The Interplay among Glucocorticoid Therapy, Platelet-Activating Factor and Endocannabinoid Release Influences the Inflammatory Response to COVID-19
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Plasma Isolation
2.3. Sample Preparation and Extraction of eCBs and PAF
2.4. Lipid Quantification by LC-MS/MS
2.5. Protein Mediator’s Measurements
2.6. RNA Extraction and Quantification
2.7. Data Transcriptome
2.8. Bioinformatics Analysis of Transcriptome Data
2.9. Statistical Analysis
3. Results
3.1. Participants Demographic Data and Clinical Characteristics
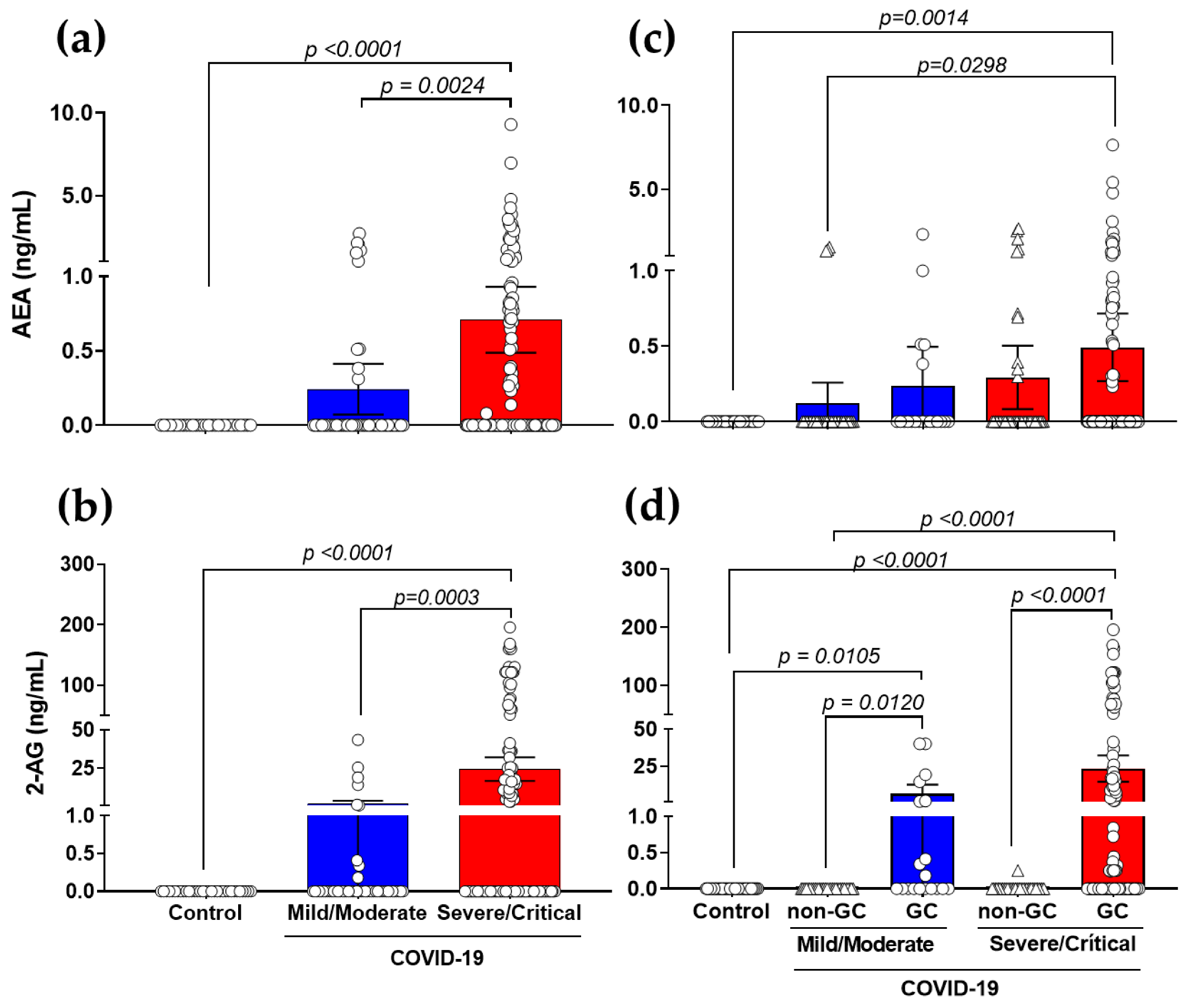
3.2. Production of AEA and 2-AG Is Elevated in Severe COVID-19 Patients with a Link to GCs-Treatment
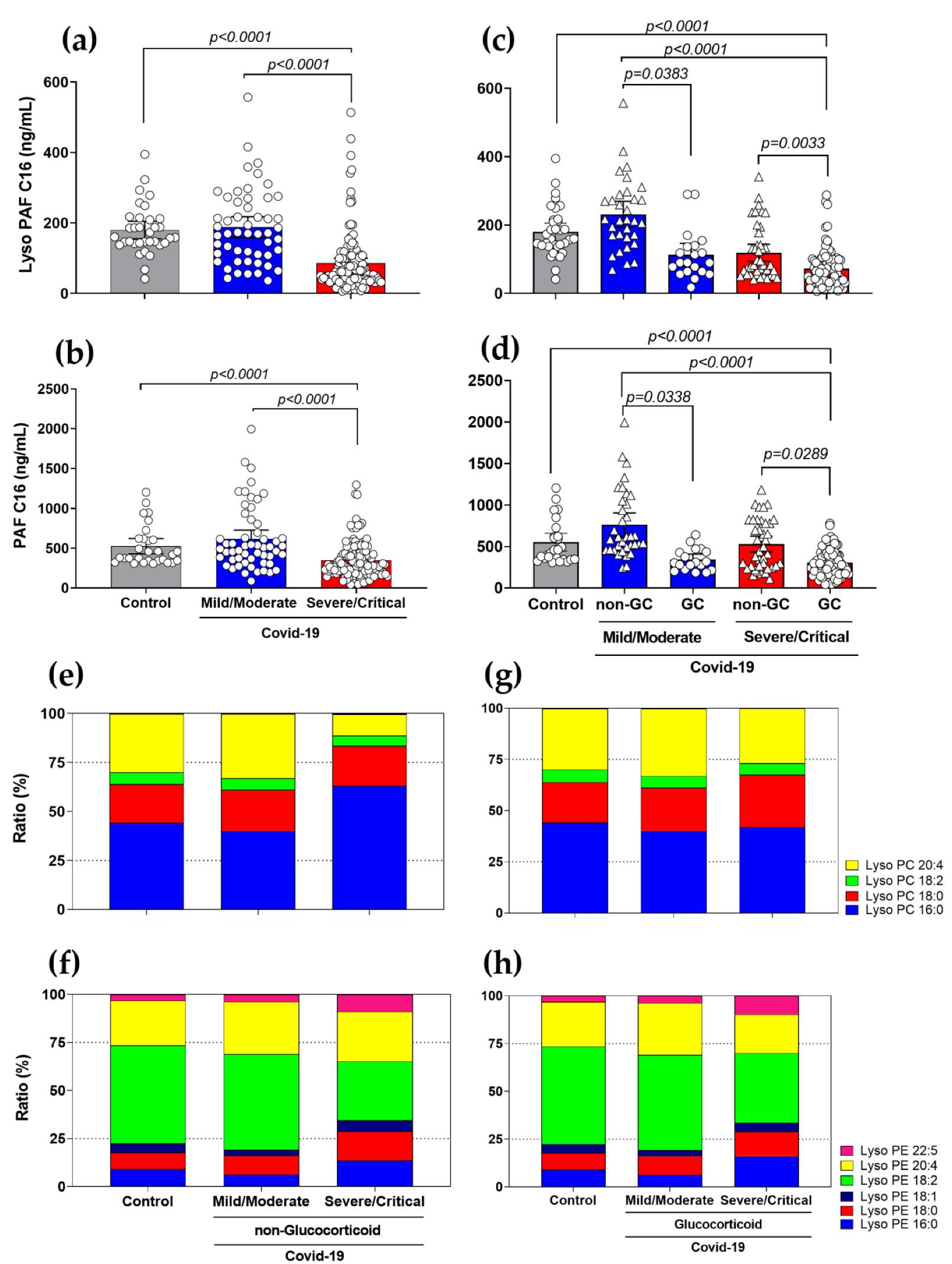
3.3. Diminished Levels of Lyso-PAF and PAF, and Different Ratios in Lyso-PC and Lyso-PE Species in Severe COVID-19 Patients with the Use of GCs
3.4. Modified Genetic Expression Profile of Metabolic Enzymes Implicated in the eCBs and PAF Pathway in COVID-19 Patients Treated with GCs
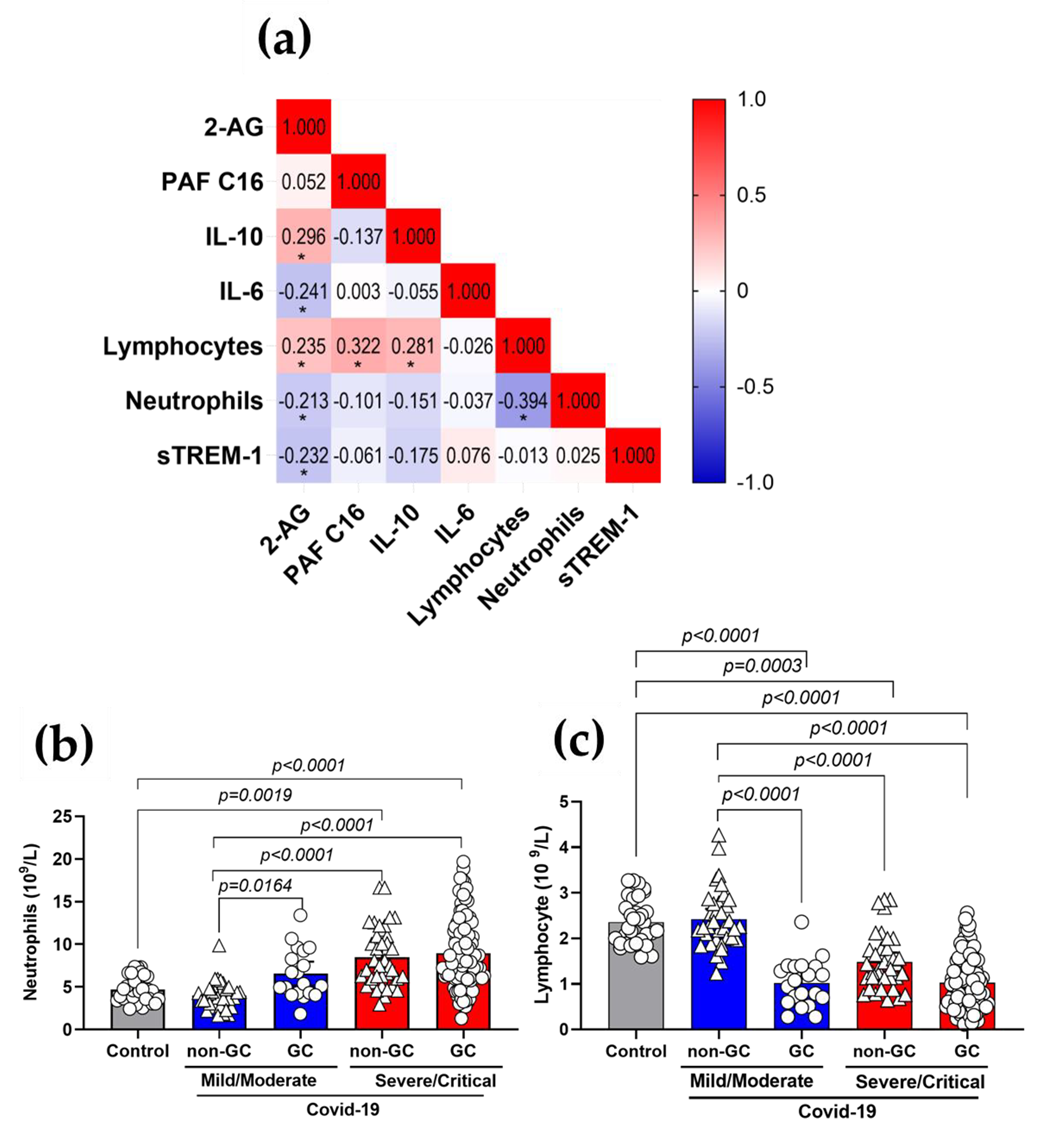
3.5. Plasma 2-AG and PAF Levels Correlated with Inflammatory Markers of COVID-19
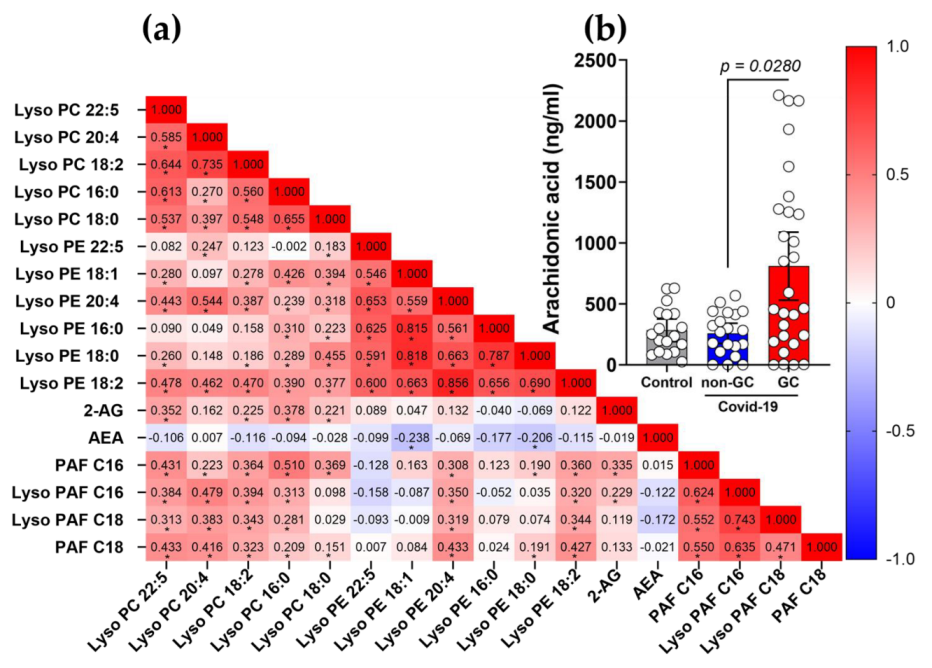
3.6. The Use of GCs Alters the Metabolism and Production of Lipid Mediators during COVID-19
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA—J. Am. Med. Assoc. 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef]
- Chiurchiù, V.; Leuti, A.; Maccarrone, M. Bioactive Lipids and Chronic Inflammation: Managing the Fire Within. Front. Immunol. 2018, 9, 38. [Google Scholar] [CrossRef]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef]
- Theoharides, T.C.; Antonopoulou, S.; Demopoulos, C.A. Coronavirus 2019, Microthromboses, and Platelet Activating Factor. Clin. Ther. 2020, 42, 1850–1852. [Google Scholar] [CrossRef]
- Lordan, R.; Tsoupras, A.; Zabetakis, I.; Demopoulos, C.A. Forty Years Since the Structural Elucidation of Platelet-Activating Factor (PAF): Historical, Current, and Future Research Perspectives. Molecules 2019, 24, 4414. [Google Scholar] [CrossRef]
- Detopoulou, P.; Nomikos, T.; Fragopoulou, E.; Chrysohoou, C.; Antonopoulou, S. Platelet Activating Factor in Heart Failure: Potential Role in Disease Progression and Novel Target for Therapy. Curr. Hear. Fail. Rep. 2013, 10, 122–129. [Google Scholar] [CrossRef]
- Kelesidis, T.; Papakonstantinou, V.; Detopoulou, P.; Fragopoulou, E.; Chini, M.; Lazanas, M.C.; Antonopoulou, S. The Role of Platelet-Activating Factor in Chronic Inflammation, Immune Activation, and Comorbidities Associated with HIV Infection. Aids Rev. 2015, 17, 191–201. [Google Scholar]
- Nagoor Meeran, M.F.; Sharma, C.; Goyal, S.N.; Kumar, S.; Ojha, S. CB2 Receptor-Selective Agonists as Candidates for Targeting Infection, Inflammation, and Immunity in SARS-CoV-2 Infections. Drug Dev. Res. 2021, 82, 7–11. [Google Scholar] [CrossRef]
- He, Q.; Xiao, F.; Yuan, Q.; Zhang, J.; Zhan, J.; Zhang, Z. Cannabinoid receptor 2: A potential novel therapeutic target for sepsis? Acta Clin. Belg. 2018, 74, 70–74. [Google Scholar] [CrossRef]
