StM171, a Stenotrophomonas maltophilia Bacteriophage That Affects Sensitivity to Antibiotics in Host Bacteria and Their Biofilm Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains Source and Culture Conditions
2.2. Phage Isolation and Purification
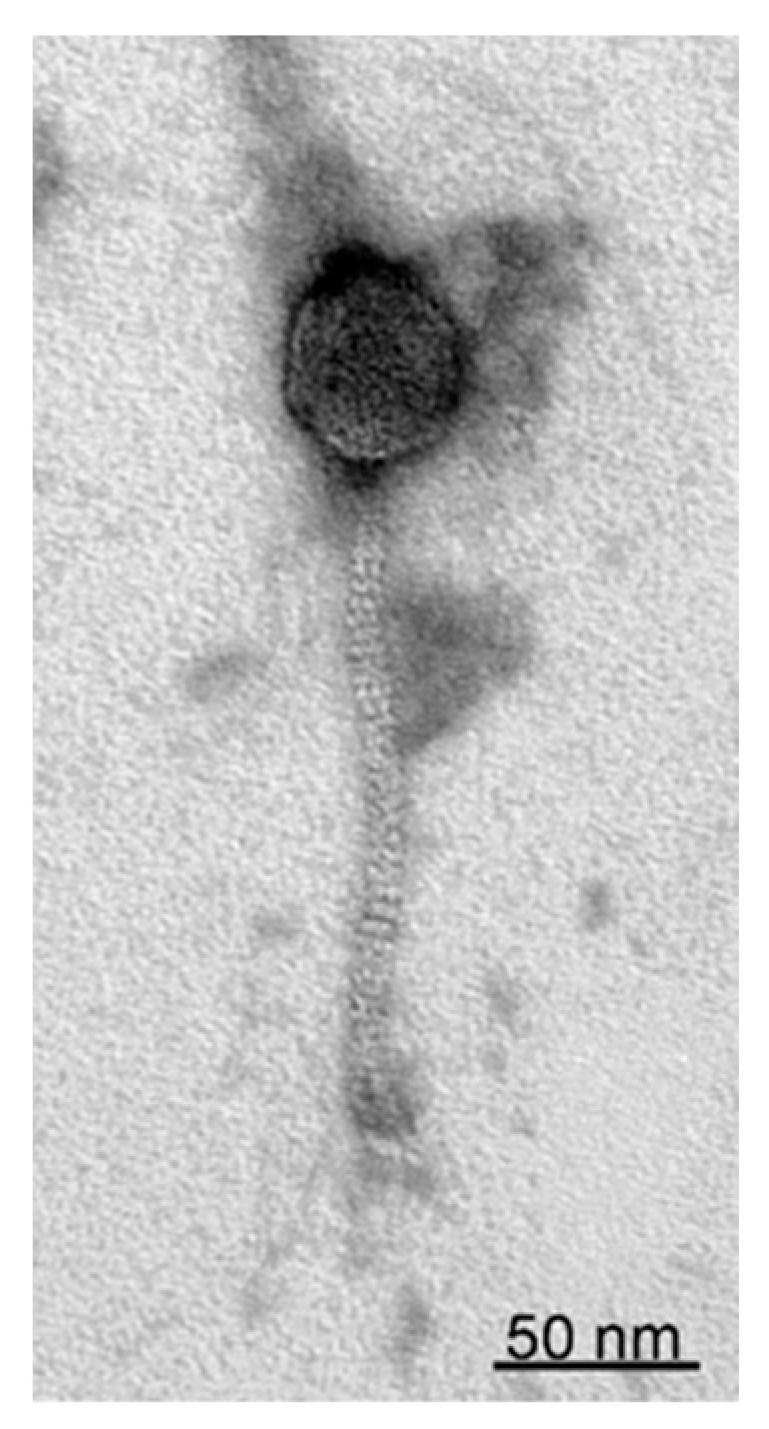
2.3. Transmission Electron Microscopy
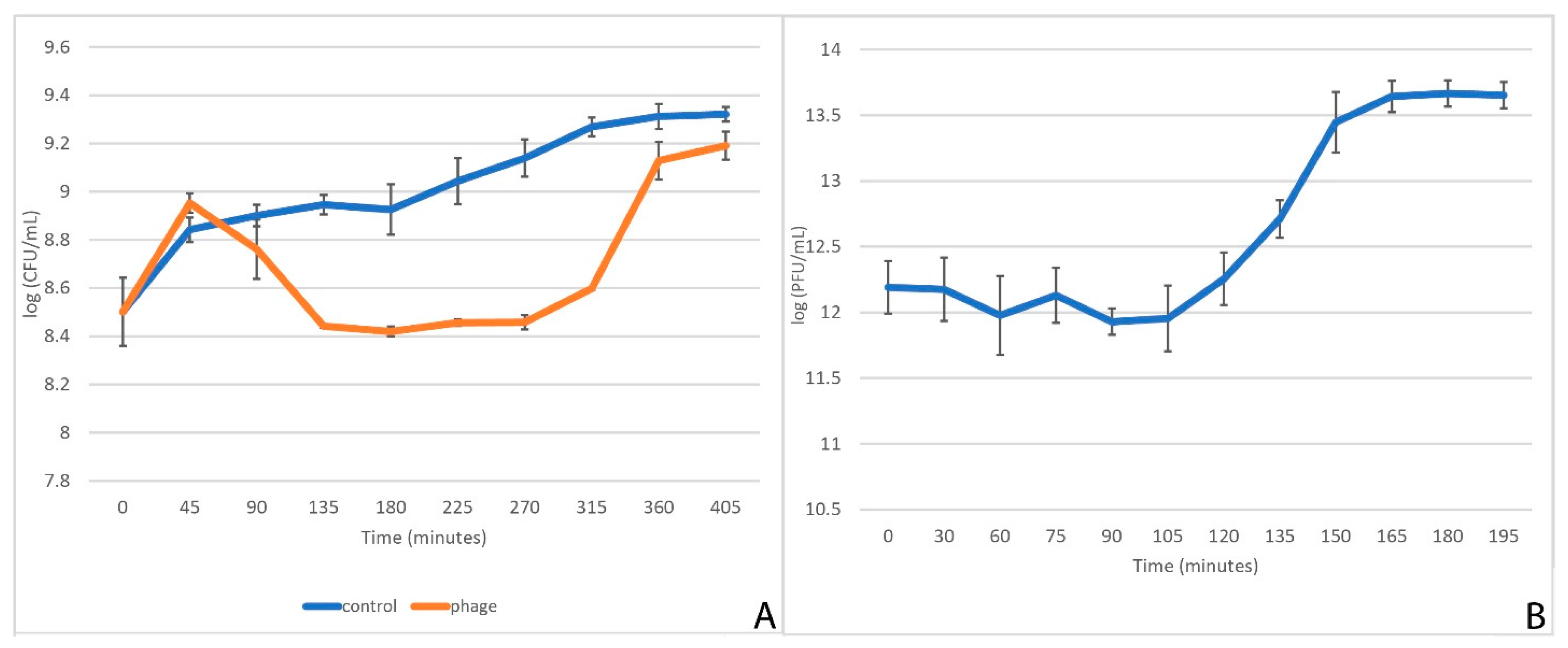
2.4. One-Step Growth Curve
2.5. Kinetics of Cell Culture Lysis
2.6. Phage Host Range Analysis
2.7. StM171’s DNA Isolation and Sequencing
2.8. Phage Genome Analysis
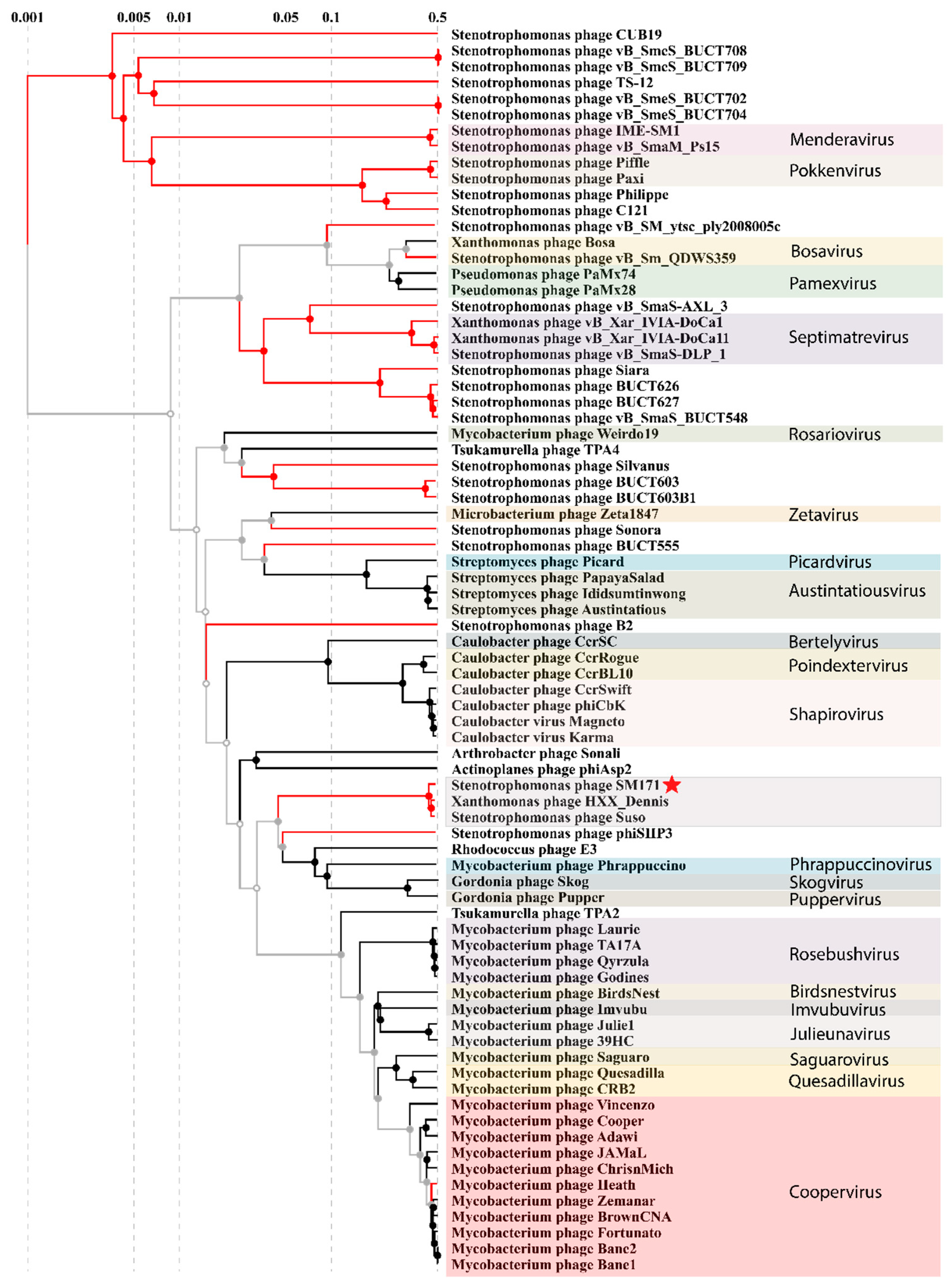
2.9. Phylogenetic Analysis
2.10. Bacterial Host Genome Sequencing and Analysis
2.11. Antibiotic Resistance of S. maltophilia Strains
2.12. Anti-Biofilm Activity of StM171 with and without Antibiotics
2.13. Propagating Bacterial Strains with Resistance to StM171 and Retesting Their Resistance to Antibiotics
2.14. Statistics
3. Results
3.1. Characteristics of StM171 and Its Growth Dynamics
3.2. StM171 Genomics and Genome Organization
