Wildebeest-Derived Malignant Catarrhal Fever: A Bovine Peripheral T Cell Lymphoma Caused by Cross-Species Transmission of Alcelaphine Gammaherpesvirus 1
Abstract
1. Wildebeest-Derived Malignant Catarrhal Fever, a Fatal Lymphoproliferative Disease
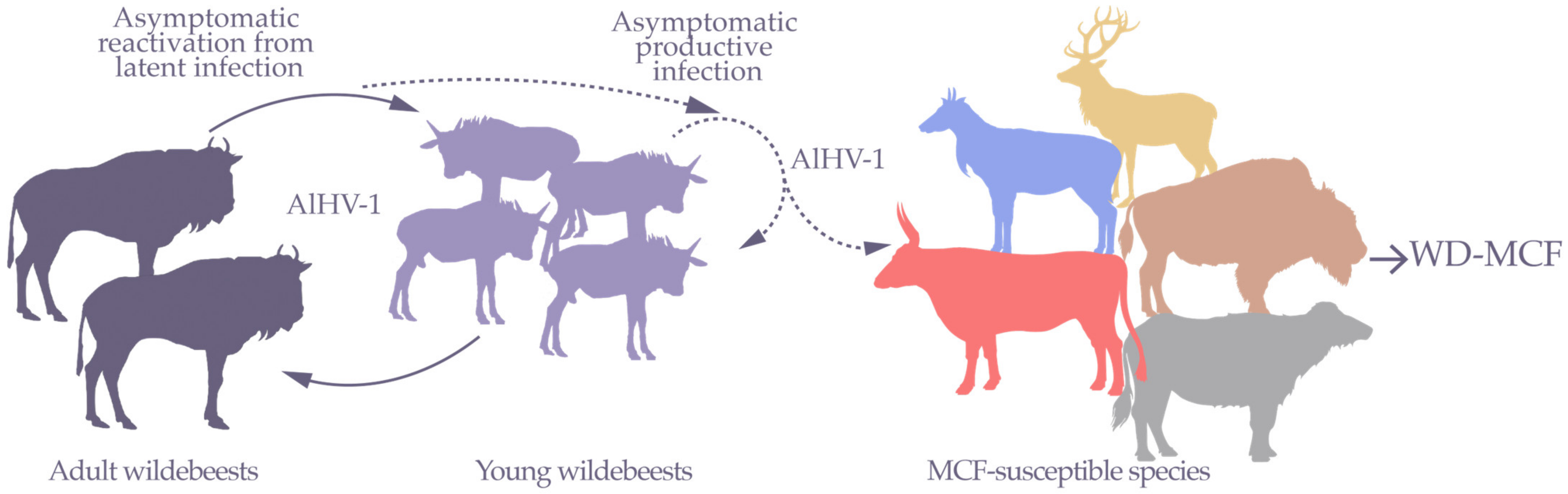
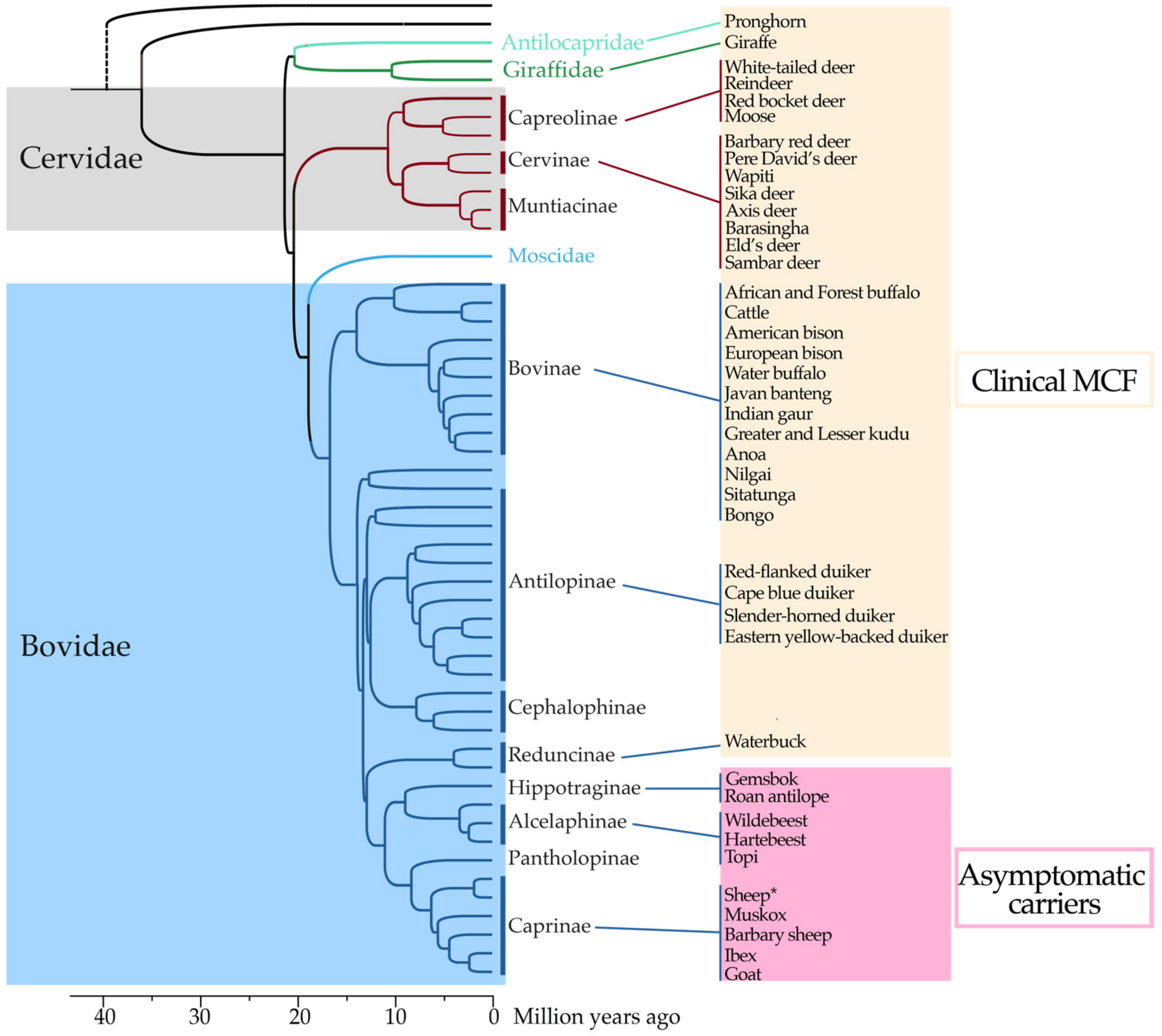
2. AlHV-1 Is a Gammaherpesvirinae, a Subfamily Responsible for Lymphoproliferative Diseases
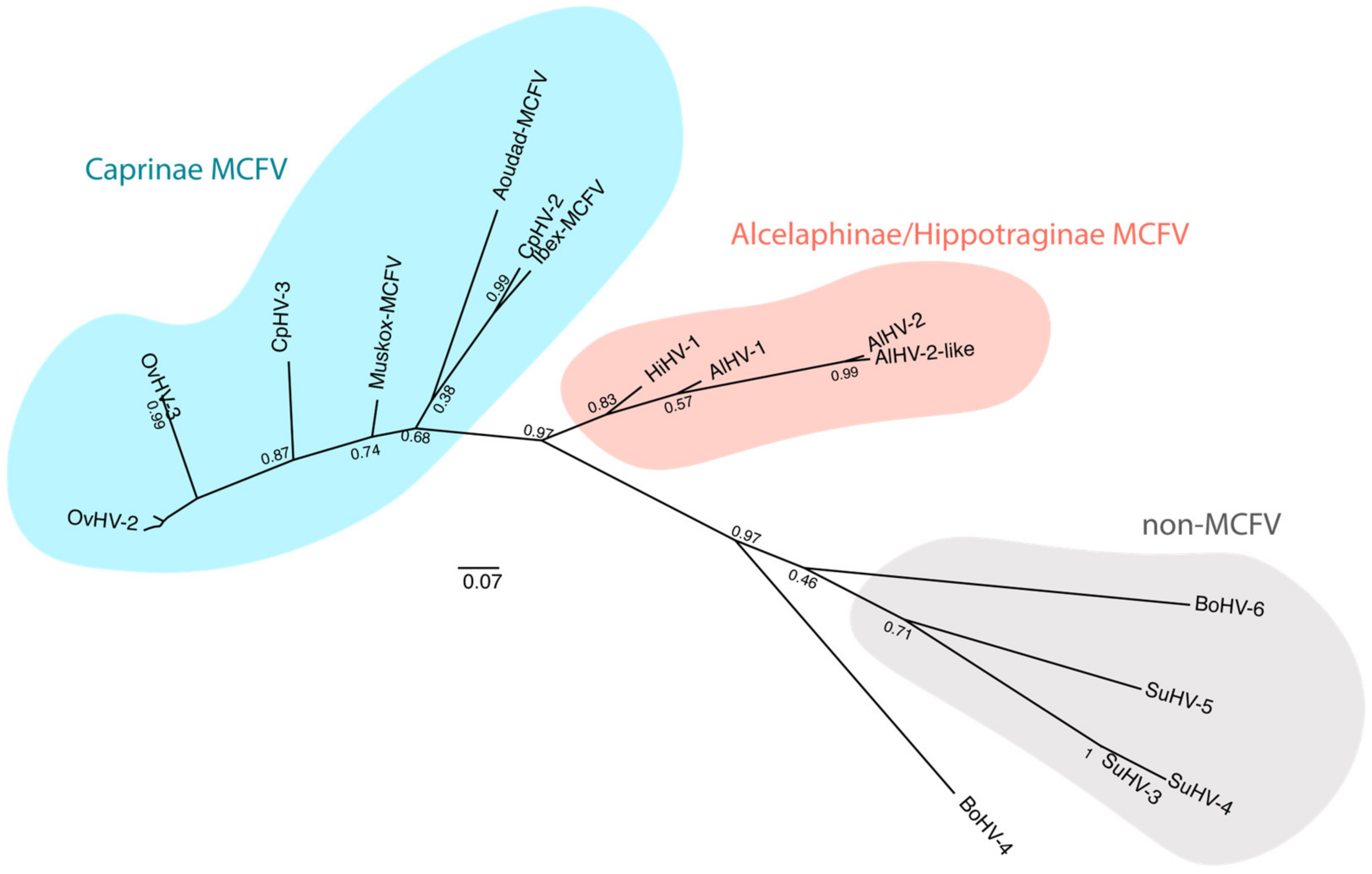
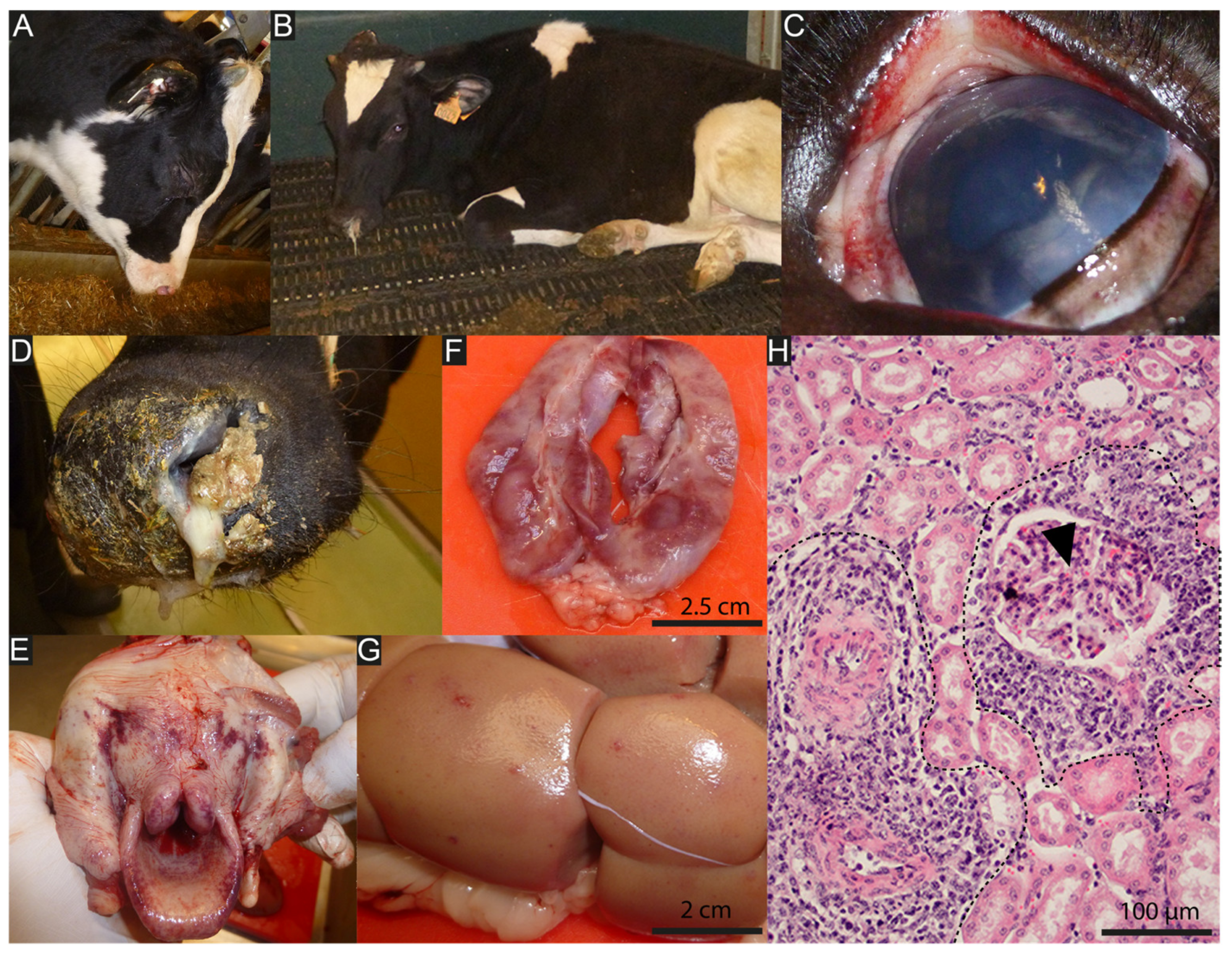
3. Unravelling WD-MCF Pathogenesis from AlHV-1 Genomic Sequence
3.1. γHV Latency Is an Essential Mechanism for WD-MCF Pathogenesis
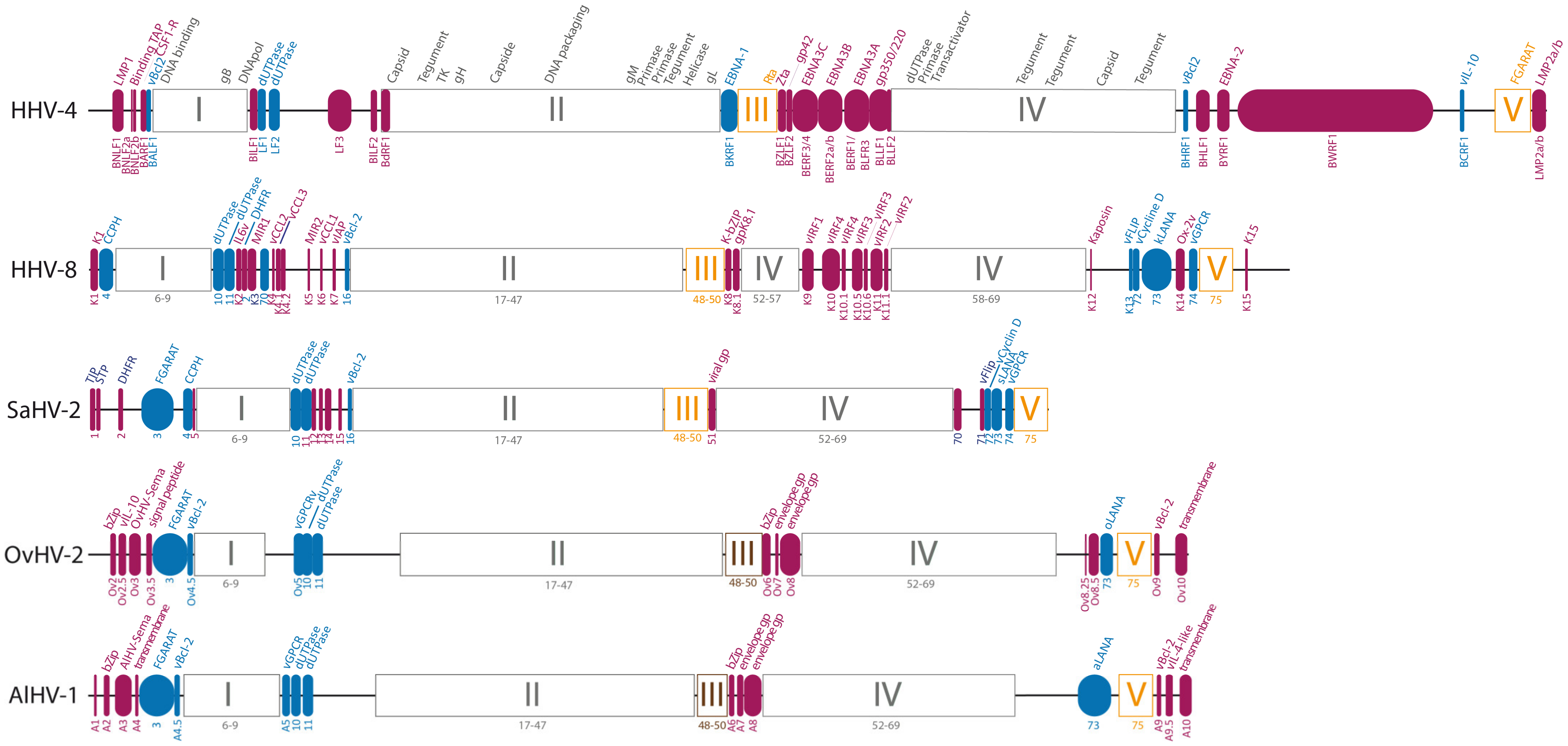
3.2. Studying AlHV-1-Specific Genes and Non-Coding RNAs to Investigate WD-MCF Pathogenesis
3.2.1. Regulation of Cell Tropism during Host Entry Is Essential for WD-MCF and Regulated by Accessory Envelope Glycoproteins A7 and A8
| AlHV-1 | OvHV-2 | % id. a | BoHV-6 | % id. a | SuHV3 | % id. a | Protein b | Description c |
|---|---|---|---|---|---|---|---|---|
| A1 | - | - | - | - | ||||
| A2 | Ov2 | 56 | Bov2 | 30 | - | - | bZIP protein | Presence of a “leucine zipper” domain, a possible transcription factor, non-essential for WD-MCF induction in rabbits [103] |
| - | Ov2.5 | - | Bov2.5 | - | - | - | viral IL-10 | Stimulates mast cell proliferation and inhibits IL-8 production by macrophages [104] |
| A3 | Ov3 | 50 | - | - | - | - | viral sema7A | Secreted, binds to plexin C1 and inhibits dendritic cell migration, non-essential for WD-MCF induction in rabbits [105,106] |
| - | Ov3.5 | - | - | - | - | - | Unknown | Signal peptide |
| A4 | - | - | - | - | - | - | SaHV-2 STP homolog | Unknown |
| A4.5 | Ov4.5 | 50 | Bov4.5 | 35 | - | - | viral Bcl-2 | Putative anti-apoptotic function |