- Cinar, R.; Iyer, M.R.; Kunos, G. Dual inhibition of CB 1 receptors and iNOS, as a potential novel approach to the pharmacological management of acute and long COVID-19. Br. J. Pharmacol. 2021, 179, 2121–2127. [Google Scholar] [CrossRef]
- Theken, K.N.; FitzGerald, G.A. Bioactive lipids in antiviral immunity. Science 2021, 371, 237–238. [Google Scholar] [CrossRef]
- Monson, E.A.; Trenerry, A.M.; Laws, J.L.; Mackenzie, J.M.; Helbig, K.J. Lipid droplets and lipid mediators in viral infection and immunity. FEMS Microbiol. Rev. 2021, 45, fuaa066. [Google Scholar] [CrossRef]
- Ho, W.S.; Zhang, R.; Tan, Y.L.; Chai, C.L.L. COVID-19 and the Promise of Small Molecule Therapeutics: Are There Lessons to Be Learnt? Pharmacol. Res. 2022, 179, 106201. [Google Scholar] [CrossRef]
- Huang, Q.; Wu, X.; Zheng, X.; Luo, S.; Xu, S.; Weng, J. Targeting inflammation and cytokine storm in COVID-19. Pharmacol. Res. 2020, 159, 105051. [Google Scholar] [CrossRef]
- Kojima, Y.; Nakakubo, S.; Kamada, K.; Yamashita, Y.; Takei, N.; Nakamura, J.; Matsumoto, M.; Horii, H.; Sato, K.; Shima, H.; et al. Combination therapy with remdesivir and immunomodulators improves respiratory status in COVID-19: A retrospective study. J. Med. Virol. 2022, 94, 5702–5712. [Google Scholar] [CrossRef]
- Ngo, D.Q.; Hamid, K.; Rana, H.; Cardinale, M.; Frenia, D.; Ghani, N.; Redel, H. A Retrospective Study of Dexamethasone, Remdesivir, and Baricitinib in Severe COVID-19. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 1–8. [Google Scholar] [CrossRef]
- Popp, M.; Stegemann, M.; Riemer, M.; Metzendorf, M.I.; Romero, C.S.; Mikolajewska, A.; Kranke, P.; Meybohm, P.; Skoetz, N.; Weibel, S. Antibiotics for the Treatment of COVID-19. Cochrane Database Syst. Rev. 2021. Issue 10. Art. No.: CD015025.. [Google Scholar] [CrossRef]
- Shi, C.; Wang, C.; Wang, H.; Yang, C.; Cai, F.; Zeng, F.; Cheng, F.; Liu, Y.; Zhou, T.; Deng, B.; et al. The Potential of Low Molecular Weight Heparin to Mitigate Cytokine Storm in Severe COVID-19 Patients: A Retrospective Cohort Study. Clin. Transl. Sci. 2020, 13, 1087–1095. [Google Scholar] [CrossRef]
- Vandewalle, J.; Luypaert, A.; de Bosscher, K.; Libert, C. Therapeutic Mechanisms of Glucocorticoids. Trends Endocrinol. Metab. 2018, 29, 42–54. [Google Scholar] [CrossRef]
- Riccardi, C.; Bruscoli, S.; Migliorati, G. Molecular mechanisms of immunomodulatory activity of glucocorticoids. Pharmacol. Res. 2002, 45, 361–368. [Google Scholar] [CrossRef]
- Sahanic, S.; Löffler-Ragg, J.; Tymoszuk, P.; Hilbe, R.; Demetz, E.; Masanetz, R.K.; Theurl, M.; Holfeld, J.; Gollmann-Tepeköylü, C.; Tzankov, A.; et al. The Role of Innate Immunity and Bioactive Lipid Mediators in COVID-19 and Influenza. Front. Physiol. 2021, 12, 688946. [Google Scholar] [CrossRef]
- Sarzani, R.; Spannella, F.; Giulietti, F.; di Pentima, C.; Giordano, P.; Giacometti, A. Possible Harm from Glucocorticoid Drugs Misuse in the Early Phase of SARS-CoV-2 Infection: A Narrative Review of the Evidence. Intern. Emerg. Med. 2022, 17, 329–338. [Google Scholar] [CrossRef]