3.3. StM171 Phylogenetic Analysis
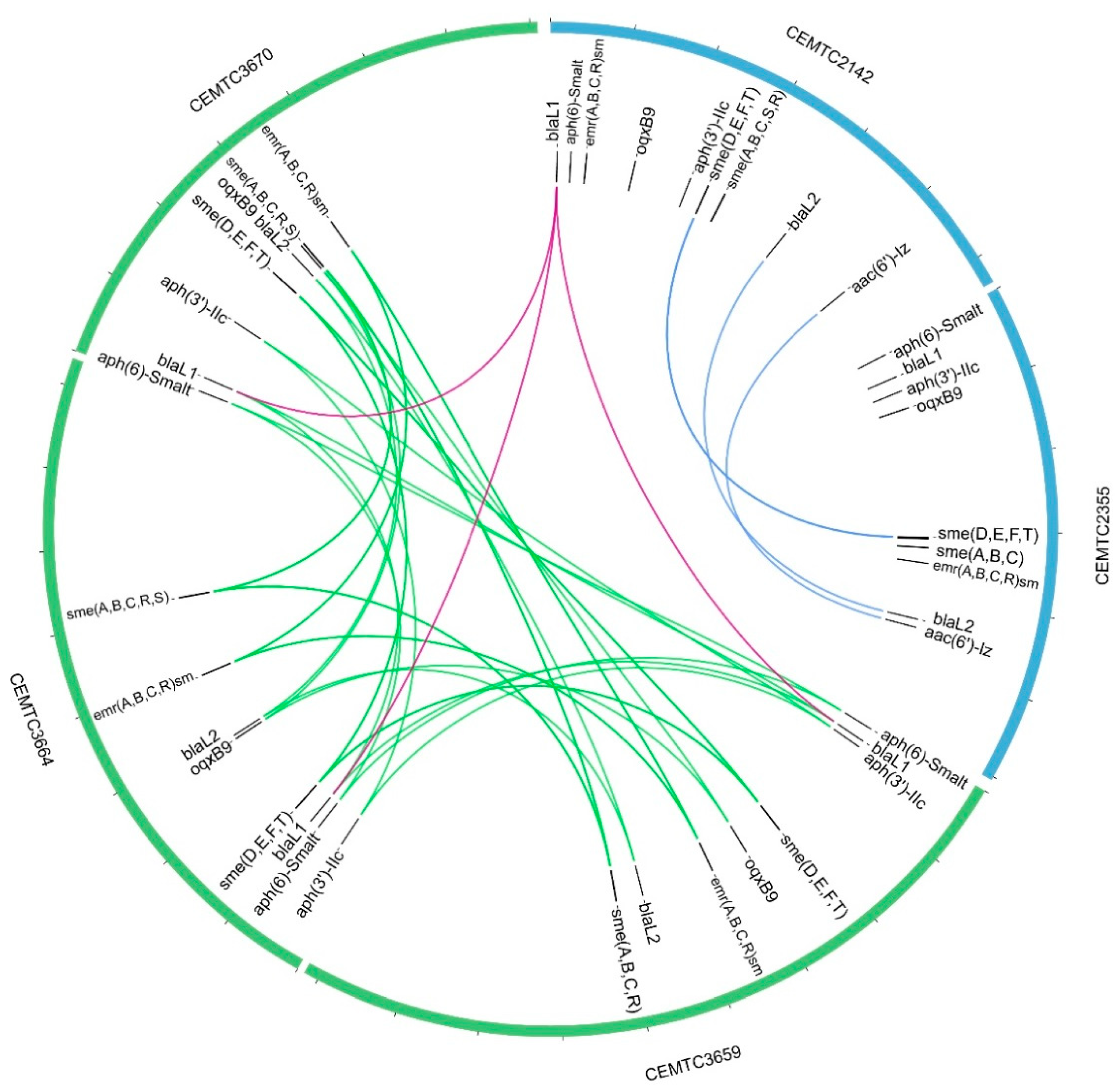
3.4. Analysis of the StM171 Bacterial Host Genomes
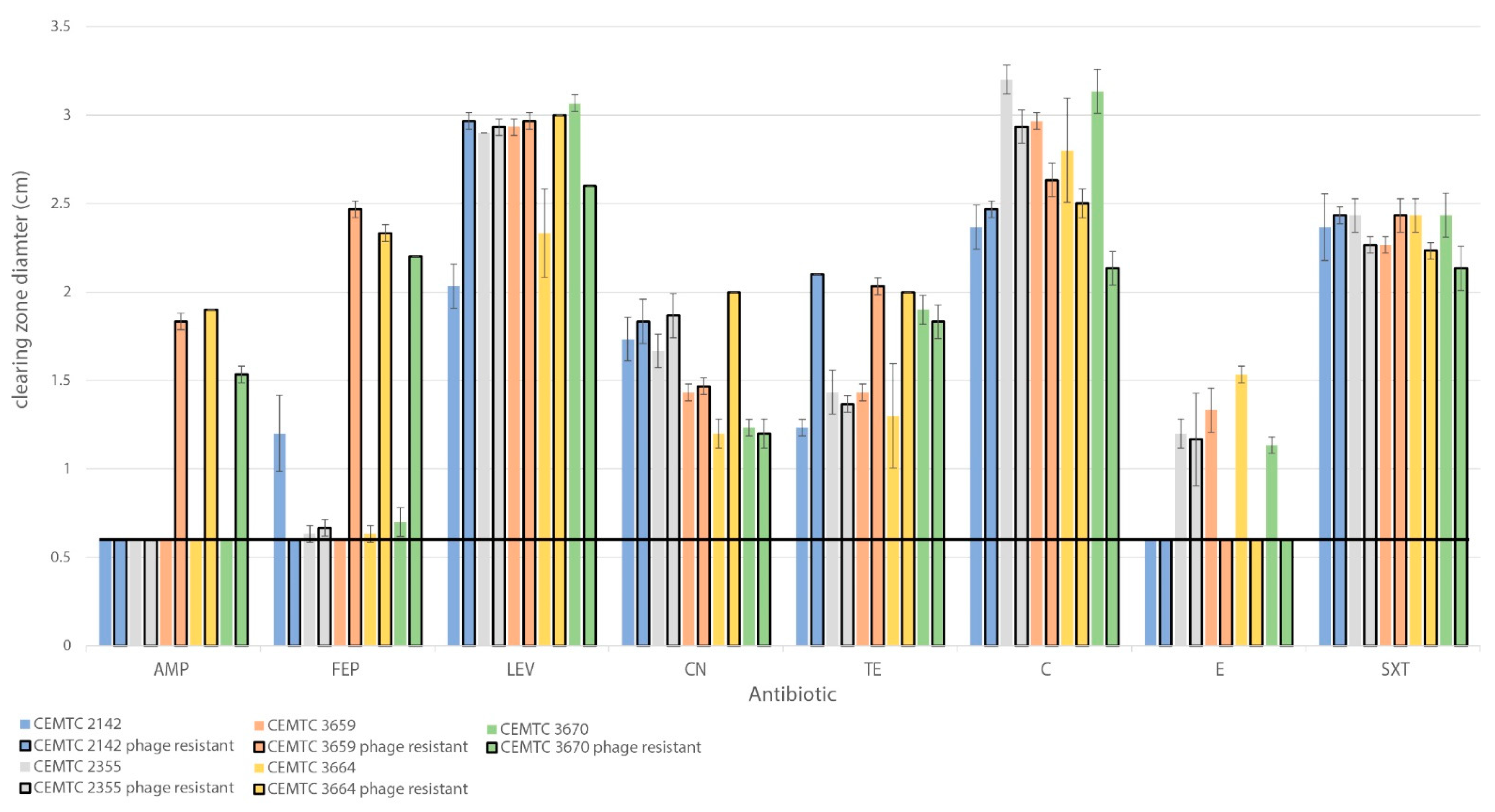
3.5. Changes in Antibiotic Sensitivity in S. maltophilia Clones That Developed Resistance to StM171
3.6. Effect of StM171 with and without Antibiotics against Biofilm Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooke, J.S. Stenotrophomonas maltophilia: An Emerging Global Opportunistic Pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Sánchez, L.E.; Vinuesa, P. Evolutionary Genetic Analysis Uncovers Multiple Species with Distinct Habitat Preferences and Antibiotic Resistance Phenotypes in the Stenotrophomonas maltophilia Complex. Front. Microbiol. 2017, 8, 1548. [Google Scholar] [CrossRef] [PubMed]
- Hugh, R.; Leifson, E. A Description of the Type Strain of Pseudomonas Maltophilia1. Int. J. Syst. Evol. Microbiol. 1963, 13, 133–138. [Google Scholar] [CrossRef]