| A5 | Ov5 | 49 | Bov5 | 41 | - | - | vGPCR | Inhibits CREB signaling pathway, non-essential for WD-MCF induction in rabbits [107] |
| A6 | Ov6 | 28 | Bov6 | - | A6/BZLF1 | 33.9 | bZIP protein (Zta homolog) | Presence of a “leucine zipper” domain, a possible transcription factor |
| A7 | Ov7 | 60 | Bov7 | 35 | A7/BZLF2 | 32.0 | EBV gp42 homolog | Virus glycoprotein, essential for WD-MCF induction in rabbits [26] |
| A8 | Ov8 | 41 | Bov8 | 25 | A8/BLLF1 | 25.3 | EBV gp350/220 homolog | Virus glycoprotein, essential for WD-MCF induction in rabbits [26] |
| - | Ov8.25 | - | - | - | - | - | Targets mitochondria | Putative role in caspase-dependent apoptosis and necrosis [108] |
| A9 | Ov9 | 50 | Bov9 | 27 | - | - | viral Bcl-2 | Putative anti-apoptotic function |
| A9.5 | - | - | - | - | - | - | viral IL-4-like | Encodes a secreted glycoprotein [109] |
| A10 | Ov10 | 22 | - | - | - | - | SaHV-2 Tip homolog | Putative oncogene |
3.2.2. Secreted AlHV-1 Semaphorin Homolog Is Dispensable for WD-MCF Induction
3.2.3. AlHV-1 Encodes Specific Protein-Coding Genes Potentially Involved in the Induction of Lymphoproliferation
3.2.4. AlHV-1 Encodes microRNAs Expressed in Lymphoblastoid Cell Lines
4. AlHV-1 Establishes a Latency-Like Infection in CD8+ T Lymphocytes to Induce Their Activation and Proliferation during WD-MCF
5. Current Status of Diagnosis and Vaccines

6. Socio-Economic Impact of Wildebeest Derived WD-MCF
7. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Mettam, R.W.M. Snotsiekte in Cattle. In 9th & 10th Reports of Director Veterinary Education & Research, Union South Africa 1923; Veterinary Education & Research: Pretoria, South Africa, 1924; pp. 395–432. Available online: https://www.cabdirect.org/cabdirect/abstract/19241000513 (accessed on 5 January 2023).
