Pathogenicity in Chickens and Turkeys of a 2021 United States H5N1 Highly Pathogenic Avian Influenza Clade 2.3.4.4b Wild Bird Virus Compared to Two Previous H5N8 Clade 2.3.4.4 Viruses
Abstract
:1. Introduction
2. Material and Methods
2.1. Viruses
2.2. Animals and Housing
2.3. Experimental Design
2.4. Viral Titration in Swabs and Tissues
2.5. Sequence Analyses
3. Results
3.1. Infectivity, Pathogenicity, and Transmission of the H5 HPAIVs in Chickens
3.2. Infectivity, Pathogenicity, and Transmission of the H5 HPAIVs in Turkeys
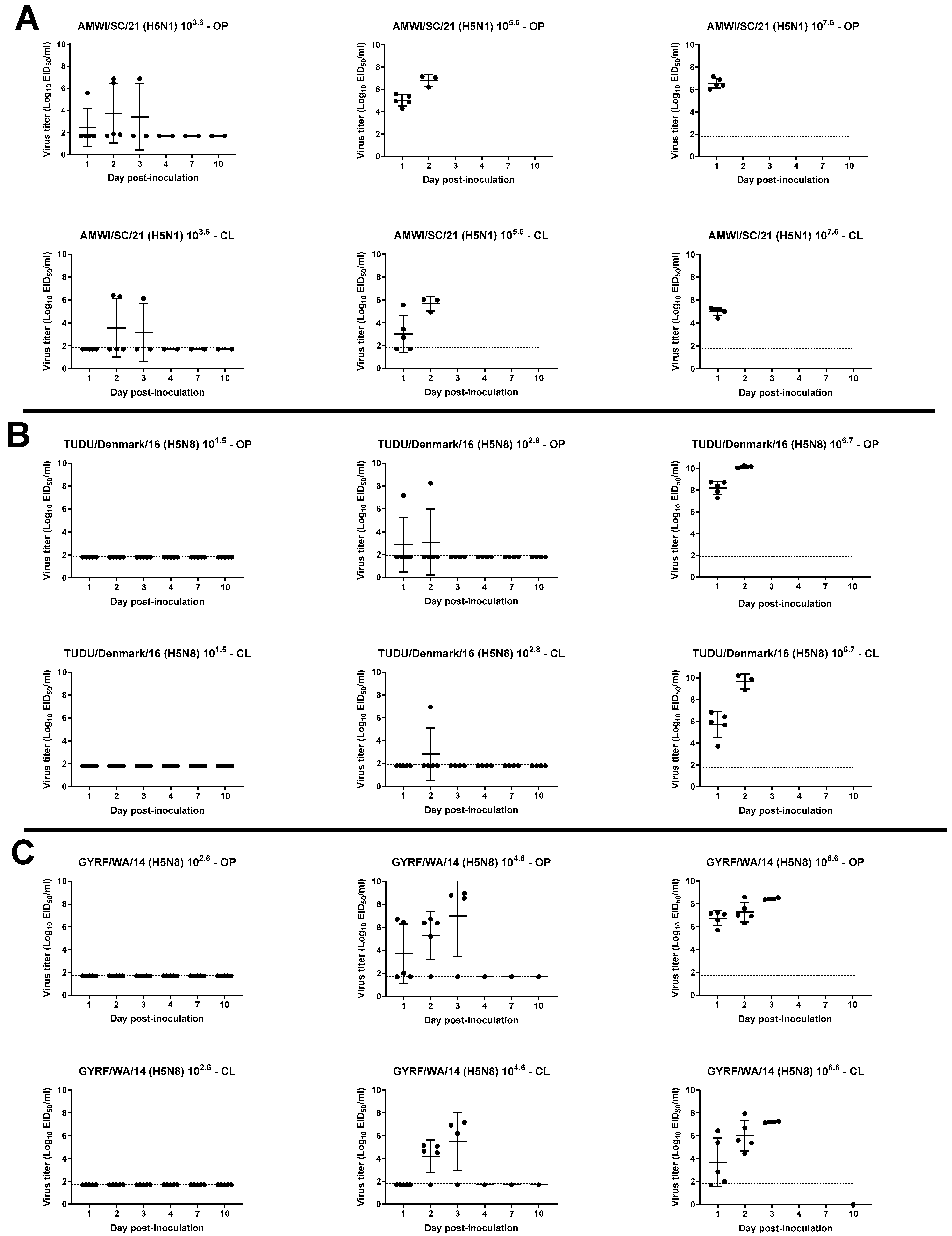
3.3. Viral Shedding in Chickens
3.4. Virus Shedding in Turkeys
3.5. Gross and Microscopic Lesions and Virus Detection in Tissues
3.6. Sequence Comparisons of the H5 HPAIVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sims, L.D.; Brown, I.H. Multi-continental panzootic of H5 highly pathogenic avian influenza (1996–2015). In Animal Influenza; John Wiley & Sons, Inc.: Ames, IA, USA, 2016; pp. 202–247. [Google Scholar]
- Sims, L.; Harder, T.; Brown, I.; Gaidet, N.; Belot, G.; von Dobschuetz, S.; Kamata, A.; Kivaria, F.; Palamara, E.; Bruni, M.; et al. Highly Pathogenic H5 Avian Influenza in 2016 and 2017—Observations and Future Perspectives; FOCUS ON; Food and Agriculture Organization of the United Nations Emergency Prevention System: Rome, Italy, 2017. [Google Scholar]
- Lee, D.H.; Bertran, K.; Kwon, J.H.; Swayne, D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 2017, 18, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.S.; Banyard, A.C.; Whittard, E.; Karibayev, T.; Al Kafagi, T.; Chvala, I.; Byrne, A.; Meruyert Akberovna, S.; King, J.; Harder, T.; et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes Infect. 2021, 10, 148–151. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control; European Reference Laboratory; Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Marangon, S.; Niqueux, E.; Staubach, C.; et al. Avian influenza overview April–June 2023. ESFA J. 2023, 21, 8191. [Google Scholar]