- Hoefel, D.; Monis, P.T.; Grooby, W.L.; Andrews, S.; Saint, C.P. Profiling Bacterial Survival through a Water Treatment Process and Subsequent Distribution System. J. Appl. Microbiol. 2005, 99, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.D.; Jeong, W.Y.; Kim, M.H.; Jung, I.Y.; Ahn, M.Y.; Ann, H.W.; Ahn, J.Y.; Han, S.H.; Choi, J.Y.; Song, Y.G.; et al. Risk Factors for Mortality in Patients with Stenotrophomonas maltophilia Bacteremia. Medicine 2016, 95, e4375. [Google Scholar] [CrossRef] [PubMed]
- Harris, N.B.; Rogers, D.G. Septicemia Associated with Stenotrophomonas maltophilia in a West African Dwarf Crocodile (Osteolaemus tetraspis Subsp. Tetraspis). J. Veter. Diagn. Investig. 2001, 13, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Roskot, N.; Smalla, K. Genotypic and Phenotypic Relationships between Clinical and Environmental Isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 1999, 37, 3594–3600. [Google Scholar] [CrossRef]
- Berg, G.; Eberl, L.; Hartmann, A. The Rhizosphere as a Reservoir for Opportunistic Human Pathogenic Bacteria. Environ. Microbiol. 2005, 7, 1673–1685. [Google Scholar] [CrossRef]
- Berg, G. Plant-Microbe Interactions Promoting Plant Growth and Health: Perspectives for Controlled Use of Microorganisms in Agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Brooke, J.S. New Strategies against Stenotrophomonas maltophilia: A Serious Worldwide Intrinsically Drug-Resistant Opportunistic Pathogen. Expert Rev. Anti-Infect. Ther. 2014, 12, 1–4. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; Confalone, P.; Nicoletti, M.; Petrucca, A.; Guarnieri, S.; Fiscarelli, E.; Savini, V.; Piccolomini, R.; Di Bonaventura, G. Adhesion to and Biofilm Formation on IB3-1 Bronchial Cells by Stenotrophomonas maltophilia Isolates from Cystic Fibrosis Patients. BMC Microbiol. 2010, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An Emerging Opportunist Human Pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Li, J. Antimicrobial Resistance in Stenotrophomonas maltophilia: Mechanisms and Clinical Implications. In Antimicrobial Drug Resistance; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 937–958. ISBN 978-3-319-47264-5. [Google Scholar]
- Trecarichi, E.M.; Tumbarello, M. Antimicrobial-Resistant Gram-Negative Bacteria in Febrile Neutropenic Patients with Cancer: Current Epidemiology and Clinical Impact. Curr. Opin. Infect. Dis. 2014, 27, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.N.; Wang, F.D.; Wang, L.S.; Liu, C.Y.; Liu, I.M. Xanthomonas Maltophilia Bacteremia: An Analysis of 32 Cases. J. Formos. Med. Assoc. 1992, 91, 1170–1176. [Google Scholar] [PubMed]
- Victor, M.A.; Arpi, M.; Bruun, B.; Jønsson, V.; Hansen, M.M. Xanthomonas Maltophilia Bacteremia in Immunocompromised Hematological Patients. Scand. J. Infect. Dis. 1994, 26, 163–170. [Google Scholar] [CrossRef]
- Sánchez, M.B. Antibiotic Resistance in the Opportunistic Pathogen Stenotrophomonas maltophilia. Front. Microbiol. 2015, 6, 658. [Google Scholar] [CrossRef]
- Pompilio, A.; Savini, V.; Fiscarelli, E.; Gherardi, G.; Di Bonaventura, G. Clonal Diversity, Biofilm Formation, and Antimicrobial Resistance among Stenotrophomonas maltophilia Strains from Cystic Fibrosis and Non-Cystic Fibrosis Patients. J. Antibiot. 2020, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Prosseda, G.; Del Chierico, F.; Cannavacciuolo, S.; Cipriani, P.; Petrucca, A.; Superti, F.; Ammendolia, M.G.; Concato, C.; Fiscarelli, E.; et al. Molecular Characterization of Virulence Determinants of Stenotrophomonas maltophilia Strains Isolated from Patients Affected by Cystic Fibrosis. Int. J. Immunopathol. Pharmacol. 2007, 20, 529–537. [Google Scholar] [CrossRef]
- Wang, A.; Wang, Q.; Kudinha, T.; Xiao, S.; Zhuo, C. Effects of Fluoroquinolones and Azithromycin on Biofilm Formation of Stenotrophomonas maltophilia. Sci. Rep. 2016, 6, 29701. [Google Scholar] [CrossRef]
- WHO|World Health Organization. Available online: https://www.who.int/drugresistance/AMR_Importance/en/ (accessed on 7 March 2021).