- Plowright, W.; Ferris, R.D.; Scott, G.R. Blue Wildebeest and the Ætiological Agent of Bovine Malignant Catarrhal Fever. Nature 1960, 188, 1167–1169. [Google Scholar] [CrossRef] [PubMed]
- Plowright, W. Chapter 14—Malignant Catarrhal Fever Virus. In Virus Infections of Ruminants; Dinter, Z., Morein, B., Eds.; Virus Infections of Vertebrates Series; Elsevier: Amsterdam, The Netherlands, 1990; pp. 123–150. ISBN 9780444873125. [Google Scholar]
- Dobson, A.P.; Borner, M.; Sinclair, A.R.E.; Hudson, P.J.; Anderson, T.M.; Bigurube, G.; Davenport, T.B.B.; Deutsch, J.; Durant, S.M.; Estes, R.D.; et al. Road Will Ruin Serengeti. Nature 2010, 467, 272–273. [Google Scholar] [CrossRef] [PubMed]
- Holdo, R.M.; Fryxell, J.M.; Sinclair, A.R.E.; Dobson, A.; Holt, R.D. Predicted Impact of Barriers to Migration on the Serengeti Wildebeest Population. PLoS ONE 2011, 6, 16370. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, J.A.; Oosthuizen, M.C.; van Vuuren, M. Gammaherpesvirus Carrier Status of Black Wildebeest (Connochaetes gnou) in South Africa. J. S. Afr. Vet. Assoc. 2008, 79, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Plowright, W. Malignant Catarrhal Fever in East Africa: II.—Observations on Wildebeest Calves at the Laboratory and Contact Transmission of the Infection to Cattle. Res. Vet. Sci. 1965, 6, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Plowright, W. Malignant Catarrhal Fever in East Africa: I.—Behaviour of the Virus in Free-Living Populations of Blue Wildebeest (Gorgon Taurinus taurinus, Burchell). Res. Vet. Sci. 1965, 6, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Plowright, W. Malignant Catarrhal Fever in East Africa: III.—Neutralizing Antibody in Free-Living Wildebeest. Res. Vet. Sci. 1967, 8, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, K.; Javaux, J.; Simon, M.; Petit, T.; Clavel, S.; Lamglait, B.; Blanc, B.; Brunet, A.; Myster, F.; Li, H.; et al. Seroprevalence of Malignant Catarrhal Fever Virus in Captive Wildebeest (Connochaetes sp.) in France. Transbound. Emerg. Dis. 2018, 65, 1697–1704. [Google Scholar] [CrossRef]
- Russell, G.C.; Stewart, J.P.; Haig, D.M. Malignant Catarrhal Fever: A Review. Vet. J. 2009, 179, 324–335. [Google Scholar] [CrossRef]
- Michel, A.L.; van der Lugt, J.J.; Bengis, R.G.; de Vos, V. Detection of AHV-1 DNA in Lung Sections from Blue Wildebeest (Connochaetes Taurinus) Calves by in Situ Hybridization. Onderstepoort J. Vet. Res. 1997, 64, 235–238. [Google Scholar]
- Mushi, E.Z.; Karstad, L.; Jessett, D. Isolation of Bovine Malignant Catarrhal Fever Virus from Ocular and Nasal Secretions of Wildebeest Calves. Res. Vet. Sci. 1980, 29, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Barnard, B.J.; Bengis, R.G.; Griessel, M.D.; de Vos, V. Excretion of Alcelaphine Herpesvirus-1 by Captive and Free-Living Wildebeest (Connochaetes taurinus). Onderstepoort J. Vet. Res. 1989, 56, 131–134. [Google Scholar] [PubMed]
- Li, H.; Cunha, C.W.; Taus, N.S.; Knowles, D.P. Malignant Catarrhal Fever: Inching toward Understanding. Annu. Rev. Anim. Biosci. 2014, 2, 209–233. [Google Scholar] [CrossRef] [PubMed]
- Meier-Trummer, C.S.; Ryf, B.; Ackermann, M. Identification of Peripheral Blood Mononuclear Cells Targeted by Ovine Herpesvirus-2 in Sheep. Vet. Microbiol. 2009, 141, 199–207. [Google Scholar] [CrossRef]
- Heuschele, W.P.; Nielsen, N.O.; Oosterhuis, J.E.; Castro, A.E. Dexamethasone-Induced Recrudescence of Malignant Catarrhal Fever and Associated Lymphosarcoma and Granulomatous Disease in a Formosan Sika Deer (Cervus nippon taiouanus). Am. J. Vet. Res. 1985, 46, 1578–1583. [Google Scholar]
- Rweyemamu, M.M.; Karstad, L.; Mushi, E.Z.; Otema, J.C.; Jessett, D.M.; Rowe, L.; Drevemo, S.; Grootenhuis, J.G. Malignant Catarrhal Fever Virus in Nasal Secretions of Wildebeest: A Probable Mechanism for Virus Transmission. J. Wildl. Dis. 1974, 10, 478–487. [Google Scholar] [CrossRef]
- Mushi, E.Z.; Rurangirwa, F.R. Epidemiology of Bovine Malignant Catarrhal Fevers, a Review. Vet. Res. Commun. 1981, 5, 127–142. [Google Scholar] [CrossRef]
- Estes, R.D.; Estes, R.K. The Birth and Survival of Wildebeest Calves. Z. Tierpsychol. 2010, 50, 45–95. [Google Scholar] [CrossRef]
- Metzler, A.E. The Malignant Catarrhal Fever Complex. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 107–124. [Google Scholar] [CrossRef]
- Traul, D.L.; Li, H.; Dasgupta, N.; O’Toole, D.; Eldridge, J.A.; Besser, T.E.; Davies, C.J. Resistance to Malignant Catarrhal Fever in American Bison (Bison bison) Is Associated with MHC Class IIa Polymorphisms. Anim. Genet. 2007, 38, 141–146. [Google Scholar] [CrossRef]
- Buxton, D.; Reid, H.W. Transmission of Malignant Catarrhal Fever to Rabbits. Vet. Rec. 1980, 106, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Dewals, B.; Boudry, C.; Farnir, F.; Drion, P.-V.; Vanderplasschen, A. Malignant Catarrhal Fever Induced by Alcelaphine Herpesvirus 1 Is Associated with Proliferation of CD8+ T Cells Supporting a Latent Infection. PLoS ONE 2008, 3, e1627. [Google Scholar] [CrossRef] [PubMed]
- Dewals, B.G.; Vanderplasschen, A. Malignant Catarrhal Fever Induced by Alcelaphine Herpesvirus 1 Is Characterized by an Expansion of Activated CD3+CD8 +CD4- T Cells Expressing a Cytotoxic Phenotype in Both Lymphoid and Non-Lymphoid Tissues. Vet. Res. 2011, 42, 95. [Google Scholar] [CrossRef] [PubMed]
- Myster, F.; Gong, M.J.; Javaux, J.; Suárez, N.M.; Wilkie, G.S.; Connelley, T.; Vanderplasschen, A.; Davison, A.J.; Dewals, B.G. Alcelaphine Herpesvirus 1 Genes A7 and A8 Regulate Viral Spread and Are Essential for Malignant Catarrhal Fever. PLoS Pathog. 2020, 16, 1008405. [Google Scholar] [CrossRef] [PubMed]
- Dewals, B.; Myster, F.; Palmeira, L.; Gillet, L.; Ackermann, M.; Vanderplasschen, A. Ex Vivo Bioluminescence Detection of Alcelaphine Herpesvirus 1 Infection during Malignant Catarrhal Fever. J. Virol. 2011, 85, 6941–6954. [Google Scholar] [CrossRef]
- Partinid, T.G.; Schrenzel, M.D.; Braun, J.; Witte, C.L.; Kubiski, S.; Lee, J.; Rideout, B.A. Herpesvirus Surveillance and Discovery in Zoo-Housed Ruminants. PLoS ONE 2021, 16, e0246162. [Google Scholar] [CrossRef]
- Crawford, T.B.; Li, H.; Rosenburg, S.R.; Norhausen, R.W.; Garner, M.M. Mural Folliculitis and Alopecia Caused by Infection with Goat-Associated Malignant Catarrhal Fever Virus in Two Sika Deer. J. Am. Vet. Med. Assoc. 2002, 221, 843–847. [Google Scholar] [CrossRef]
- Heuschele, W.P.; Oosterhuis, J.; Anderson, M.P.; Swansen, M.; Fletcher, H.R. Malignant Catarrhal Fever in Wild Ruminants. In One Medicine; Ryder, O.A., Byrd, M.L., Eds.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 296–308. [Google Scholar] [CrossRef]
- O’Toole, D.; Taus, N.S.; Montgomery, D.L.; Oaks, J.L.; Crawford, T.B.; Li, H. Intra-Nasal Inoculation of American Bison (Bison bison) with Ovine Herpesvirus-2 (OvHV-2) Reliably Reproduces Malignant Catarrhal Fever. Vet. Pathol. 2007, 44, 655–662. [Google Scholar] [CrossRef]
- Hoffman, D.; Soeripto, S.; Sobironingsih, S.; Campbell, R.S.F.; Clarke, B.C. The Clinico-Pathology of a Malignant Catarrhal Fever Syndrome in the Indonesian Swamp Buffalo (Bubalus bubalis). Aust. Vet. J. 1984, 61, 108–112. [Google Scholar] [CrossRef]
