Epidemiological Insights into the Omicron Outbreak via MeltArray-Assisted Real-Time Tracking of SARS-CoV-2 Variants
Abstract
:1. Introduction
2. Methods
2.1. Plasmids and Virus Strains
2.2. Sample Collection and Processing
2.3. MeltArray Assay
2.4. Analytical Evaluation of MeltArray Assay
2.5. NGS
2.6. ddPCR Assays for XBB.1.16 and XBB.1.9
2.7. Statistical Analysis
3. Results
3.1. MeltArray Assay Design
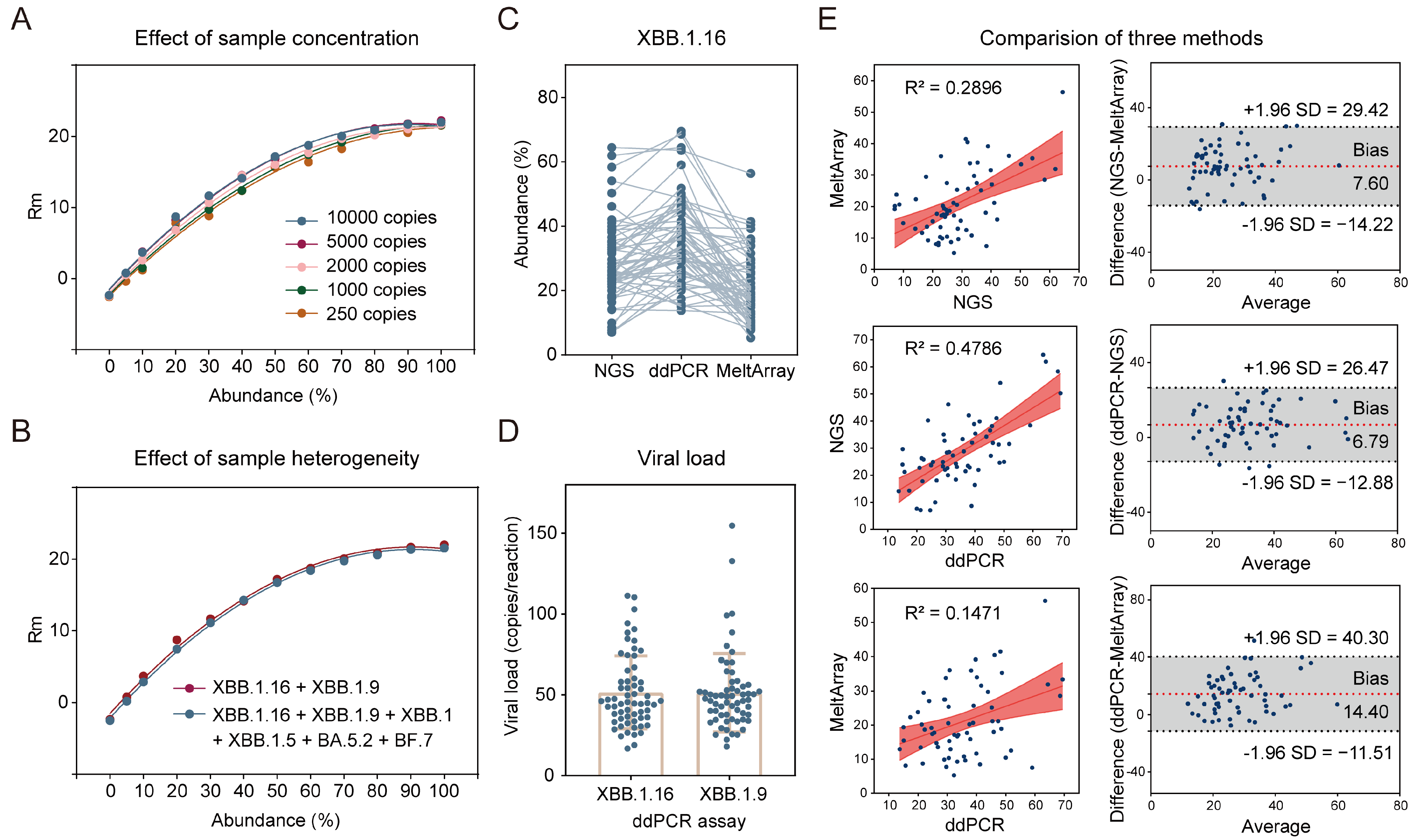
3.2. Analytical Performance of MeltArray Assay
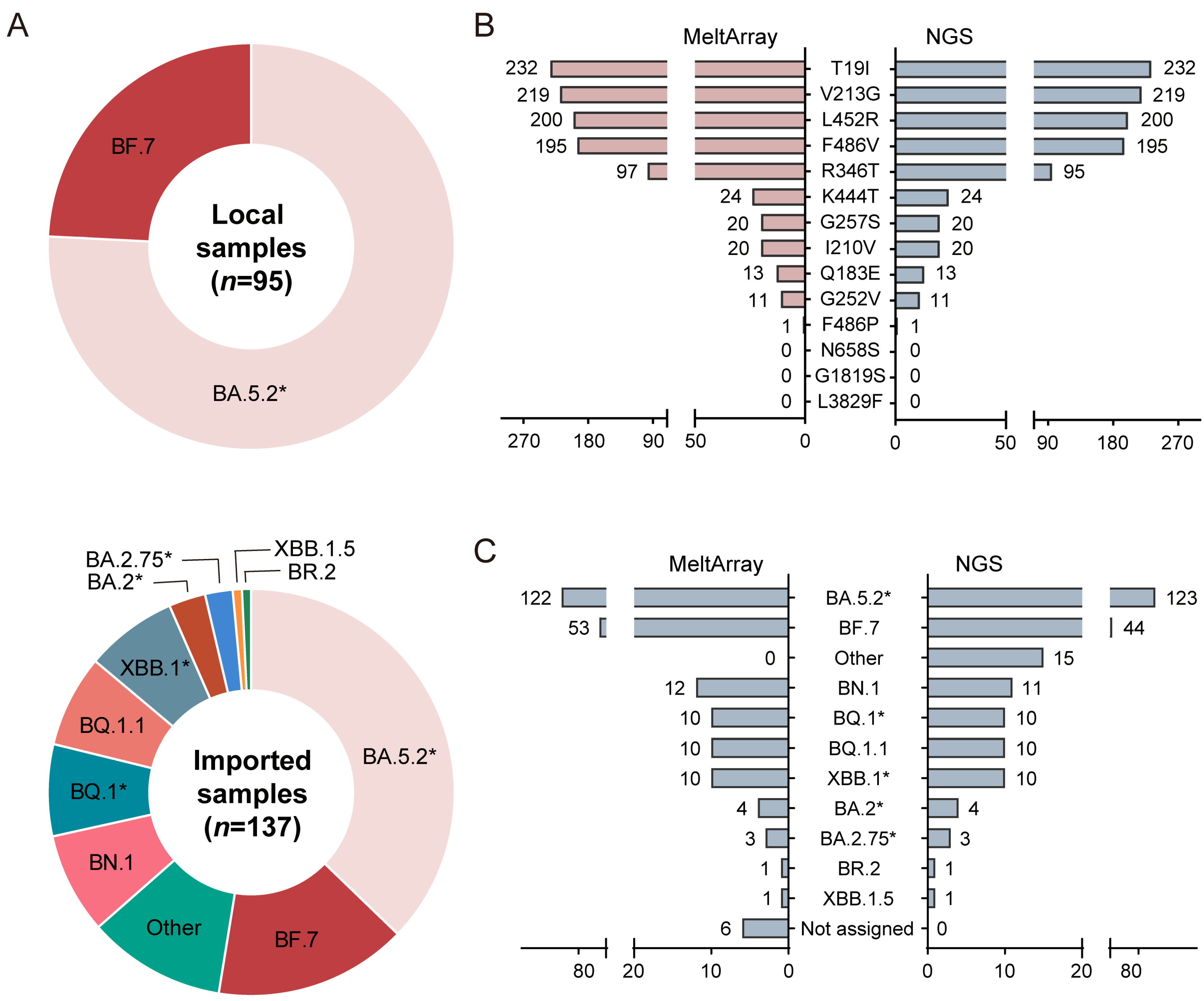
3.3. Clinical Evaluation of MeltArray Assay
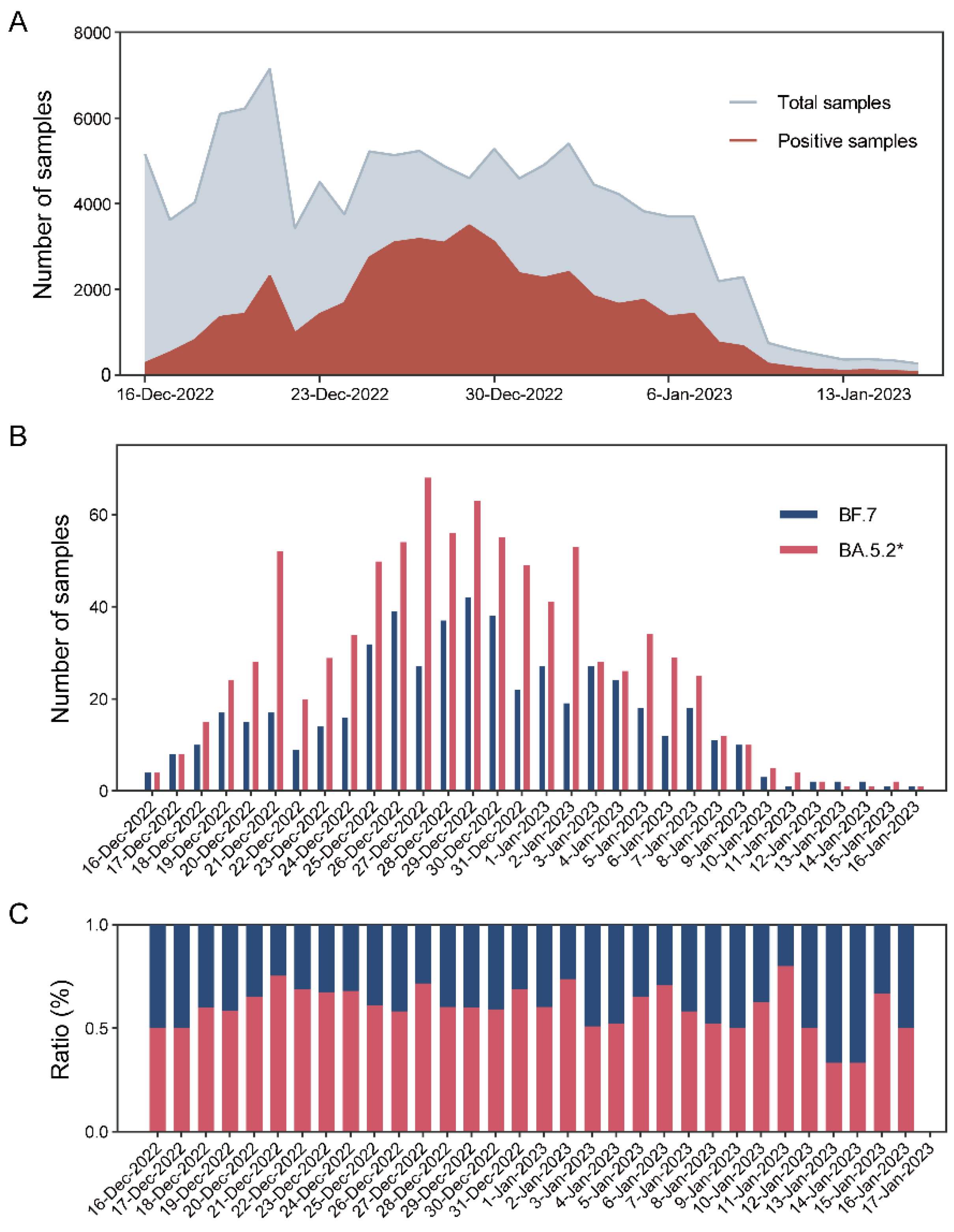
3.4. Local Epidemic Surveillance during the Omicron Outbreak
3.5. Potential for Estimation of Variant Abundance via MeltArray Assay
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.; et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Wang, L.; Jiao, F.; Yu, X.; Xu, W.; Huang, Z.; Li, X.; Wang, Q.; Zhu, Y.; Man, Q.; et al. SARS-CoV-2 Omicron subvariants exhibit distinct fusogenicity, but similar sensitivity, to pan-CoV fusion inhibitors. Emerg. Microbes Infect. 2023, 12, 2178241. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Evans, J.P.; Faraone, J.N.; Zheng, Y.-M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Lozanski, G.; et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 2023, 31, 9–17. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, S.; Lu, F. Impact of National Omicron Outbreak at the end of 2022 on the future outlook of COVID-19 in China. Emerg. Microbes Infect. 2023, 12, 2191738. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, L.; Feng, Z.; Xu, H.; Li, F.; Shen, Y.; Zhang, D.; Liu, W.J.; Gao, G.F.; Wang, Q. Characterisation of SARS-CoV-2 variants in Beijing during 2022: An epidemiological and phylogenetic analysis. Lancet 2023, 401, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Adikari, T.N.; Ferguson, J.M.; Hammond, J.M.; Stevanovski, I.; Beukers, A.G.; Naing, Z.; Yeang, M.; Verich, A.; Gamaarachchi, H.; et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020, 11, 6272. [Google Scholar] [CrossRef]
- Chiara, M.; D’Erchia, A.M.; Gissi, C.; Manzari, C.; Parisi, A.; Resta, N.; Zambelli, F.; Picardi, E.; Pavesi, G.; Horner, D.S.; et al. Next generation sequencing of SARS-CoV-2 genomes: Challenges, applications and opportunities. Brief. Bioinform. 2021, 22, 616–630. [Google Scholar] [CrossRef]
- Vega-Magaña, N.; Sánchez-Sánchez, R.; Hernández-Bello, J.; Venancio-Landeros, A.A.; Peña-Rodríguez, M.; Vega-Zepeda, R.A.; Galindo-Ornelas, B.; Díaz-Sánchez, M.; García-Chagollán, M.; Macedo-Ojeda, G.; et al. RT-qPCR assays for rapid detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 mutations: A screening strategy to identify variants with clinical impact. Front. Cell. Infect. Microbiol. 2021, 11, 672562. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, G.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Iaconelli, M.; Lucentini, L.; Bonadonna, L.; Brusaferro, S.; Brandtner, D.; Fasanella, A.; et al. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021, 197, 117104. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, D.; Du, C.; Liu, Q.; Lin, S.; Liang, L.; Xu, Y.; Liao, Y.; Li, Q. Highly multiplex PCR assays by coupling the 5′-flap endonuclease activity of Taq DNA polymerase and molecular beacon reporters. Proc. Natl. Acad. Sci. USA 2022, 119, e2110672119. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, M.; Deng, Y.; Xu, X.; Lin, D.; Zhang, Y.; Li, S.; Ding, J.; Shi, X.; Yau, C.I.; et al. A rapid, high-throughput, and sensitive PEG-precipitation method for SARS-CoV-2 wastewater surveillance. Water Res. 2023, 230, 119560. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Du, C.; Lv, Z.; Wang, F.; Zhou, L.; Peng, Y.; Li, W.; Fu, Y.; Song, J.; Jia, C.; et al. Longitudinal wastewater surveillance addressed public health priorities during the transition from “dynamic COVID-zero” to “opening up” in China: A population-based study. medRxiv 2023. [Google Scholar] [CrossRef]
- Yan, T.; Xu, Y.; Zheng, R.; Zeng, X.; Chen, Z.; Lin, S.; Xia, Z.; Liao, Y.; Zhang, Y.; Li, Q. Accessible and adaptable multiplexed real-time PCR approaches to identify SARS-CoV-2 variants of concern. Microbiol. Spectr. 2022, 10, e0322222. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Wang, L.; Zhu, Y.; Lu, L.; Jiang, S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct. Target. Ther. 2022, 7, 241. [Google Scholar] [CrossRef]
- Wang, H.; Miller, J.A.; Verghese, M.; Sibai, M.; Solis, D.; Mfuh, K.O.; Jiang, B.; Iwai, N.; Mar, M.; Huang, C.; et al. Multiplex SARS-CoV-2 Genotyping Reverse Transcriptase PCR for Population-Level Variant Screening and Epidemiologic Surveillance. J. Clin. Microbiol. 2021, 59, e0085921. [Google Scholar] [CrossRef]
- Babiker, A.; Immergluck, K.; Stampfer, S.D.; Rao, A.; Bassit, L.; Su, M.; Nguyen, V.; Stittleburg, V.; Ingersoll, J.M.; Bradley, H.L.; et al. Single-Amplicon Multiplex Real-Time Reverse Transcription-PCR with Tiled Probes To Detect SARS-CoV-2 spike Mutations Associated with Variants of Concern. J. Clin. Microbiol. 2021, 59, e0144621. [Google Scholar] [CrossRef]
- Banada, P.; Green, R.; Banik, S.; Chopoorian, A.; Streck, D.; Jones, R.; Chakravorty, S.; Alland, D. A simple RT-PCR melting temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. J Clin Microbiol. 2021, 59, e0084521. [Google Scholar] [CrossRef]