- Peters, D.L.; McCutcheon, J.G.; Dennis, J.J. Characterization of Novel Broad-Host-Range Bacteriophage DLP3 Specific to Stenotrophomonas maltophilia as a Potential Therapeutic Agent. Front. Microbiol. 2020, 11, 1358. [Google Scholar] [CrossRef]
- Peters, D.L.; McCutcheon, J.G.; Stothard, P.; Dennis, J.J. Novel Stenotrophomonas maltophilia Temperate Phage DLP4 Is Capable of Lysogenic Conversion. BMC Genom. 2019, 20, 300. [Google Scholar] [CrossRef] [PubMed]
- Morozova, V.; Babkin, I.; Kozlova, Y.; Baykov, I.; Bokovaya, O.; Tikunov, A.; Ushakova, T.; Bardasheva, A.; Ryabchikova, E.; Zelentsova, E.; et al. Isolation and Characterization of a Novel Klebsiella Pneumoniae N4-like Bacteriophage KP8. Viruses 2019, 11, 1115. [Google Scholar] [CrossRef] [PubMed]
- Morozova, V.; Fofanov, M.; Tikunova, N.; Babkin, I.; Morozov, V.V.; Tikunov, A. First crAss-Like Phage Genome Encoding the Diversity-Generating Retroelement (DGR). Viruses 2020, 12, 573. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Pajunen, M.; Kiljunen, S.; Skurnik, M. Bacteriophage φYeO3-12, Specific for Yersinia Enterocolitica Serotype O:3, Is Related to Coliphages T3 and T7. J. Bacteriol. 2000, 182, 5114–5120. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Coffey, A.; Edwards, R.; Meaney, W.; Fitzgerald, G.F.; Ross, R.P. Genome of Staphylococcal Phage K: A New Lineage of Myoviridae Infecting Gram-Positive Bacteria with a Low G+C Content. J. Bacteriol. 2004, 186, 2862–2871. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- States, D.J.; Gish, W. Combined Use of Sequence Similarity and Codon Bias for Coding Region Identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at Its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Gene Prediction; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1962, pp. 1–14. [Google Scholar] [CrossRef]
- Proksee—Genome Analysis. Available online: https://proksee.ca/ (accessed on 23 April 2023).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Grant, J.R.; Van Domselaar, G. Visualizing and Comparing Circular Genomes Using the CGView Family of Tools. Brief. Bioinform. 2019, 20, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- GC Content Calculator|VectorBuilder. Available online: https://en.vectorbuilder.com/tool/gc-content-calculator.html (accessed on 2 May 2023).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic Assignment of Uncultivated Prokaryotic Virus Genomes Is Enabled by Gene-Sharing Networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef]
- Petit, R.A.; Read, T.D. Bactopia: A Flexible Pipeline for Complete Analysis of Bacterial Genomes. mSystems 2020, 5, e00190-20. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Shen, W. Csvtk—A Cross-Platform, Efficient and Practical CSV/TSV Toolkit. Available online: https://github.com/shenwei356/csvtk (accessed on 20 April 2023).
- Seemann, T. ABRicate. Available online: https://github.com/tseemann/abricate (accessed on 20 April 2023).
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A Database for Classification of Antimicrobial Drug, Biocide and Metal Resistance Determinants in Metagenomic Sequence Data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Titus Brown, C.; Irber, L. Sourmash: A Library for MinHash Sketching of DNA. JOSS 2016, 1, 27. [Google Scholar] [CrossRef]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate Paired Shotgun Read Merging via Overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Song, L.; Florea, L.; Langmead, B. Lighter: Fast and Memory-Efficient Sequencing Error Correction without Counting. Genome Biol. 2014, 15, 509. [Google Scholar] [CrossRef]
- Andrews, S. FastQC. Available online: https://github.com/s-andrews/FastQC (accessed on 20 April 2023).
- Petit, R. Fastq-Scan. Available online: https://github.com/rpetit3/fastq-scan (accessed on 20 April 2023).