- Clark, K.A.; Robinson, R.M.; Weishuhn, L.L.; McConnell, S. Further Observations on Malignant Catarrhal Fever in Texas Deer. J. Wildl. Dis. 1972, 8, 72–74. [Google Scholar] [CrossRef]
- Castro, A.E.; Daley, G.G.; Zimmer, M.A.; Whitenack, D.L.; Jensen, J. Malignant Catarrhal Fever in an Indian Gaur and Greater Kudu: Experimental Transmission, Isolation, and Identification of a Herpesvirus. Am. J. Vet. Res. 1982, 43, 5–11. [Google Scholar] [PubMed]
- Boever, W.J.; Kurka, B. Malignant Catarrhal Fever in Greater Kudus. J. Am. Vet. Med. Assoc. 1974, 165, 817–819. [Google Scholar] [PubMed]
- Phillips, I.L.; Cunha, C.W.; Galbraith, D.; Highland, M.A.; Bildfell, R.J.; Li, H. High Copy Number of Ovine Gammaherpesvirus 2 DNA Associated with Malignant Catarrhal Fever-like Syndrome in a Lamb. J. Vet. Diagn. Invest. 2018, 30, 623–627. [Google Scholar] [CrossRef]
- Pesavento, P.A.; Dange, R.B.; Ferreras, M.C.; Dasjerdi, A.; Pérez, V.; LaRoca, A.; Silván, J.B.; Diab, S.; Jackson, K.; Phillips, I.L.; et al. Systemic Necrotizing Vasculitis in Sheep Is Associated with Ovine Herpesvirus 2. Vet. Pathol. 2019, 56, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Kalunda, M.; Ferris, D.H.; Dardiri, A.H.; Lee, K.M. Malignant Catarrhal Fever III. Experimental Infection of Sheep, Domestic Rabbits and Laboratory Animals with Malignant Catarrhal Fever Virus. Can. J. Comp. Med. 1981, 45, 310–314. [Google Scholar]
- Chen, L.; Qiu, Q.; Jiang, Y.; Wang, K.; Lin, Z.; Li, Z.; Bibi, F.; Yang, Y.; Wang, J.; Nie, W.; et al. Large-Scale Ruminant Genome Sequencing Provides Insights into Their Evolution and Distinct Traits. Science 2019, 364, eaav6202. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Malignant Catarrhal Fever. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2018; Volume 8, pp. 1172–1183. [Google Scholar]
- Palmeira, L.; Sorel, O.; van Campe, W.; Boudry, C.; Roels, S.; Myster, F.; Reschner, A.; Coulie, P.G.; Kerkhofs, P.; Vanderplasschen, A.; et al. An Essential Role for γ-Herpesvirus Latency-Associated Nuclear Antigen Homolog in an Acute Lymphoproliferative Disease of Cattle. Proc. Natl. Acad. Sci. USA 2013, 110, E1933–E1942. [Google Scholar] [CrossRef]
- Lankester, F.; Russell, G.C.; Lugelo, A.; Ndabigaye, A.; Mnyambwa, N.; Keyyu, J.; Kazwala, R.; Grant, D.; Percival, A.; Deane, D.; et al. A Field Vaccine Trial in Tanzania Demonstrates Partial Protection against Malignant Catarrhal Fever in Cattle. Vaccine 2016, 34, 831–838. [Google Scholar] [CrossRef]
- Russell, G.C.; Benavides, J.; Grant, D.; Todd, H.; Deane, D.; Percival, A.; Thomson, J.; Connelly, M.; Haig, D.M. Duration of Protective Immunity and Antibody Responses in Cattle Immunised against Alcelaphine Herpesvirus-1-Induced Malignant Catarrhal Fever. Vet. Res. 2012, 43, 1–11. [Google Scholar] [CrossRef]
- Pierson, R.E.; Hamdy, F.M.; Dardiri, A.H.; Ferris, D.H.; Schloer, G.M. Comparison of African and American Forms of Malignant Catarrhal Fever: Transmission and Clinical Signs. Am. J. Vet. Res. 1979, 40, 1091–1095. [Google Scholar]
- Hunt, R.D.; Billups, L.H. Wildebeest-Associated Malignant Catarrhal Fever in Africa: A Neoplastic Disease of Cattle Caused by an Oncogenic Herpesvirus? Comp. Immunol. Microbiol. Infect. Dis. 1979, 2, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Liggitt, H.D.; DeMartini, J.C. The Pathomorphology of Malignant Catarrhal Fever. Vet. Pathol. 1980, 17, 58–72. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, D.; Li, H. The Pathology of Malignant Catarrhal Fever, With an Emphasis on Ovine Herpesvirus 2. Vet. Pathol. 2014, 51, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Schock, A.; Reid, H.W. Characterisation of the Lymphoproliferation in Rabbits Experimentally Affected with Malignant Catarrhal Fever. Vet. Microbiol. 1996, 53, 111–119. [Google Scholar] [CrossRef]
- Simon, S.; Li, H.; O’Toole, D.; Crawford, T.B.; Oaks, J.L. The Vascular Lesions of a Cow and Bison with Sheep-Associated Malignant Catarrhal Fever Contain Ovine Herpesvirus 2-Infected CD8(+) T Lymphocytes. J. Gen. Virol. 2003, 84, 2009–2013. [Google Scholar] [CrossRef]
- Nelson, D.D.; Davis, W.C.; Brown, W.C.; Li, H.; O’Toole, D.; Oaks, J.L. CD8(+)/Perforin(+)/WC1(-) Gamma Delta T Cells, Not CD8(+) Alpha Beta T Cells, Infiltrate Vasculitis Lesions of American Bison (Bison bison) with Experimental Sheep-Associated Malignant Catarrhal Fever. Vet. Immunol. Immunopathol. 2010, 136, 284–291. [Google Scholar] [CrossRef]
- Nakajima, Y.; Momotani, E.; Ishikawa, Y.; Murakami, T.; Shimura, N.; Onuma, M. Phenotyping of Lymphocyte Subsets in the Vascular and Epithelial Lesions of a Cow with Malignant Catarrhal Fever. Vet. Immunol. Immunopathol. 1992, 33, 279–284. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The Order Herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benkő, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef]
- Krug, L.T.; Pellett, P.E. The Family Herpesviridae. In Fied’s Virology; Howley, P.M., Knipe, D.M., Eds.; Wolter Kluwer: Philadelphia, PA, USA, 2021; pp. 212–234. [Google Scholar]
- Damania, B.; Kenney, S.C.; Raab-Traub, N. Epstein-Barr Virus: Biology and Clinical Disease. Cell 2022, 185, 3652–3670. [Google Scholar] [CrossRef]
- Fleckenstein, B.; Ensser, A. Gammaherpesviruses of New World primates. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P., Roizman, B., Whitley, R., Eds.; Cambridge University Press: Cambridge, UK, 2007; pp. 1076–1092. [Google Scholar] [CrossRef]
- Verma, S.C.; Robertson, E.S. Molecular Biology and Pathogenesis of Kaposi Sarcoma-Associated Herpesvirus. FEMS Microbiol. Lett. 2003, 222, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Grabar, S.; Costagliola, D. Epidemiology of Kaposi’s Sarcoma. Cancers 2021, 13, 5692. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s Sarcoma and Its Associated Herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Oksenhendler, E.; Meignin, V. HHV-8 Associated Lymphoma. Curr. Opin. Oncol. 2022, 34, 432–438. [Google Scholar] [CrossRef]
- Wang, Y.; Tibbetts, S.A.; Krug, L.T. Conquering the Host: Determinants of Pathogenesis Learned from Murine Gammaherpesvirus 68. Annu. Rev. Virol. 2021, 8, 349–371. [Google Scholar] [CrossRef]
- Lange, P.T.; Damania, B. Modeling Oncogenic Herpesvirus Infections in Humanized Mice. Curr. Opin. Virol. 2020, 44, 90–96. [Google Scholar] [CrossRef]
- Cunha, C.W.; Slater, O.M.; Macbeth, B.; Duignan, P.J.; Warren, A.; Highland, M.A.; Li, H. Domestic Sheep and Bighorn Sheep Carry Distinct Gammaherpesviruses Belonging to the Genus Macavirus. Virus Res. 2019, 272, 197729. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Li, H.; Gailbreath, K.; Flach, E.J.; Taus, N.S.; Cooley, J.; Keller, J.; Russell, G.C.; Knowles, D.P.; Haig, D.M.; Oaks, J.L.; et al. A Novel Subgroup of Rhadinoviruses in Ruminants. J. Gen. Virol. 2005, 86, 3021–3026. [Google Scholar] [CrossRef]