- Wei, P.-F. Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7). Chin. Med. J. 2020, 133, 1087–1095. [Google Scholar] [CrossRef]
- Wan, S.; Xiang, Y.; Fang, W.; Zheng, Y.; Li, B.; Hu, Y.; Lang, C.; Huang, D.; Sun, Q.; Xiong, Y.; et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J. Med. Virol. 2020, 92, 797–806. [Google Scholar] [CrossRef]
- The RECOVERY Collaborative Group. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Godzien, J.; Ciborowski, M.; Martínez-Alcázar, M.P.; Samczuk, P.; Kretowski, A.; Barbas, C. Rapid and Reliable Identification of Phospholipids for Untargeted Metabolomics with LC–ESI–QTOF–MS/MS. J. Proteome Res. 2015, 14, 3204–3216. [Google Scholar] [CrossRef]
- R Core Team. R a Language and Environment for Statistical Computing: Reference Index; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. [Google Scholar]
- Allaire, J.J. RStudio: Integrated Development Environment for R; RStudio Integrated Development Environment for R: Boston, MA, USA, 2019. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Benjaminit, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Epskamp, S.; Cramer, A.O.J.; Waldorp, L.J.; Schmittmann, V.D.; Borsboom, D. Qgraph: Network Visualizations of Relationships in Psychometric Data. J. Stat. Softw. 2012, 48, 1–18. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Gallo, C.G.; Fiorino, S.; Posabella, G.; Antonacci, D.; Tropeano, A.; Pausini, E.; Pausini, C.; Guarniero, T.; Hong, W.; Giampieri, E.; et al. The Function of Specialized Pro-Resolving Endogenous Lipid Mediators, Vitamins, and Other Micronutrients in the Control of the Inflammatory Processes: Possible Role in Patients with SARS-CoV-2 Related Infection. Prostaglandins Other Lipid Mediat. 2022, 159, 106619. [Google Scholar] [CrossRef]
- Casari, I.; Manfredi, M.; Metharom, P.; Falasca, M. Dissecting Lipid Metabolism Alterations in SARS-CoV-2. Prog. Lipid Res. 2021, 82, 101092. [Google Scholar] [CrossRef]
- Devane, W.A.; Hanus, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef]
- Sugiura, T.; Kondo, S.; Sukagawa, A.; Nakane, S.; Shinoda, A.; Itoh, K.; Yamashita, A.; Waku, K. 2-Arachidonoylgylcerol: A Possible Endogenous Cannabinoid Receptor Ligand in Brain. Biochem. Biophys. Res. Commun. 1995, 215, 89–97. [Google Scholar] [CrossRef]
- Cascio, M.G.; Marini, P. Biosynthesis and Fate of Endocannabinoids. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 231. [Google Scholar]
- Gui, H.; Tong, Q.; Qu, W.; Mao, C.-M.; Dai, S.-M. The endocannabinoid system and its therapeutic implications in rheumatoid arthritis. Int. Immunopharmacol. 2015, 26, 86–91. [Google Scholar] [CrossRef]
- Ligresti, A.; De Petrocellis, L.; Di Marzo, V. From phytocannabinoids to cannabinoid receptors and endocannabinoids: Pleiotropic physiological and pathological roles through complex pharmacology. Physiol. Rev. 2016, 96, 1593–1659. [Google Scholar] [CrossRef]