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- The Galaxy Community. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2022 Update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Azimi, A.; Aslanimehr, M.; Yaseri, M.; Shadkam, M.; Douraghi, M. Distribution of Smf-1, rmlA, spgM and rpfF Genes among Stenotrophomonas maltophilia Isolates in Relation to Biofilm-Forming Capacity. J. Glob. Antimicrob. Resist. 2020, 23, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.S.; Malamud, F.; Rigano, L.A.; Russo, D.M.; Marano, M.R.; Castagnaro, A.P.; Zorreguieta, A.; Bouarab, K.; Dow, J.M.; Vojnov, A.A. Controlled Synthesis of the DSF Cell–Cell Signal Is Required for Biofilm Formation and Virulence in Xanthomonas Campestris. Environ. Microbiol. 2007, 9, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z. Role of the Acetyltransferase AAC(6’)-Iz Modifying Enzyme in Aminoglycoside Resistance in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2003, 51, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Lewis, K.; Matin, A. EmrR Is a Negative Regulator of the Escherichia Coli Multidrug Resistance Pump EmrAB. J. Bacteriol. 1995, 177, 2328–2334. [Google Scholar] [CrossRef]
- Li, X.-Z.; Zhang, L.; Poole, K. SmeC, an Outer Membrane Multidrug Efflux Protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2002, 46, 333–343. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.Z.; Poole, K. SmeDEF Multidrug Efflux Pump Contributes to Intrinsic Multidrug Resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001, 45, 3497–3503. [Google Scholar] [CrossRef]
- Materon, I.C.; Queenan, A.M.; Koehler, T.M.; Bush, K.; Palzkill, T. Biochemical Characterization of Beta-Lactamases Bla1 and Bla2 from Bacillus Anthracis. Antimicrob. Agents Chemother. 2003, 47, 2040–2042. [Google Scholar] [CrossRef]
- Kim, S.-K.; Demuth, M.; Schlesinger, S.R.; Kim, S.J.; Urbanczyk, J.; Shaw, R.W.; Shin, H. Inhibition of Bacillus Anthracis Metallo-β-Lactamase by Compounds with Hydroxamic Acid Functionality. J. Enzym. Inhib. Med. Chem. 2016, 31, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Succi, J.; Tenover, F.C.; Koehler, T.M. Beta-Lactamase Genes of the Penicillin-Susceptible Bacillus Anthracis Sterne Strain. J. Bacteriol. 2003, 185, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, A.; Avison, M.B. Aph(3′)-IIc, an Aminoglycoside Resistance Determinant from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2007, 51, 359–360. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Wang, M.; Park, C.H.; Kim, E.-C.; Jacoby, G.A.; Hooper, D.C. oqxAB Encoding a Multidrug Efflux Pump in Human Clinical Isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 3582–3584. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.-Z.; Poole, K. Multiple Antibiotic Resistance in Stenotrophomonas maltophilia: Involvement of a Multidrug Efflux System. Antimicrob. Agents Chemother. 2000, 44, 287–293. [Google Scholar] [CrossRef]
- Barber, O.W.; Miramontes, I.M.; Jain, M.; Ozer, E.A.; Hartmann, E.M. The Future of Bacteriophage Therapy Will Promote Antimicrobial Susceptibility. mSystems 2021, 6, e00218-21. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jdeed, G.; Morozova, V.; Kozlova, Y.; Tikunov, A.; Ushakova, T.; Bardasheva, A.; Manakhov, A.; Mitina, M.; Zhirakovskaya, E.; Tikunova, N. StM171, a Stenotrophomonas maltophilia Bacteriophage That Affects Sensitivity to Antibiotics in Host Bacteria and Their Biofilm Formation. Viruses 2023, 15, 2455. https://doi.org/10.3390/v15122455
Jdeed G, Morozova V, Kozlova Y, Tikunov A, Ushakova T, Bardasheva A, Manakhov A, Mitina M, Zhirakovskaya E, Tikunova N. StM171, a Stenotrophomonas maltophilia Bacteriophage That Affects Sensitivity to Antibiotics in Host Bacteria and Their Biofilm Formation. Viruses. 2023; 15(12):2455. https://doi.org/10.3390/v15122455
Chicago/Turabian StyleJdeed, Ghadeer, Vera Morozova, Yuliya Kozlova, Artem Tikunov, Tatyana Ushakova, Alevtina Bardasheva, Andrey Manakhov, Maria Mitina, Elena Zhirakovskaya, and Nina Tikunova. 2023. "StM171, a Stenotrophomonas maltophilia Bacteriophage That Affects Sensitivity to Antibiotics in Host Bacteria and Their Biofilm Formation" Viruses 15, no. 12: 2455. https://doi.org/10.3390/v15122455
APA StyleJdeed, G., Morozova, V., Kozlova, Y., Tikunov, A., Ushakova, T., Bardasheva, A., Manakhov, A., Mitina, M., Zhirakovskaya, E., & Tikunova, N. (2023). StM171, a Stenotrophomonas maltophilia Bacteriophage That Affects Sensitivity to Antibiotics in Host Bacteria and Their Biofilm Formation. Viruses, 15(12), 2455. https://doi.org/10.3390/v15122455