- Dry, I.; Todd, H.; Deane, D.; Percival, A.; McLean, K.; Inglis, N.F.; Manson, E.D.; Haig, D.M.; Nayuni, S.; Hutt-Fletcher, L.M.; et al. Alcelaphine Herpesvirus 1 Glycoprotein B: Recombinant Expression and Antibody Recognition. Arch. Virol. 2016, 161, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Dry, I.; Haig, D.M.; Inglis, N.F.; Imrie, L.; Stewart, J.P.; Russell, G.C. Proteomic Analysis of Pathogenic and Attenuated Alcelaphine Herpesvirus 1. J. Virol. 2008, 82, 5390–5397. [Google Scholar] [CrossRef] [PubMed]
- Plowright, W.; Macadam, R.F.; Armstrong, J.A. Growth and Characterization of the Virus of Bovine Malignant Catarrhal Fever in East Africa. J. Gen. Microbiol. 1963, 39, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Plowright, W. Malignant Catarrhal Fever Virus. Virus Infect. Rumin. 1990, 3, 123–150. [Google Scholar]
- Castro, A.E.; Daley, G.G. Electron Microscopic Study of the African Strain of Malignant Catarrhal Fever Virus in Bovine Cell Cultures. Am. J. Vet. Res. 1982, 43, 576–582. [Google Scholar]
- Bridgen, A.; Herring, A.J.; Inglis, N.F.; Reid, H.W. Preliminary Characterization of the Alcelaphine Herpesvirus 1 Genome. J. Gen. Virol. 1989, 70 Pt 5, 1141–1150. [Google Scholar] [CrossRef]
- Ensser, A.; Pflanz, R.; Fleckenstein, B. Primary Structure of the Alcelaphine Herpesvirus 1 Genome. J. Virol. 1997, 71, 6517–6525. [Google Scholar] [CrossRef]
- Plowright, W.; Herniman, K.A.; Jessett, D.M.; Kalunda, M.; Rampton, C.S. Immunisation of Cattle against the Herpesvirus of Malignant Catarrhal Fever: Failure of Inactivated Culture Vaccines with Adjuvant. Res. Vet. Sci. 1975, 19, 159–166. [Google Scholar] [CrossRef]
- Myster, F.; van Beurden, S.J.S.J.; Sorel, O.; Suárez, N.M.; Vanderplasschen, A.; Davison, A.J.A.J.; Dewals, B.G.B.G.; Suarez, N.M.; Vanderplasschen, A.; Davison, A.J.A.J.; et al. Genomic Duplication and Translocation of Reactivation Transactivator and BZIP-Homolog Genes Is a Conserved Event in Alcelaphine Herpesvirus 1. Sci. Rep. 2016, 6, 38607. [Google Scholar] [CrossRef]
- Bridgen, A.; Munro, R.; Reid, H.W. The Detection of Alcelaphine Herpesvirus-1 DNA by in Situ Hybridization of Tissues from Rabbits Affected with Malignant Catarrhal Fever. J. Comp. Pathol. 1992, 106, 351–359. [Google Scholar] [CrossRef]
- Ensser, A. Genome Sequence of the Alcelaphine Gammaherpesvirus 1 Attenuated Laboratory Strain WC11. Genome Announc. 2017, 5, e01219-17. [Google Scholar] [CrossRef] [PubMed]
- Coulter, L.J.; Wright, H.; Reid, H.W. Molecular Genomic Characterization of the Viruses of Malignant Catarrhal Fever. J. Comp. Pathol. 2001, 124, 2–19. [Google Scholar] [CrossRef] [PubMed]
- Leenadevi, T.; Dalziel, R.G. Alcelaphine Herpesvirus-1 Open Reading Frame 57 Encodes an Immediate-Early Protein with Regulatory Function. Vet. Res. Commun. 2009, 33, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Staudt, M.R.; Dittmer, D.P. The Rta/Orf50 Transactivator Proteins of the Gamma-Herpesviridae. Curr. Top. Microbiol. Immunol. 2007, 312, 71–100. [Google Scholar] [CrossRef]
- Hart, J.; Ackermann, M.; Jayawardane, G.; Russell, G.; Haig, D.M.; Reid, H.; Stewart, J.P. Complete Sequence and Analysis of the Ovine Herpesvirus 2 Genome. J. Gen. Virol. 2007, 88, 28–39. [Google Scholar] [CrossRef]
- Albrecht, J.-C.; Nicholas, J.; Biller, D.; Cameron, K.R.; Biesinger, B.; Newman, C.; Wiitmann, S.; Craxton, M.A.; Coleman, H.; Fleckenstein, B.; et al. Primary Structure of the Herpesvirus Saimiri Genome. J. Virol. 1992, 66, 5047–5058. [Google Scholar] [CrossRef]
- Glenn, M.; Rainbow, L.; Auradé, F.; Davison, A.; Schulz, T.F. Identification of a Spliced Gene from Kaposi’s Sarcoma-Associated Herpesvirus Encoding a Protein with Similarities to Latent Membrane Proteins 1 and 2A of Epstein-Barr Virus. J. Virol. 1999, 73, 6953–6963. [Google Scholar] [CrossRef]
- De Jesus, O.; Smith, P.R.; Spender, L.C.; Elgueta Karstegl, C.; Niller, H.H.; Huang, D.; Farrell, P.J. Updated Epstein–Barr Virus (EBV) DNA Sequence and Analysis of a Promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 2003, 84, 1443–1450. [Google Scholar] [CrossRef]
- Frame, F.M.; Dalziel, R.G. Transcriptional Control by the R-Transactivator Protein of Alcelaphine Herpesvirus-1. Vet. Res. Commun. 2008, 32, 215–223. [Google Scholar] [CrossRef]
- Dewals, B.; Boudry, C.; Gillet, L.; Markine-Goriaynoff, N.; de Leval, L.; Haig, D.M.; Vanderplasschen, A. Cloning of the Genome of Alcelaphine Herpesvirus 1 as an Infectious and Pathogenic Bacterial Artificial Chromosome. J. Gen. Virol. 2006, 87, 509–517. [Google Scholar] [CrossRef]
- Sorel, O.; Dewals, B.G. The Critical Role of Genome Maintenance Proteins in Immune Evasion During Gammaherpesvirus Latency. Front. Microbiol. 2019, 9, 3315. [Google Scholar] [CrossRef] [PubMed]
- Blake, N. Immune Evasion by Gammaherpesvirus Genome Maintenance Proteins. J. Gen. Virol. 2010, 91, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Kwun, H.J.; da Silva, S.R.; Shah, I.M.; Blake, N.; Moore, P.S.; Chang, Y. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen 1 Mimics Epstein-Barr Virus EBNA1 Immune Evasion through Central Repeat Domain Effects on Protein Processing. J. Virol. 2007, 81, 8225–8235. [Google Scholar] [CrossRef] [PubMed]
- Apcher, S.; Daskalogianni, C.; Manoury, B.; Fåhraeus, R. Epstein Barr Virus-Encoded EBNA1 Interference with MHC Class I Antigen Presentation Reveals a Close Correlation between MRNA Translation Initiation and Antigen Presentation. PLoS Pathog. 2010, 6, 1001151. [Google Scholar] [CrossRef] [PubMed]
- Bennett, N.J.; May, J.S.; Stevenson, P.G. Gamma-Herpesvirus Latency Requires T Cell Evasion during Episome Maintenance. PLoS Biol. 2005, 3, 0638–0649. [Google Scholar] [CrossRef] [PubMed]
- Sorel, O.; Chen, T.; Myster, F.; Javaux, J.; Vanderplasschen, A.; Dewals, B.G. Macavirus Latency-Associated Protein Evades Immune Detection through Regulation of Protein Synthesis in Cis Depending upon Its Glycin/Glutamate-Rich Domain. PLoS Pathog. 2017, 13, 1006691. [Google Scholar] [CrossRef]
- Bravo Cruz, A.G.; Damania, B. In Vivo Models of Oncoproteins Encoded by Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2019, 93, e01053-18. [Google Scholar] [CrossRef]
- Hutt-Fletcher, L.M. EBV Glycoproteins: Where Are We Now? Future Virol. 2015, 10, 1155–1162. [Google Scholar] [CrossRef]
- Gillet, L.; Frederico, B.; Stevenson, P.G. Host Entry by Gamma-Herpesviruses--Lessons from Animal Viruses? Curr. Opin. Virol. 2015, 15, 34–40. [Google Scholar] [CrossRef]
- Borza, C.M.; Hutt-Fletcher, L.M. Alternate Replication in B Cells and Epithelial Cells Switches Tropism of Epstein-Barr Virus. Nat. Med. 2002, 8, 594–599. [Google Scholar] [CrossRef]
- Möhl, B.S.; Chen, J.; Sathiyamoorthy, K.; Jardetzky, T.S.; Longnecker, R. Structural and Mechanistic Insights into the Tropism of Epstein-Barr Virus. Mol. Cells 2016, 39, 286–291. [Google Scholar] [CrossRef]
- Jean-Pierre, V.; Lupo, J.; Buisson, M.; Morand, P.; Germi, R. Main Targets of Interest for the Development of a Prophylactic or Therapeutic Epstein-Barr Virus Vaccine. Front. Microbiol. 2021, 12, 701611. [Google Scholar] [CrossRef] [PubMed]
- Machiels, B.; Lété, C.; de Fays, K.; Mast, J.; Dewals, B.; Stevenson, P.G.; Vanderplasschen, A.; Gillet, L. The Bovine Herpesvirus 4 Bo10 Gene Encodes a Nonessential Viral Envelope Protein That Regulates Viral Tropism through Both Positive and Negative Effects. J. Virol. 2011, 85, 1011–1024. [Google Scholar] [CrossRef]