- Cabral, G.; Griffin-Thomas, G.A.C.A.L. Cannabinoids as Therapeutic Agents for Ablating Neuroinflammatory Disease. Endocr. Metab. Immune Disord. Drug Targets 2008, 8, 159–172. [Google Scholar] [CrossRef]
- Herkenham, M.; Lynn, A.; Johnson, M.R.; Melvin, L.; de Costa, B.; Rice, K. Characterization and localization of cannabinoid receptors in rat brain: A quantitative in vitro autoradiographic study. J. Neurosci. 1991, 11, 563–583. [Google Scholar] [CrossRef]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef]
- Cabral, G.A.; Griffin-Thomas, L. Emerging Role of the CB2 Cannabinoid Receptor in Immune Regulation and Therapeutic Prospects. Expert Rev. Mol. Med. 2009, 11, e3. [Google Scholar] [CrossRef]
- Lucaciu, O.; Aghiorghiesei, O.; Petrescu, N.B.; Mirica, I.C.; Benea, H.R.C.; Apostu, D. In quest of a new therapeutic approach in COVID-19: The endocannabinoid system. Drug Metab. Rev. 2021, 53, 478–490. [Google Scholar] [CrossRef]
- Pandey, R.; Mousawy, K.; Nagarkatti, M.; Nagarkatti, P. Endocannabinoids and immune regulation. Pharmacol. Res. 2009, 60, 85–92. [Google Scholar] [CrossRef]
- Nichols, J.M.; Kaplan, B.L.F. Immune Responses Regulated by Cannabidiol. Cannabis Cannabinoid Res. 2020, 5, 12–31. [Google Scholar] [CrossRef]
- Mackie, K. CANNABINOID RECEPTORS AS THERAPEUTIC TARGETS. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 101–122. [Google Scholar] [CrossRef]
- Nomura, D.K.; Morrison, B.E.; Blankman, J.L.; Long, J.Z.; Kinsey, S.G.; Marcondes, M.C.G.; Ward, A.M.; Hahn, Y.K.; Lichtman, A.H.; Conti, B.; et al. Endocannabinoid Hydrolysis Generates Brain Prostaglandins That Promote Neuroinflammation. Science 2011, 334, 809–813. [Google Scholar] [CrossRef]
- Sultan, M.; Alghetaa, H.; Mohammed, A.; Abdulla, O.A.; Wisniewski, P.J.; Singh, N.; Nagarkatti, P.; Nagarkatti, M. The Endocannabinoid Anandamide Attenuates Acute Respiratory Distress Syndrome by Downregulating miRNA that Target Inflammatory Pathways. Front. Pharmacol. 2021, 12, 644281. [Google Scholar] [CrossRef]
- Sultan, M.; Wilson, K.; Abdulla, O.A.; Busbee, P.B.; Hall, A.; Carter, T.; Singh, N.; Chatterjee, S.; Nagarkatti, P.; Nagarkatti, M. Endocannabinoid Anandamide Attenuates Acute Respiratory Distress Syndrome through Modulation of Microbiome in the Gut-Lung Axis. Cells 2021, 10, 3305. [Google Scholar] [CrossRef]
- Krishnan, G.; Chatterjee, N. Endocannabinoids affect innate immunity of Muller glia during HIV-1 Tat cytotoxicity. Mol. Cell. Neurosci. 2014, 59, 10–23. [Google Scholar] [CrossRef]
- Xie, J.; Wu, W.; Li, S.; Hu, Y.; Hu, M.; Li, J.; Yang, Y.; Huang, T.; Zheng, K.; Wang, Y.; et al. Clinical characteristics and outcomes of critically ill patients with novel coronavirus infectious disease (COVID-19) in China: A retrospective multicenter study. Intensiv. Care Med. 2020, 46, 1863–1872. [Google Scholar] [CrossRef]