- Zhao, Y.; Lee, A.; Composto, K.; Cunningham, M.H.; Mediavilla, J.R.; Fennessey, S.; Corvelo, A.; Chow, K.F.; Zody, M.; Chen, L.; et al. A novel diagnostic test to screen SARS-CoV-2 variants containing E484K and N501Y mutations. Emerg. Microbes Infect. 2021, 10, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-Y.; Jian, M.-J.; Chang, C.-K.; Lin, J.-C.; Yeh, K.-M.; Chen, C.-W.; Hsieh, S.-S.; Hung, K.-S.; Tang, S.-H.; Perng, C.-L.; et al. Emergency SARS-CoV-2 Variants of Concern: Novel Multiplex Real-Time RT-PCR Assay for Rapid Detection and Surveillance. Microbiol. Spectr. 2022, 10, e0251321. [Google Scholar] [CrossRef]
- Rybicka, M.; Miłosz, E.; Bielawski, K.P. Superiority of MALDI-TOF Mass Spectrometry over Real-Time PCR for SARS-CoV-2 RNA Detection. Viruses 2021, 13, 730. [Google Scholar] [CrossRef]
- Lanyon, H.E.; Hoyle, J.S.; Downard, K.M. Resolving omicron sub-variants of SARS CoV-2 coronavirus with MALDI mass spectrometry. Analyst 2023, 148, 966–972. [Google Scholar] [CrossRef]
- Zhao, F.; Lu, J.; Lu, B.; Qin, T.; Wang, X.; Hou, X.; Meng, F.; Xu, X.; Li, T.; Zhou, H.; et al. A Novel Strategy for the Detection of SARS-CoV-2 Variants Based on Multiplex PCR-Mass Spectrometry Minisequencing Technology. Microbiol. Spectr. 2021, 9, e0126721. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Hirunpatrawong, P.; Petcharat, S.; Torvorapanit, P.; Jitsatja, A.; Thippamom, N.; Ninwattana, S.; Phanlop, C.; Buathong, R.; Tangwangvivat, R.; et al. Simultaneous detection of omicron and other SARS-CoV-2 variants by multiplex PCR MassARRAY technology. Sci. Rep. 2023, 13, 2089. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, M.; Lin, W.; Dong, W.; Xu, J. Evolutionary analysis of Omicron variant BF.7 and BA.5.2 pandemic in China. J. Biosaf. Biosecur. 2023, 5, 14–20. [Google Scholar] [CrossRef]