- World Organisation for Animal Health (WOAH). HPAI SITUATION—Update. Available online: https://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/OIE_AI_situation_report/HPAI_asof14012021.pdf (accessed on 15 August 2023).
- Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science 2016, 354, 213–217. [Google Scholar] [CrossRef]
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly. Epidemiol. Rec. 2022, 95, 525–539. [Google Scholar]
- Beerens, N.; Heutink, R.; Bergervoet, S.A.; Harders, F.; Bossers, A.; Koch, G. Multiple Reassorted Viruses as Cause of Highly Pathogenic Avian Influenza A(H5N8) Virus Epidemic, the Netherlands, 2016. Emerg. Infect. Dis. 2017, 23, 1966–1973. [Google Scholar] [CrossRef]
- Pohlmann, A.; Starick, E.; Harder, T.; Grund, C.; Höper, D.; Globig, A.; Staubach, C.; Dietze, K.; Strebelow, G.; Ulrich, R.G.; et al. Outbreaks among Wild Birds and Domestic Poultry Caused by Reassorted Influenza A(H5N8) Clade 2.3.4.4 Viruses, Germany, 2016. Emerg. Infect. Dis. 2017, 23, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.; Monne, I.; Mulatti, P.; Zecchin, B.; Bonfanti, L.; Ormelli, S.; Milani, A.; Cecchettin, K.; Lemey, P.; Moreno, A.; et al. Genetic Diversity of Highly Pathogenic Avian Influenza A(H5N8/H5N5) Viruses in Italy, 2016–2017. Emerg. Infect. Dis. 2017, 23, 1543–1547. [Google Scholar] [CrossRef]
- Lycett, S.J.; Pohlmann, A.; Staubach, C.; Caliendo, V.; Woolhouse, M.; Beer, M.; Kuiken, T.; Global Consortium for H5N8 and Related Influenza Viruses. Genesis and spread of multiple reassortants during the 2016/2017 H5 avian influenza epidemic in Eurasia. Proc. Natl. Acad. Sci. USA 2020, 117, 20814–20825. [Google Scholar] [CrossRef]
- Khomenko, S.; Abolnik, C.; Roberts, L.; Waller, L.; Shaw, K.; Monne, I.; Taylor, J.; Dhingra, M.; Pittiglio, C.; Mugyeom, M.; et al. 2016–2018 Spread of H5N8 Highly Pathogenic Avian Influenza (HPAI) in Sub-Saharan Africa: Epidemiological and Ecological Observations; FOCUS ON 12; Food and Agriculture Organization of the United Nations Emergency Prevention System: Rome, Italy, 2018. [Google Scholar]