- da Silva-Neto, P.V.; de Carvalho, J.C.S.; Pimentel, V.E.; Pérez, M.M.; Toro, D.M.; Fraga-Silva, T.F.C.; Fuzo, C.A.; Oliveira, C.N.S.; Rodrigues, L.C.; Argolo, J.G.M.; et al. sTREM-1 Predicts Disease Severity and Mortality in COVID-19 Patients: Involvement of Peripheral Blood Leukocytes and MMP-8 Activity. Viruses 2021, 13, 2521. [Google Scholar] [CrossRef]
- Prescott, S.M.; Zimmerman, G.A.; Stafforini, D.M.; McIntyre, T.M. Platelet-Activating Factor and Related Lipid Mediators. Annu. Rev. Biochem. 2000, 69, 419–445. [Google Scholar] [CrossRef]
- Zimmerman, G.A.; McIntyre, T.M.; Prescott, S.M.; Stafforini, D.M. The platelet-activating factor signaling system and its regulators in syndromes of inflammation and thrombosis. Crit. Care Med. 2002, 30, S294–S301. [Google Scholar] [CrossRef]
- Ivanov, A.I.; Patel, S.; Kulchitsky, V.A.; Romanovsky, A.A. Platelet-Activating Factor: A Previously Unrecognized Mediator of Fever. J. Physiol. 2003, 553, 221–228. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Sawa, H.; Yasuda, H. Mechanism of increased angiotensin-converting enzyme activity stimulated by platelet-activating factor. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 1990, 1052, 503–508. [Google Scholar] [CrossRef]
- Datta, P.K.; Liu, F.; Fischer, T.; Rappaport, J.; Qin, X. SARS-CoV-2 Pandemic and Research Gaps: Understanding SARS-CoV-2 Interaction with the ACE2 Receptor and Implications for Therapy. Theranostics 2020, 10, 7448. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Nookala, V. Biochemistry, Platelet Activating Factor. Clin. Rev. Allergy 2020, 1, 353–367. [Google Scholar]
- Demopoulos, C.; Antonopoulou, S.; Theoharides, T.C. COVID-19, Microthromboses, Inflammation, and Platelet Activating Factor. BioFactors 2020, 46, 927–933. [Google Scholar] [CrossRef]
- Benmansour, N.C.; Carvelli, J.; Vivier, E. Complement cascade in severe forms of COVID-19: Recent advances in therapy. Eur. J. Immunol. 2021, 51, 1652–1659. [Google Scholar] [CrossRef]
- Soy, M.; Keser, G.; Atagündüz, P.; Tabak, F.; Atagündüz, I.; Kayhan, S. Cytokine storm in COVID-19: Pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020, 39, 2085–2094. [Google Scholar] [CrossRef]
- Wong, R.S.Y. Inflammation in COVID-19: From pathogenesis to treatment. Int. J. Clin. Exp. Pathol. 2021, 14, 831–844. [Google Scholar]
- Caiazzo, E.; Rezig, A.O.; Bruzzese, D.; Ialenti, A.; Cicala, C.; Cleland, J.G.; Guzik, T.J.; Maffia, P.; Pellicori, P. Systemic administration of glucocorticoids, cardiovascular complications and mortality in patients hospitalised with COVID-19, SARS, MERS or influenza: A systematic review and meta-analysis of randomised trials. Pharmacol. Res. 2021, 176, 106053. [Google Scholar] [CrossRef]
- Oba, Y. The Use of Corticosteroids in SARS. New Engl. J. Med. 2003, 348, 2034–2035. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, Y.; Zhang, Z.-L.; Liu, Y.-X.; Le, K.-J.; Cui, M.; Yu, Y.-T.; Gu, Z.-C.; Gao, Y.; Lin, H.-W. Efficacy and safety of current therapeutic options for COVID-19—Lessons to be learnt from SARS and MERS epidemic: A systematic review and meta-analysis. Pharmacol. Res. 2020, 157, 104872. [Google Scholar] [CrossRef]
