, 1, 149–161. [Google Scholar] [CrossRef]
- Boehmer, P.E.; Lehman, I.R. Herpes simplex virus type 1 ICP8: Helix-destabilizing properties. J. Virol. 1993, 67, 711–715. [Google Scholar] [CrossRef]
- Harrow, S.; Papanastassiou, V.; Harland, J.; Mabbs, R.; Petty, R.; Fraser, M.; Hadley, D.; Patterson, J.; Brown, S.M.; Rampling, R. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: Safety data and long-term survival. Gene Ther. 2004, 11, 1648–1658. [Google Scholar] [CrossRef]
- Zlei, M.; Egert, S.; Wider, D.; Ihorst, G.; Wäsch, R.; Engelhardt, M. Characterization of in vitro growth of multiple myeloma cells. Exp. Hematol. 2007, 35, 1550–1561. [Google Scholar] [CrossRef]
- Huang, Y.H.; Almowaled, M.; Li, J.; Venner, C.; Sandhu, I.; Peters, A.; Lavasanifar, A.; Lai, R. Three-Dimensional Reconstructed Bone Marrow Matrix Culture Improves the Viability of Primary Myeloma Cells In-Vitro via a STAT3-Dependent Mechanism. Curr. Issues Mol. Biol. 2021, 43, 313–323. [Google Scholar] [CrossRef]
- Chaidos, A.; Barnes, C.; Cowan, G.; May, P.; Melo, V.; Hatjiharissi, E.; Papaioannou, M.; Harrington, H.; Doolittle, H.; Terpos, E.; et al. Clinical drug resistance linked to interconvertible phenotypic and functional states of tumor-propagating cells in multiple myeloma. Blood 2013, 121, 318–328. [Google Scholar] [CrossRef]
- Matsui, W.; Wang, Q.; Barber, J.P.; Brennan, S.; Smith, B.D.; Borrello, I.; McNiece, I.; Lin, L.; Ambinder, R.F.; Peacock, C.; et al. Clonogenic Multiple Myeloma Progenitors, Stem Cell Properties, and Drug Resistance. Cancer Res 2008, 68, 190–197. [Google Scholar] [CrossRef]
- Coukos, G.; Makrigiannakis, A.; Kang, E.H.; Rubin, S.C.; Albelda, S.M.; Molnar-Kimber, K.L. Oncolytic Herpes Simplex Virus-1 Lacking ICP34.5 Induces p53-independent Death and Is Efficacious against Chemotherapy-resistant Ovarian Cancer. Clin. Cancer Res. 2000, 6, 3342–3353. [Google Scholar]
- Streby, K.A.; Currier, M.A.; Triplet, M.; Ott, K.; Dishman, D.J.; Vaughan, M.R.; Ranalli, M.A.; Setty, B.; Skeens, M.A.; Whiteside, S.; et al. First-in-Human Intravenous Seprehvir in Young Cancer Patients: A Phase 1 Clinical Trial. Mol. Ther. 2019, 27, 1930–1938. [Google Scholar] [CrossRef]
- Zhu, W.; Wei, L.; Zhang, H.; Chen, J.; Qin, X. Oncolytic adenovirus armed with IL-24 Inhibits the growth of breast cancer in vitro and in vivo. J. Exp. Clin. Cancer Res. 2012, 31, 1–10. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tazzyman, S.; Stewart, G.R.; Yeomans, J.; Linford, A.; Lath, D.; Conner, J.; Muthana, M.; Chantry, A.D.; Lawson, M.A. HSV1716 Prevents Myeloma Cell Regrowth When Combined with Bortezomib In Vitro and Significantly Reduces Systemic Tumor Growth in Mouse Models. Viruses 2023, 15, 603. https://doi.org/10.3390/v15030603
Tazzyman S, Stewart GR, Yeomans J, Linford A, Lath D, Conner J, Muthana M, Chantry AD, Lawson MA. HSV1716 Prevents Myeloma Cell Regrowth When Combined with Bortezomib In Vitro and Significantly Reduces Systemic Tumor Growth in Mouse Models. Viruses. 2023; 15(3):603. https://doi.org/10.3390/v15030603
Chicago/Turabian StyleTazzyman, Simon, Georgia R. Stewart, James Yeomans, Adam Linford, Darren Lath, Joe Conner, Munitta Muthana, Andrew D. Chantry, and Michelle A. Lawson. 2023. "HSV1716 Prevents Myeloma Cell Regrowth When Combined with Bortezomib In Vitro and Significantly Reduces Systemic Tumor Growth in Mouse Models" Viruses 15, no. 3: 603. https://doi.org/10.3390/v15030603
APA StyleTazzyman, S., Stewart, G. R., Yeomans, J., Linford, A., Lath, D., Conner, J., Muthana, M., Chantry, A. D., & Lawson, M. A. (2023). HSV1716 Prevents Myeloma Cell Regrowth When Combined with Bortezomib In Vitro and Significantly Reduces Systemic Tumor Growth in Mouse Models. Viruses, 15(3), 603. https://doi.org/10.3390/v15030603