- Poen, M.J.; Venkatesh, D.; Bestebroer, T.M.; Vuong, O.; Scheuer, R.D.; Oude Munnink, B.B.; de Meulder, D.; Richard, M.; Kuiken, T.; Koopmans, M.P.G.; et al. Co-circulation of genetically distinct highly pathogenic avian influenza A clade 2.3.4.4 (H5N6) viruses in wild waterfowl and poultry in Europe and East Asia, 2017–2018. Virus Evol. 2019, 5, vez004. [Google Scholar] [CrossRef]
- Alarcon, P.; Brouwer, A.; Venkatesh, D.; Duncan, D.; Dovas, C.I.; Georgiades, G.; Monne, I.; Fusaro, A.; Dan, A.; Śmietanka, K.; et al. Comparison of 2016–17 and Previous Epizootics of Highly Pathogenic Avian Influenza H5 Guangdong Lineage in Europe. Emerg. Infect. Dis. 2018, 24, 2270–2283. [Google Scholar] [CrossRef]
- Molini, U.; Aikukutu, G.; Roux, J.-P.; Kemper, J.; Ntahonshikira, C.; Marruchella, G.; Khaiseb, S.; Cattoli, G.; Dundon, W.G. Avian influenza H5N8 outbreak in African penguins (Spheniscus demersus), Namibia, 2019. J. Wildl. Dis. 2019, 56, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, D.; Brouwer, A.; Goujgoulova, G.; Ellis, R.; Seekings, J.; Brown, I.H.; Lewis, N.S. Regional Transmission and Reassortment of 2.3.4.4b Highly Pathogenic Avian Influenza (HPAI) Viruses in Bulgarian Poultry 2017/18. Viruses 2020, 12, 605. [Google Scholar] [CrossRef]
- Mine, J.; Uchida, Y.; Sharshov, K.; Sobolev, I.; Shestopalov, A.; Saito, T. Phylogeographic evidence for the inter- and intracontinental dissemination of avian influenza viruses via migration flyways. PLoS ONE 2019, 14, e0218506. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.; Zecchin, B.; Vrancken, B.; Abolnik, C.; Ademun, R.; Alassane, A.; Arafa, A.; Awuni, J.A.; Couacy-Hymann, E.; Coulibaly, M.B.; et al. Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza. Nat. Commun. 2019, 10, 5310. [Google Scholar] [CrossRef]
- Baek, Y.-G.; Lee, Y.-N.; Lee, D.-H.; Cheon, S.-H.; Kye, S.-J.; Park, Y.-R.; Si, Y.-J.; Lee, M.-H.; Lee, Y.-J. A novel reassortant clade 2.3.4.4 highly pathogenic avian influenza H5N6 virus identified in South Korea in 2018. Infect. Genet. Evol. 2020, 78, 104056. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Avian Influenza Portal. 2020. Available online: http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/ (accessed on 5 January 2021).