- Inagaki, N.; Miura, T.; Nakajima, T.; Yoshida, K.; Nagai, H.; Koda, A. Studies on the Anti-allergic Mechanism of Glucocorticoids in Mice. J. Pharm. -Dyn. 1992, 15, 581–587. [Google Scholar] [CrossRef]
- Löwenberg, M.; Verhaar, A.P.; Bilderbeek, J.; van Marle, J.; Buttgereit, F.; Peppelenbosch, M.P.; Van Deventer, S.J.; Hommes, D.W. Glucocorticoids cause rapid dissociation of a T-cell-receptor-associated protein complex containing LCK and FYN. EMBO Rep. 2006, 7, 1023–1029. [Google Scholar] [CrossRef]
- Malcher-Lopes, R.; Buzzi, M. Chapter 11 Glucocorticoid-Regulated Crosstalk Between Arachidonic Acid and Endocannabinoid Biochemical Pathways Coordinates Cognitive-, Neuroimmune-, and Energy Homeostasis-Related Adaptations to Stress. Vitam. Horm. 2009, 81, 263–313. [Google Scholar]
- Jang, B.-C.; Lim, K.-J.; Suh, M.-H.; Park, J.-G.; Suh, S.-I. Dexamethasone suppresses interleukin-1Î2-induced human Î2-defensin 2 mRNA expression: Involvement of p38 MAPK, JNK, MKP-1, and NF-κB transcriptional factor in A549 cells. FEMS Immunol. Med. Microbiol. 2007, 51, 171–184. [Google Scholar] [CrossRef]
- Chantong, B.; Kratschmar, D.V.; Nashev, L.G.; Balazs, Z.; Odermatt, A. Mineralocorticoid and glucocorticoid receptors differentially regulate NF-kappaB activity and pro-inflammatory cytokine production in murine BV-2 microglial cells. J. Neuroinflammation 2012, 9, 260. [Google Scholar] [CrossRef]
- Leslie, C.C. Properties and Regulation of Cytosolic Phospholipase A2. J. Biol. Chem. 1997, 272, 16709–16712. [Google Scholar] [CrossRef]
- Mitchell, J.A.; Belvisi, M.G.; Akarasereenont, P.; Robbins, R.A.; Kwon, O.-J.; Croxtall, J.J.; Barnes, P.J.; Vane, J.R. Induction of cyclo-oxygenase-2 by cytokines in human pulmonary epithelial cells: Regulation by dexamethasone. Br. J. Pharmacol. 1994, 113, 1008–1014. [Google Scholar] [CrossRef]
- Castillo, P.E.; Younts, T.J.; Chávez, A.E.; Hashimotodani, Y. Endocannabinoid Signaling and Synaptic Function. Neuron 2012, 76, 70–81. [Google Scholar] [CrossRef]
- Katona, I.; Freund, T.F. Endocannabinoid signaling as a synaptic circuit breaker in neurological disease. Nat. Med. 2008, 14, 923–930. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Harvey-White, J.; Huang, B.X.; Kim, H.-Y.; Luquet, S.; Palmiter, R.D.; Krystal, G.; Rai, R.; Mahadevan, A.; et al. Multiple pathways involved in the biosynthesis of anandamide. Neuropharmacology 2008, 54, 1–7. [Google Scholar] [CrossRef]
- Simon, G.M.; Cravatt, B.F. Anandamide Biosynthesis Catalyzed by the Phosphodiesterase GDE1 and Detection of Glycerophospho-N-acyl Ethanolamine Precursors in Mouse Brain. J. Biol. Chem. 2008, 283, 9341–9349. [Google Scholar] [CrossRef]
- Murataeva, N.; Straiker, A.; Mackie, K. Parsing the players: 2-arachidonoylglycerol synthesis and degradation in the CNS. Br. J. Pharmacol. 2014, 171, 1379–1391. [Google Scholar] [CrossRef]
- Pertwee, R.G. Endocannabinoids and Their Pharmacological Actions. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 231. [Google Scholar]