- Lu, G.; Ling, Y.; Jiang, M.; Tan, Y.; Wei, D.; Jiang, L.; Yu, S.; Jiang, F.; Wang, S.; Dai, Y.; et al. Primary assessment of the diversity of Omicron sublineages and the epidemiologic features of autumn/winter 2022 COVID-19 wave in Chinese mainland. Front. Med. 2023, 17, 1–10. [Google Scholar] [CrossRef]
- La Rosa, G.; Brandtner, D.; Ferraro, G.B.; Veneri, C.; Mancini, P.; Iaconelli, M.; Lucentini, L.; Del Giudice, C.; Orlandi, L.; Suffredini, E.; et al. Wastewater surveillance of SARS-CoV-2 variants in October–November 2022 in Italy: Detection of XBB.1, BA.2.75 and rapid spread of the BQ.1 lineage. Sci. Total. Environ. 2023, 873, 162339. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome sequencing of sewage detects re-gionally prevalent SARS-CoV-2 variants. mBio 2021, 12, e02703-20. [Google Scholar] [CrossRef]
- Van Poelvoorde, L.A.E.; Delcourt, T.; Coucke, W.; Herman, P.; De Keersmaecker, S.C.J.; Saelens, X.; Roosens, N.H.C.; Vanneste, K. Strategy and Performance Evaluation of Low-Frequency Variant Calling for SARS-CoV-2 Using Targeted Deep Illumina Sequencing. Front. Microbiol. 2021, 12, 747458. [Google Scholar] [CrossRef]
- Safford, H.R.; Shapiro, K.; Bischel, H.N. Opinion: Wastewater analysis can be a powerful public health tool-if it’s done sensibly. Proc. Natl. Acad. Sci. USA 2022, 119, e2119600119. [Google Scholar] [CrossRef]
- Dächert, C.; Muenchhoff, M.; Graf, A.; Autenrieth, H.; Bender, S.; Mairhofer, H.; Wratil, P.R.; Thieme, S.; Krebs, S.; Grzimek-Koschewa, N.; et al. Rapid and sensitive identification of omicron by variant-specific PCR and nanopore sequencing: Paradigm for diagnostics of emerging SARS-CoV-2 variants. Med. Microbiol. Immunol. 2022, 211, 71–77. [Google Scholar] [CrossRef] [PubMed]

| Gene Target | Codon Change | Channel | Tm (℃) | CV (%) | Limit of Detection |
|---|---|---|---|---|---|