- Dollery, S.J. Towards Understanding KSHV Fusion and Entry. Viruses 2019, 11, 1073. [Google Scholar] [CrossRef] [PubMed]
- Zeippen, C.; Javaux, J.; Xiao, X.; Ledecq, M.; Mast, J.; Farnir, F.; Vanderplasschen, A.; Stevenson, P.; Gillet, L. The Major Envelope Glycoprotein of Murid Herpesvirus 4 Promotes Sexual Transmission. J. Virol. 2017, 91, e00235-17. [Google Scholar] [CrossRef] [PubMed]
- AlHajri, S.M.; Cunha, C.W.; Nicola, A.; Aguilar, H.C.; Li, H.; Taus, N.S. Ovine Herpesvirus 2 Glycoproteins B, H, and L Are Sufficient for, and Viral Glycoprotein Ov8 Can Enhance, Cell-Cell Membrane Fusion. J. Virol. 2017, 91, e02454-16. [Google Scholar] [CrossRef]
- Parameswaran, N.; Dewals, B.G.; Giles, T.C.; Deppmann, C.; Blythe, M.; Vanderplasschen, A.; Emes, R.D.; Haig, D. The A2 Gene of Alcelaphine Herpesvirus-1 Is a Transcriptional Regulator Affecting Cytotoxicity in Virus-Infected T Cells but Is Not Required for Malignant Catarrhal Fever Induction in Rabbits. Virus Res. 2014, 188, 68–80. [Google Scholar] [CrossRef]
- Jayawardane, G.; Russell, G.C.; Thomson, J.; Deane, D.; Cox, H.; Gatherer, D.; Ackermann, M.; Haig, D.M.; Stewart, J.P. A Captured Viral Interleukin 10 Gene with Cellular Exon Structure. J. Gen. Virol. 2008, 89, 2447–2455. [Google Scholar] [CrossRef]
- Myster, F.; Palmeira, L.; Sorel, O.; Bouillenne, F.; DePauw, E.; Schwartz-Cornil, I.; Vanderplasschen, A.; Dewals, B.G. Viral Semaphorin Inhibits Dendritic Cell Phagocytosis and Migration but Is Not Essential for Gammaherpesvirus-Induced Lymphoproliferation in Malignant Catarrhal Fever. J. Virol. 2015, 89, 3630–3647. [Google Scholar] [CrossRef]
- Ensser, A.; Fleckenstein, B. Alcelaphine Herpesvirus Type 1 Has a Semaphorin-like Gene. J. Gen. Virol. 1995, 76, 1063–1067. [Google Scholar] [CrossRef]
- Boudry, C.; Markine-Goriaynoff, N.; Delforge, C.; Springael, J.-Y.; de Leval, L.; Drion, P.; Russell, G.; Haig, D.M.; Vanderplasschen, A.F.; Dewals, B. The A5 Gene of Alcelaphine Herpesvirus 1 Encodes a Constitutively Active G-Protein-Coupled Receptor That Is Non-Essential for the Induction of Malignant Catarrhal Fever in Rabbits. J. Gen. Virol. 2007, 88, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Tobler, K.; Uster, S.; Sigrist-Nagy, R.; Hierweger, M.M.; Ackermann, M. Ovine Herpesvirus 2 Encodes a Previously Unrecognized Protein, POv8.25, That Targets Mitochondria and Triggers Apoptotic Cell Death. J. Virol. 2020, 94, e01536-19. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.C.; Todd, H.; Deane, D.; Percival, A.; Dagleish, M.P.; Haig, D.M.; Stewart, J.P. A Novel Spliced Gene in Alcelaphine Herpesvirus 1 Encodes a Glycoprotein Which Is Secreted in Vitro. J. Gen. Virol. 2013, 94, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, H.; Kumanogoh, A. Diverse Roles for Semaphorin-Plexin Signaling in the Immune System. Trends Immunol. 2012, 33, 127–135. [Google Scholar] [CrossRef]
- Suzuki, K.; Kumanogoh, A.; Kikutani, H. Semaphorins and Their Receptors in Immune Cell Interactions. Nat. Immunol. 2008, 9, 17–23. [Google Scholar] [CrossRef]
- Fard, D.; Tamagnone, L. Semaphorins in Health and Disease. Cytokine Growth Factor Rev. 2021, 57, 55–63. [Google Scholar] [CrossRef]
- Suzuki, K.; Okuno, T.; Yamamoto, M.; Pasterkamp, R.J.; Takegahara, N.; Takamatsu, H.; Kitao, T.; Takagi, J.; Rennert, P.D.; Kolodkin, A.L.; et al. Semaphorin 7A Initiates T-Cell-Mediated Inflammatory Responses through A1β1 Integrin. Nature 2007, 446, 680–684. [Google Scholar] [CrossRef]
- Comeau, M.R.; Johnson, R.; DuBose, R.F.; Petersen, M.; Gearing, P.; VandenBos, T.; Park, L.; Farrah, T.; Buller, R.M.; Cohen, J.I.; et al. A Poxvirus-Encoded Semaphorin Induces Cytokine Production from Monocytes and Binds to a Novel Cellular Semaphorin Receptor, VESPR. Immunity 1998, 8, 473–482. [Google Scholar] [CrossRef]
- Vischer, H.F.; Vink, C.; Smit, M.J. A Viral Conspiracy: Hijacking the Chemokine System through Virally Encoded Pirated Chemokine Receptors. Curr. Top. Microbiol. Immunol. 2006, 303, 121–154. [Google Scholar] [CrossRef]
- Vischer, H.F.; Siderius, M.; Leurs, R.; Smit, M.J. Herpesvirus-Encoded GPCRs: Neglected Players in Inflammatory and Proliferative Diseases? Nat. Rev. Drug Discov. 2014, 13, 123–139. [Google Scholar] [CrossRef]
- Beisser, P.S.; Verzijl, D.; Gruijthuijsen, Y.K.; Beuken, E.; Smit, M.J.; Leurs, R.; Bruggeman, C.A.; Vink, C. The Epstein-Barr Virus BILF1 Gene Encodes a G Protein-Coupled Receptor That Inhibits Phosphorylation of RNA-Dependent Protein Kinase. J. Virol. 2005, 79, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.; Philpott, N.J.; Cesarman, E. The Kaposi’s Sarcoma-Associated Herpesvirus G Protein-Coupled Receptor Has Broad Signaling Effects in Primary Effusion Lymphoma Cells. J. Virol. 2003, 77, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Stolz, M.L.; McCormick, C. The BZIP Proteins of Oncogenic Viruses. Viruses 2020, 12, 757. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Uppal, T.; Strahan, R.; Dabral, P.; Verma, S.C. The Modulation of Apoptotic Pathways by Gammaherpesviruses. Front. Microbiol. 2016, 7, 585. [Google Scholar] [CrossRef]
- Altmann, M.; Hammerschmidt, W. Epstein-Barr Virus Provides a New Paradigm: A Requirement for the Immediate Inhibition of Apoptosis. PLoS Biol. 2005, 3, e404. [Google Scholar] [CrossRef] [PubMed]
- Bellows, D.S.; Howell, M.; Pearson, C.; Hazlewood, S.A.; Hardwick, J.M. Epstein-Barr Virus BALF1 Is a BCL-2-Like Antagonist of the Herpesvirus Antiapoptotic BCL-2 Proteins. J. Virol. 2002, 76, 2469–2479. [Google Scholar] [CrossRef]
- Marshall, W.L.; Yim, C.; Gustafson, E.; Graf, T.; Sage, D.R.; Hanify, K.; Williams, L.; Fingeroth, J.; Finberg, R.W. Epstein-Barr Virus Encodes a Novel Homolog of the Bcl-2 Oncogene That Inhibits Apoptosis and Associates with Bax and Bak. J. Virol. 1999, 73, 5181–5185. [Google Scholar] [CrossRef]
- Kvansakul, M.; Wei, A.H.; Fletcher, J.I.; Willis, S.N.; Chen, L.; Roberts, A.W.; Huang, D.C.S.; Colman, P.M. Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1. PLoS Pathog. 2010, 6, 1001236. [Google Scholar] [CrossRef]
- Damania, B. Oncogenic γ-Herpesviruses: Comparison of Viral Proteins Involved in Tumorigenesis. Nat. Rev. Microbiol. 2004, 2, 656–668. [Google Scholar] [CrossRef]
- Cho, N.-H.; Feng, P.; Lee, S.-H.; Lee, B.-S.; Liang, X.; Chang, H.; Jung, J.U. Inhibition of T Cell Receptor Signal Transduction by Tyrosine Kinase–Interacting Protein of Herpesvirus Saimiri. J. Exp. Med. 2004, 200, 681–687. [Google Scholar] [CrossRef]
- Kjellen, P.; Amdjadi, K.; Lund, T.C.; Medveczky, P.G.; Sefton, B.M. The Herpesvirus Saimiri Tip484 and Tip488 Proteins Both Stimulate Lck Tyrosine Protein Kinase Activity in Vivo and in Vitro. Virology 2002, 297, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Isakov, N.; Biesinger, B. Lck Protein Tyrosine Kinase Is a Key Regulator of T-Cell Activation and a Target for Signal Intervention by Herpesvirus Saimiri and Other Viral Gene Products. Eur. J. Biochem. 2000, 267, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Duboise, S.M.; Guo, J.; Czajak, S.; Desrosiers, R.C.; Jung, J.U. STP and Tip Are Essential for Herpesvirus Saimiri Oncogenicity. J. Virol. 1998, 72, 1308. [Google Scholar] [CrossRef] [PubMed]