- Blankman, J.L.; Simon, G.M.; Cravatt, B.F. A Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol. 2007, 14, 1347–1356. [Google Scholar] [CrossRef]
- Marrs, W.R.; Blankman, J.L.; A Horne, E.; Thomazeau, A.; Lin, Y.H.; Coy, J.; Bodor, A.L.; Muccioli, G.; Hu, S.S.-J.; Woodruff, G.; et al. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat. Neurosci. 2010, 13, 951–957. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Endocannabinoid Oxygenation by Cyclooxygenases, Lipoxygenases, and Cytochromes P450: Cross-Talk between the Eicosanoid and Endocannabinoid Signaling Pathways. Chem. Rev. 2011, 111, 5899–5921. [Google Scholar] [CrossRef]
- Demopoulos, C.; Pinckard, R.; Hanahan, D. Platelet-activating factor. Evidence for 1-O-alkyl-2-acetyl-sn-glyceryl-3-phosphorylcholine as the active component (a new class of lipid chemical mediators). J. Biol. Chem. 1979, 254, 9355–9358. [Google Scholar] [CrossRef]
- Singh, P.; Singh, I.N.; Mondal, S.C.; Singh, L.; Garg, V.K. Platelet-activating factor (PAF)-antagonists of natural origin. Fitoterapia 2012, 84, 180–201. [Google Scholar] [CrossRef]
- Snyder, F. Platelet-activating factor and related acetylated lipids as potent biologically active cellular mediators. Am. J. Physiol. Physiol. 1990, 259, C697–C708. [Google Scholar] [CrossRef]
- Uemura, Y.; Lee, T.; Snyder, F. A coenzyme A-independent transacylase is linked to the formation of platelet-activating factor (PAF) by generating the lyso-PAF intermediate in the remodeling pathway. J. Biol. Chem. 1991, 266, 8268–8272. [Google Scholar] [CrossRef]
- Shindou, H.; Hishikawa, D.; Nakanishi, H.; Harayama, T.; Ishii, S.; Taguchi, R.; Shimizu, T. A Single Enzyme Catalyzes Both Platelet-Activating Factor Production and Membrane Biogenesis of Inflammatory Cells: Cloning and Characterization of Acetyl-CoA:Lyso-PAF Acetyltransferase. J. Biol. Chem. 2007, 282, 6532–6539. [Google Scholar] [CrossRef]
- Harayama, T.; Shindou, H.; Ogasawara, R.; Suwabe, A.; Shimizu, T. Identification of a Novel Noninflammatory Biosynthetic Pathway of Platelet-activating Factor*. J. Biol. Chem. 2008, 283, 11097–11106. [Google Scholar] [CrossRef]
- Chen, J.; Yang, L.; Foulks, J.M.; Weyrich, A.S.; Marathe, G.K.; McIntyre, T.M. Intracellular PAF catabolism by PAF acetylhydrolase counteracts continual PAF synthesis. J. Lipid Res. 2007, 48, 2365–2376. [Google Scholar] [CrossRef]
- Marathe, G.K.; Prescott, S.M.; A Zimmerman, G.; McIntyre, T.M. Oxidized LDL contains inflammatory PAF-like phospholipids. Trends Cardiovasc. Med. 2001, 11, 139–142. [Google Scholar] [CrossRef]
- Shamizadeh, S.; Brockow, K.; Ring, J. Rupatadine: Efficacy and safety of a non-sedating antihistamine with PAF-antagonist effects. Allergo J. Int. 2014, 23, 87–95. [Google Scholar] [CrossRef]
- Papakonstantinou, V.D.; Lagopati, N.; Tsilibary, E.C.; Demopoulos, C.A.; Philippopoulos, A.I. A Review on Platelet Activating Factor Inhibitors: Could a New Class of Potent Metal-Based Anti-Inflammatory Drugs Induce Anticancer Properties? Bioinorg. Chem. Appl. 2017, 2017, 6947034. [Google Scholar] [CrossRef]