| (Mean ± 3SD) | (Copies/Reaction) | ||||
| T19I | ACA > ATA | ROX | 66.02 ± 0.24 | 0.12 | 50 |
| Q183E | CAG > GAG | Cy5 | 82.69 ± 0.21 | 0.08 | 50 |
| I210V | ATT > GTT | HEX | 81.76 ± 0.24 | 0.10 | 50 |
| V213G | GTG > GGG | FAM | 80.53 ± 0.23 | 0.10 | 50 |
| G252V | GGT > GTT | Cy5 | 71.87 ± 0.22 | 0.10 | 50 |
| G257S | GGT > AGT | FAM | 73.40 ± 0.21 | 0.10 | 50 |
| R346T | AGA > ACA | HEX | 71.51 ± 0.22 | 0.10 | 50 |
| K444T | AAG > ACG | Cy5 | 67.98 ± 0.16 | 0.08 | 50 |
| L452R | CTG > CGG | ROX | 81.55 ± 0.24 | 0.10 | 50 |
| F486V | TTT > GTT | Cy5 | 77.63 ± 0.23 | 0.10 | 50 |
| F486P | TTT > CCT | ROX | 71.22 ± 0.24 | 0.11 | 50 |
| N658S | AAC > AGC | HEX | 66.71 ± 0.18 | 0.09 | 50 |
| G1819S | GGT > AGT | Quasar 705 | 70.90 ± 0.23 | 0.11 | 50 |
| L3829F | CTC > TTC | Atto 425 | 70.98 ± 0.30 | 0.14 | 50 |
| Variant | MeltArray Assay/NGS | Sensitivity% (95% CI) | Specificity% (95% CI) | PPV% (95% CI) | NPV% (95% CI) | Accuracy% (95% CI) | Kappa | |||
|---|---|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/+ | −/− | |||||||
| BA.5.2* | 117 | 5 | 6 | 104 | 95.12 (89.77–97.75) | 95.41 (89.71–98.02) | 95.90 (90.76–98.24) | 94.55 (88.61–97.48) | 95.26 (91.71–97.33) | 0.90 (0.78–1.03) |
| BF.7 | 44 | 9 | 0 | 179 | 100 (91.97–100) | 95.21 (91.15–97.46) | 83.02 (70.77–90.80) | 100 (97.90–100) | 96.12 (92.79–97.95) | 0.88 (0.76–1.01) |
| BN.1 | 11 | 1 | 0 | 220 | 100 (74.12–100) | 99.55 (97.48–99.92) | 91.67 (64.61–98.51) | 100 (98.28–100) | 99.57 (97.60–99.92) | 0.95 (0.83–1.08) |
| BQ.1* | 10 | 0 | 0 | 222 | 100 (72.25–100) | 100 (98.30–100) | 100 (72.25–100) | 100 (98.30–100) | 100 (98.37–100) | 1 (0.87–1.13) |
| BQ.1.1 | 10 | 0 | 0 | 222 | 100 (72.25–100) | 100 (98.30–100) | 100 (72.25–100) | 100 (98.30–100) | 100 (98.37–100) | 1 (0.87–1.13) |