- Walz, N.; Christalla, T.; Tessmer, U.; Grundhoff, A. A Global Analysis of Evolutionary Conservation among Known and Predicted Gammaherpesvirus MicroRNAs. J. Virol. 2010, 84, 716–728. [Google Scholar] [CrossRef]
- Sorel, O.; Dewals, B.G.B.G. MicroRNAs in Large Herpesvirus DNA Genomes: Recent Advances. Biomol. Concepts 2016, 7, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Herpesvirus MicroRNAs: Phenotypes and Functions. Curr. Opin. Virol. 2011, 1, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Bellare, P.; Ganem, D. Regulation of KSHV Lytic Switch Protein Expression by a Virus-Encoded MicroRNA: An Evolutionary Adaptation That Fine-Tunes Lytic Reactivation. Cell Host Microbe 2009, 6, 570–575. [Google Scholar] [CrossRef]
- Levy, C.S.; Hopkins, J.; Russell, G.C.; Dalziel, R.G. Novel Virus-Encoded MicroRNA Molecules Expressed by Ovine Herpesvirus 2-Immortalized Bovine T-Cells. J. Gen. Virol. 2012, 93, 150–154. [Google Scholar] [CrossRef]
- Sorel, O.; Tuddenham, L.; Myster, F.; Palmeira, L.; Kerkhofs, P.; Pfeffer, S.; Vanderplasschen, A.; Dewals, B.G. Small RNA Deep Sequencing Identifies Viral MicroRNAs during Malignant Catarrhal Fever Induced by Alcelaphine Herpesvirus 1. J. Gen. Virol. 2015, 96, 3360–3372. [Google Scholar] [CrossRef]
- Cook, C.G.; Splitter, G.A. Lytic Function of Bovine Lymphokine-Activated Killer Cells from a Normal and a Malignant Catarrhal Fever Virus-Infected Animal. Vet. Immunol. Immunopathol. 1988, 19, 105–118. [Google Scholar] [CrossRef]
- Swa, S.; Wright, H.; Thomson, J.; Reid, H.; Haig, D. Constitutive Activation of Lck and Fyn Tyrosine Kinases in Large Granular Lymphocytes Infected with the γ-Herpesvirus Agents of Malignant Catarrhal Fever. Immunology 2001, 102, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, A.; Herrmann, F.; Oster, W.; Mertelsmann, R. Lymphokine Activated Killer Cells. Blut 1989, 59, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Anderson, I.E.; Buxton, D.; Campbell, I.; Russell, G.; Davis, W.C.; Hamilton, M.J.; Haig, D.M. Immunohistochemical Study of Experimental Malignant Catarrhal Fever in Rabbits. J. Comp. Pathol. 2007, 136, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.C.; Benavides, J.; Grant, D.M.; Todd, H.; Thomson, J.; Puri, V.; Nath, M.; Haig, D.M. Host Gene Expression Changes in Cattle Infected with Alcelaphine Herpesvirus 1. Virus Res. 2012, 169, 246–254. [Google Scholar] [CrossRef]
- Patel, J.R.; Edington, N. The Detection and Behaviour of the Herpesvirus of Malignant Catarrhal Fever in Bovine Lymphocytes. Arch. Virol. 1981, 68, 321–326. [Google Scholar] [CrossRef]
- Li, H.; Shen, D.T.; Knowles, D.P.; Gorham, J.R.; Crawford, T.B. Competitive Inhibition Enzyme-Linked Immunosorbent Assay for Antibody in Sheep and Other Ruminants to a Conserved Epitope of Malignant Catarrhal Fever Virus. J. Clin. Microbiol. 1994, 32, 1674–1679. [Google Scholar] [CrossRef]
- Tham, K.M.; Ng, K.; Young, L.W. Polymerase Chain Reaction Amplification of Wildebeest-Associated and Cervine-Derived Malignant Catarrhal Fever Virus DNA. Arch. Virol. 1994, 135, 355–364. [Google Scholar] [CrossRef]
- Fraser, S.J.; Nettleton, P.F.; Dutia, B.M.; Haig, D.M.; Russell, G.C. Development of an Enzyme-Linked Immunosorbent Assay for the Detection of Antibodies against Malignant Catarrhal Fever Viruses in Cattle Serum. Vet. Microbiol. 2006, 116, 21–28. [Google Scholar] [CrossRef]
- Dewals, B.G.; Gillet, L.; Gerdes, T.; Taracha, E.L.N.; Thiry, E.; Vanderplasschen, A. Antibodies against Bovine Herpesvirus 4 Are Highly Prevalent in Wild African Buffaloes throughout Eastern and Southern Africa. Vet. Microbiol. 2005, 110, 209–220. [Google Scholar] [CrossRef]
- Rossiter, P.B. Immunoglobulin Response of Rabbits Infected with Malignant Catarrhal Fever Virus. Res. Vet. Sci. 1982, 33, 120–122. [Google Scholar] [CrossRef]
- Rossiter, P.B.; Mushi, E.Z.; Plowright, W. The Development of Antibodies in Rabbits and Cattle Infected Experimentally with an African Strain of Malignant Catarrhal Fever Virus. Vet. Microbiol. 1977, 2, 57–66. [Google Scholar] [CrossRef]
- Taus, N.S.; Cunha, C.W.; Marquard, J.; O’Toole, D.; Li, H. Cross-Reactivity of Neutralizing Antibodies among Malignant Catarrhal Fever Viruses. PLoS ONE 2015, 10, 145073. [Google Scholar] [CrossRef] [PubMed]
- Traul, D.L.; Elias, S.; Taus, N.S.; Herrmann, L.M.; Oaks, J.L.; Li, H. A Real-Time PCR Assay for Measuring Alcelaphine Herpesvirus-1 DNA. J. Virol. Methods 2005, 129, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.; Seal, B.; Ridpath, J. Molecular Diagnosis of Alcelaphine Herpesvirus (Malignant Catarrhal Fever) Infections by Nested Amplification of Viral DNA in Bovine Blood Buffy Coat Specimens. J. Vet. Diag. Invest. 1991, 3, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Bianchessi, L.; Rocchi, M.S.; Maley, M.; Piccinini, R.; Turin, L. Molecular Tools to Identify and Characterize Malignant Catarrhal Fever Viruses (MCFV) of Ruminants and Captive Artiodactyla. Viruses 2022, 14, 2697. [Google Scholar] [CrossRef] [PubMed]
- Edington, N.; Plowright, W. The Protection of Rabbits against the Herpesvirus of Malignant Catarrhal Fever by Inactivated Vaccines. Res. Vet. Sci. 1980, 28, 384–386. [Google Scholar] [CrossRef]
- Mirangi, P.K. Attempts to Immunize Cattle against Virulent African Malignant Catarrhal Fever Virus (Alcelaphine Herpesvirus-1) with a Herpesvirus Isolated from American Cattle. Vet. Microbiol. 1991, 28, 129–139. [Google Scholar] [CrossRef]
- Rossiter, P.B.; Gumm, I.D.; Mirangi, P.K. Immunological Relationships between Malignant Catarrhal Fever Virus (Alcelaphine Herpesvirus 1) and Bovine Cytomegalovirus (Bovine Herpesvirus 3). Vet. Microbiol. 1988, 16, 211–218. [Google Scholar] [CrossRef]
- Haig, D.M.; Grant, D.; Deane, D.; Campbell, I.; Thomson, J.; Jepson, C.; Buxton, D.; Russell, G.C. An Immunisation Strategy for the Protection of Cattle against Alcelaphine Herpesvirus-1-Induced Malignant Catarrhal Fever. Vaccine 2008, 26, 4461–4468. [Google Scholar] [CrossRef]
- Russell, G.C.; Haig, D.M.; Dagleish, M.P.; Todd, H.; Percival, A.; Grant, D.M.; Thomson, J.; Karagianni, A.E.; Benavides, J. Analysis of Immune Responses to Attenuated Alcelaphine Herpesvirus 1 Formulated with and without Adjuvant. Vaccine X 2021, 8, 100090. [Google Scholar] [CrossRef]
- Lankester, F.; Lugelo, A.; Werling, D.; Mnyambwa, N.; Keyyu, J.; Kazwala, R.; Grant, D.; Smith, S.; Parameswaran, N.; Cleaveland, S.; et al. The Efficacy of Alcelaphine Herpesvirus-1 (AlHV-1) Immunization with the Adjuvants Emulsigen® and the Monomeric TLR5 Ligand FliC in Zebu Cattle against AlHV-1 Malignant Catarrhal Fever Induced by Experimental Virus Challenge. Vet. Microbiol. 2016, 195, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Cook, E.; Russell, G.; Grant, D.; Mutisya, C.; Omoto, L.; Dobson, E.; Lankester, F.; Nene, V. A Randomised Vaccine Field Trial in Kenya Demonstrates Protection against Wildebeest-Associated Malignant Catarrhal Fever in Cattle. Vaccine 2019, 37, 5946–5953. [Google Scholar] [CrossRef]