- Peplow, P. Regulation of platelet-activating factor (PAF) activity in human diseases by phospholipase A2inhibitors, PAF acetylhydrolases, PAF receptor antagonists and free radical scavengers. Prostaglandins Leukot. Essent. Fat. Acids 1999, 61, 65–82. [Google Scholar] [CrossRef]
- Hariyanto, T.I.; Kurniawan, A. Dyslipidemia is associated with severe coronavirus disease 2019 (COVID-19) infection. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1463–1465. [Google Scholar] [CrossRef]
- Yanai, H. Metabolic Syndrome and COVID-19. Cardiol. Res. 2020, 11, 360–365. [Google Scholar] [CrossRef]
- Doaei, S.; Gholami, S.; Rastgoo, S.; Gholamalizadeh, M.; Bourbour, F.; Bagheri, S.E.; Samipoor, F.; Akbari, M.E.; Shadnoush, M.; Ghorat, F.; et al. The effect of omega-3 fatty acid supplementation on clinical and biochemical parameters of critically ill patients with COVID-19: A randomized clinical trial. J. Transl. Med. 2021, 19, 1–9. [Google Scholar] [CrossRef]
- Williams, D.M. Clinical Pharmacology of Corticosteroids. Respir. Care 2018, 63, 655–670. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Carvalho, J.C.S.; da Silva-Neto, P.V.; Toro, D.M.; Fuzo, C.A.; Nardini, V.; Pimentel, V.E.; Pérez, M.M.; Fraga-Silva, T.F.C.; Oliveira, C.N.S.; Degiovani, A.M.; et al. The Interplay among Glucocorticoid Therapy, Platelet-Activating Factor and Endocannabinoid Release Influences the Inflammatory Response to COVID-19. Viruses 2023, 15, 573. https://doi.org/10.3390/v15020573
de Carvalho JCS, da Silva-Neto PV, Toro DM, Fuzo CA, Nardini V, Pimentel VE, Pérez MM, Fraga-Silva TFC, Oliveira CNS, Degiovani AM, et al. The Interplay among Glucocorticoid Therapy, Platelet-Activating Factor and Endocannabinoid Release Influences the Inflammatory Response to COVID-19. Viruses. 2023; 15(2):573. https://doi.org/10.3390/v15020573
Chicago/Turabian Stylede Carvalho, Jonatan C. S., Pedro V. da Silva-Neto, Diana M. Toro, Carlos A. Fuzo, Viviani Nardini, Vinícius E. Pimentel, Malena M. Pérez, Thais F. C. Fraga-Silva, Camilla N. S. Oliveira, Augusto M. Degiovani, and et al. 2023. "The Interplay among Glucocorticoid Therapy, Platelet-Activating Factor and Endocannabinoid Release Influences the Inflammatory Response to COVID-19" Viruses 15, no. 2: 573. https://doi.org/10.3390/v15020573
APA Stylede Carvalho, J. C. S., da Silva-Neto, P. V., Toro, D. M., Fuzo, C. A., Nardini, V., Pimentel, V. E., Pérez, M. M., Fraga-Silva, T. F. C., Oliveira, C. N. S., Degiovani, A. M., Ostini, F. M., Feitosa, M. R., Parra, R. S., da Rocha, J. J. R., Feres, O., Vilar, F. C., Gaspar, G. G., Santos, I. K. F. M., Fernandes, A. P. M., ... on behalf of the ImmunoCovid Study Group. (2023). The Interplay among Glucocorticoid Therapy, Platelet-Activating Factor and Endocannabinoid Release Influences the Inflammatory Response to COVID-19. Viruses, 15(2), 573. https://doi.org/10.3390/v15020573