| XBB.1* | 10 | 0 | 0 | 222 | 100 (72.25–100) | 100 (98.30–100) | 100 (72.25–100) | 100 (98.30–100) | 100 (98.37–100) | 1 (0.87–1.13) |
| BA.2* | 4 | 0 | 0 | 228 | 100 (51.01–100) | 100 (98.34–100) | 100 (51.01–100) | 100 (98.34–100) | 100 (98.37–100) | 1 (0.87–1.13) |
| BA.2.75* | 3 | 0 | 0 | 229 | 100 (43.85–100) | 100 (98.35–100) | 100 (43.85–100) | 100 (98.35–100) | 100 (98.37–100) | 1 (0.87–1.13) |
| XBB.1.5 | 1 | 0 | 0 | 231 | 100 (20.65–100) | 100 (98.36–100) | 100 (20.65–100) | 100 (98.36–100) | 100 (98.37–100) | 1 (0.87–1.13) |
| BR.2 | 1 | 0 | 0 | 231 | 100 (20.65–100) | 100 (98.36–100) | 100 (20.65–100) | 100 (98.36–100) | 100 (98.37–100) | 1 (0.87–1.13) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, T.; Zheng, R.; Li, Y.; Sun, S.; Zeng, X.; Yue, Z.; Liao, Y.; Hu, Q.; Xu, Y.; Li, Q. Epidemiological Insights into the Omicron Outbreak via MeltArray-Assisted Real-Time Tracking of SARS-CoV-2 Variants. Viruses 2023, 15, 2397. https://doi.org/10.3390/v15122397
Yan T, Zheng R, Li Y, Sun S, Zeng X, Yue Z, Liao Y, Hu Q, Xu Y, Li Q. Epidemiological Insights into the Omicron Outbreak via MeltArray-Assisted Real-Time Tracking of SARS-CoV-2 Variants. Viruses. 2023; 15(12):2397. https://doi.org/10.3390/v15122397
Chicago/Turabian StyleYan, Ting, Rongrong Zheng, Yinghui Li, Siyang Sun, Xiaohong Zeng, Zhijiao Yue, Yiqun Liao, Qinghua Hu, Ye Xu, and Qingge Li. 2023. "Epidemiological Insights into the Omicron Outbreak via MeltArray-Assisted Real-Time Tracking of SARS-CoV-2 Variants" Viruses 15, no. 12: 2397. https://doi.org/10.3390/v15122397
APA StyleYan, T., Zheng, R., Li, Y., Sun, S., Zeng, X., Yue, Z., Liao, Y., Hu, Q., Xu, Y., & Li, Q. (2023). Epidemiological Insights into the Omicron Outbreak via MeltArray-Assisted Real-Time Tracking of SARS-CoV-2 Variants. Viruses, 15(12), 2397. https://doi.org/10.3390/v15122397