- Cunha, C.W.; Baker, K.N.; O’Toole, D.; Cole, E.; Shringi, S.; Dewals, B.G.; Vanderplasschen, A.; Li, H. A Vaccine Targeting Ovine Herpesvirus 2 Glycoprotein B Protects against Sheep-Associated Malignant Catarrhal Fever. Vaccines 2022, 10, 2156. [Google Scholar] [CrossRef] [PubMed]
- Cunha, C.W.C.W.; Taus, N.S.N.S.; Dewals, B.G.B.G.; Vanderplasschen, A.; Knowles, D.P.D.P.; Li, H. Replacement of Glycoprotein B in Alcelaphine Herpesvirus 1 by Its Ovine Herpesvirus 2 Homolog: Implications in Vaccine Development for Sheep-Associated Malignant Catarrhal Fever. mSphere 2016, 1, e00108-16. [Google Scholar] [CrossRef] [PubMed]
- Swai, E.S.; Kapaga, A.M.; Sudi, F.; Loomu, P.M.; Joshua, G. Malignant Catarrhal Fever in Pastoral Maasai Herds Caused by Wildebeest Associated Alcelaphine Herpesvirus-1: An Outbreak Report. Vet. Res. Forum. 2013, 4, 133–136. [Google Scholar]
- Cleaveland, S.; Kusiluka, L.; Ole Kuwai, J.; Bell, C.; Kazwala, R. Assessing the Impact of Malignant Catarrhal Fever in Ngorongoro District, Tanzania. In Report to the DFID Animal Health Programme, Centre for Tropical Veterinary Medicine; University of Edinburgh: Edinburgh, UK, 2001. [Google Scholar]
- Bedelian, C.; Nkedianye, D.; Herrero, M. Maasai Perception of the Impact and Incidence of Malignant Catarrhal Fever (MCF) in Southern Kenya. Prev. Vet. Med. 2007, 78, 296–316. [Google Scholar] [CrossRef]
- Lankester, F.; Lugelo, A.; Kazwala, R.; Keyyu, J.; Cleaveland, S.; Yoder, J. The Economic Impact of Malignant Catarrhal Fever on Pastoralist Livelihoods. PLoS ONE 2015, 10, 116059. [Google Scholar] [CrossRef]
- Mlilo, D.; Mhlanga, M.; Mwembe, R.; Sisito, G.; Moyo, B.; Sibanda, B. The Epidemiology of Malignant Catarrhal Fever (MCF) and Contribution to Cattle Losses in Farms around Rhodes Matopos National Park, Zimbabwe. Trop. Anim. Health Prod. 2015, 47, 989–994. [Google Scholar] [CrossRef]
- Wambua, L.; Wambua, P.N.; Ramogo, A.M.; Mijele, D.; Otiende, M.Y. Wildebeest-Associated Malignant Catarrhal Fever: Perspectives for Integrated Control of a Lymphoproliferative Disease of Cattle in Sub-Saharan Africa. Arch. Virol. 2016, 161, 1–10. [Google Scholar] [CrossRef]
- Nthiwa, D.; Alonso, S.; Odongo, D.; Kenya, E.; Bett, B. A Participatory Epidemiological Study of Major Cattle Diseases amongst Maasai Pastoralists Living in Wildlife-Livestock Interfaces in Maasai Mara, Kenya. Trop. Anim. Health Prod. 2019, 51, 1097–1103. [Google Scholar] [CrossRef]
- Castro, A.E.; Heuschele, W.P. Conference on Malignant Catarrhal Fever. Bov. Pract. 1985, 20, 162–168. [Google Scholar]
- Srinivas, K.P.; Depledge, D.P.; Abebe, J.S.; Rice, S.A.; Mohr, I.; Wilson, A.C. Widespread Remodeling of the M6A RNA-Modification Landscape by a Viral Regulator of RNA Processing and Export. Proc. Natl. Acad. Sci. USA 2021, 118, e2104805118. [Google Scholar] [CrossRef] [PubMed]
- Baquero-Perez, B.; Antanaviciute, A.; Yonchev, I.D.; Carr, I.M.; Wilson, S.A.; Whitehouse, A. The Tudor SND1 Protein Is an M6A RNA Reader Essential for Replication of Kaposi’s Sarcoma-Associated Herpesvirus. eLife 2019, 8, e47261. [Google Scholar] [CrossRef] [PubMed]
| Genus | Virus Species | Abbreviations | Target Cells | Host Target | Associated Diseases |
|---|---|---|---|---|---|
| Lymphocryptovirus | Epstein-Barr virus, Human gammaherpesvirus 4 | EBV, HHV-4 | B cells | Human | Infectious mononucleosis, Nasopharyngeal carcinoma, Hodgkin’s and Burkitt’s lymphomas |
| Rhadinovirus | Kaposi’s sarcoma-associated herpesvirus, Human gammaherpesvirus 8 | KSHV, HHV-8 | B cells | Human | Kaposi’s sarcoma, Multicentric Castleman ‘s disease, Primary effusion lymphoma |
| Murine gammaherpesvirus 68, Murid gammaherpesvirus 4 | MHV-68, MuHV-4 | B cells/Macrophages | Rodents | B cell lymphomas | |
| Herpesvirus saimiri, Saimiriine gammaherpesvirus 2 | HVS, SaHV-2 | T cells | Squirrel monkey/non-human primates | T cell lymphomas | |
| Percavirus | Equid gammaherpesvirus 2 | EHV-2 | B cells | Horse | Respiratory symptoms |
| Macavirusa | Alcelaphine gammaherpesvirus 1 | AlHV-1 | T cells | Wildebeest/Other ruminants | Malignant Catarrhal Fever |
| Alcelaphine gammaherpesvirus 2 | AlHV-2 | Unknow | Hartebeest/Barbary red deer/Bison/Topi | Malignant Catarrhal Fever | |
| Alcelaphine gammaherpesvirus 2-like | AlHV-2-like | Unknown | Barbary red deer | Malignant Catarrhal Fever | |
| WD-MCFV-oryx | WD-MCFV-oryx | Unknown | Oryx | No disease | |
| Alcelaphine gammaherpesvirus 2 | AlHV-2 | Unknow | Hartebeest/Barbary red deer/Bison/Topi | Malignant Catarrhal Fever | |
| Ovine gammaherpesvirus 2 | OvHV-2 | T cells | Sheep/Other ruminants | Malignant Catarrhal Fever | |
| Caprine gammaherpesvirus 2 | CpHV-2 | Unknow | Goats/White-tailed deer/Sika deer | Malignant Catarrhal Fever | |
| WD-MCFV–white-tailed deer, Caprine gammaherpesvirus 3 | CpHV-3 | Unknow | Goat/White-tailed deer/ Red brocket deer /Reindeer | Malignant Catarrhal Fever | |
| Ibex-WD-MCFV | Ibex-WD-MCFV | Unknown | Ibex/Bongo/Anoa/Pronghorn | Malignant Catarrhal Fever | |
| Muskox-WD-MCFV | Muskox-WD-MCFV | Unknown | Muskox | No diseases | |
| Aoudad-WD-MCFV | Aoudad-WD-MCFV | Unknown | Aoudad | No diseases | |
| Bovine lymphotropic herpesvirus, Bovine gammaherpesvirus 6 | BoHV6 | B cells | Cattle | No disease | |
| Porcine lymphotropic herpesvirus 1, Suid gammaherpesvirus 3 | SuHV3 | Unknown | Swine | Post-transplantation lymphoproliferative disorder (PTLD) | |
| Porcine lymphotropic herpesvirus 2, Suid gammaherpesvirus 4 | SuHV4 | Unknown | Swine | Post-transplantation lymphoproliferative disorder (PTLD) | |
| Porcine lymphotropic herpesvirus 3, Suid gammaherpesvirus 5 | SuHV5 | Unknown | Swine | Post-transplantation lymphoproliferative disorder (PTLD) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, M.; Myster, F.; van Campe, W.; Roels, S.; Mostin, L.; van den Berg, T.; Vanderplasschen, A.; Dewals, B.G. Wildebeest-Derived Malignant Catarrhal Fever: A Bovine Peripheral T Cell Lymphoma Caused by Cross-Species Transmission of Alcelaphine Gammaherpesvirus 1. Viruses 2023, 15, 526. https://doi.org/10.3390/v15020526
Gong M, Myster F, van Campe W, Roels S, Mostin L, van den Berg T, Vanderplasschen A, Dewals BG. Wildebeest-Derived Malignant Catarrhal Fever: A Bovine Peripheral T Cell Lymphoma Caused by Cross-Species Transmission of Alcelaphine Gammaherpesvirus 1. Viruses. 2023; 15(2):526. https://doi.org/10.3390/v15020526
Chicago/Turabian StyleGong, Meijiao, Françoise Myster, Willem van Campe, Stefan Roels, Laurent Mostin, Thierry van den Berg, Alain Vanderplasschen, and Benjamin G. Dewals. 2023. "Wildebeest-Derived Malignant Catarrhal Fever: A Bovine Peripheral T Cell Lymphoma Caused by Cross-Species Transmission of Alcelaphine Gammaherpesvirus 1" Viruses 15, no. 2: 526. https://doi.org/10.3390/v15020526
APA StyleGong, M., Myster, F., van Campe, W., Roels, S., Mostin, L., van den Berg, T., Vanderplasschen, A., & Dewals, B. G. (2023). Wildebeest-Derived Malignant Catarrhal Fever: A Bovine Peripheral T Cell Lymphoma Caused by Cross-Species Transmission of Alcelaphine Gammaherpesvirus 1. Viruses, 15(2), 526. https://doi.org/10.3390/v15020526