- Isoda, N.; Twabela, A.T.; Bazarragchaa, E.; Ogasawara, K.; Hayashi, H.; Wang, Z.J.; Kobayashi, D.; Watanabe, Y.; Saito, K.; Kida, H.; et al. Re-Invasion of H5N8 High Pathogenicity Avian Influenza Virus Clade 2.3.4.4b in Hokkaido, Japan, 2020. Viruses 2020, 12, 1439. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Lee, D.-H.; Kwon, J.-H.; Kim, Y.-J.; Lee, S.-H.; Cho, A.Y.; Kim, T.-H.; Park, J.-E.; Lee, S.-I.; Song, C.-S. Highly Pathogenic Avian Influenza Clade 2.3.4.4b Subtype H5N8 Virus Isolated from Mandarin Duck in South Korea, 2020. Viruses 2020, 12, 1389. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture, Animal and Plant Health Inspection Service (USDA-APHIS), 2022–2023 Confirmations of Highly Pathogenic Avian Influenza in Commercial and Backyard Flocks. Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/avian/avian-influenza/hpai-2022/2022-hpai-commercial-backyard-flocks (accessed on 31 July 2023).
- U.S. Department of Agriculture, Animal and Plant Health Inspection Service (USDA-APHIS), 2022–2023 Detections of Hhighly Pathogenic Avian Influenza in Wild Birds. Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/avian/avian-influenza/hpai-2022/2022-hpai-wild-birds (accessed on 31 July 2023).
- Bevins, S.N.; Shriner, S.A.; Cumbee, J.C., Jr.; Dilione, K.E.; Douglass, K.E.; Ellis, J.W.; Killian, M.L.; Torchetti, M.K.; Lenoch, J.B. Intercontinental Movement of Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4 Virus to the United States, 2021. Emerg. Infect. Dis. 2022, 28, 1006–1011. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture, Animal and Plant Health Inspection Service (USDA-APHIS), 2023. Epidemiologic and Other Analyses of HPAI Affected Poultry Flocks July 2022 Interim Report. USDA:APHIS:VS: Center for Epidemiology and Animal Health, Fort Collins, CO, p. 74. July 2022. Available online: https://www.aphis.usda.gov/animal_health/downloads/animal_diseases/ai/epi-analyses-hpai-poultry-july2022.pdf (accessed on 31 July 2023).
- Youk, S.; Torchetti, M.K.; Lantz, K.; Lenoch, J.B.; Killian, M.L.; Leyson, C.; Bevins, S.N.; Dilione, K.; Ip, H.S.; Stallknecht, D.E.; et al. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: Introductions into the United States and reassortments, December 2021–April 2022. Virology 2023, 587, 109860. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture, Animal and Plant Health Inspection Service (USDA-APHIS), 2022–2023. Detections of Highly Pathogenic Avian Influenza in Mammals. Available online: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/avian/avian-influenza/hpai-2022/2022-hpai-mammals (accessed on 31 July 2023).
- Spackman, E.; Pantin-Jackwood, M.J.; Lee, S.A.; Prosser, D. The pathogenesis of a 2022 North American highly pathogenic clade 2.3.4.4b H5N1 avian influenza virus in mallards (Anas platyrhynchos). Avian Pathol. 2023, 52, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Pantin-Jackwood, M.J.; Kapczynski, D.R.; Swayne, D.E.; Suarez, D.L. H5N2 Highly Pathogenic Avian Influenza Viruses from the US 2014–2015 outbreak have an unusually long pre-clinical period in turkeys. BMC Vet.Res. 2016, 12, 260. [Google Scholar] [CrossRef]
- Leyson, C.; Youk, S.S.; Smith, D.; Dimitrov, K.; Lee, D.H.; Larsen, L.E.; Swayne, D.E.; Pantin-Jackwood, M.J. Pathogenicity and genomic changes of a 2016 European H5N8 highly pathogenic avian influenza virus (clade 2.3.4.4) in experimentally infected mallards and chickens. Virology 2019, 537, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Killian, M.L. Avian Influenza Virus Isolation, Propagation, and Titration in Embryonated Chicken Eggs. Methods Mol. Biol. 2020, 2123, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Swayne, D.E.; Slemons, R.D. Using Mean Infectious Dose of High- and Low-Pathogenicity Avian Influenza Viruses Originating from Wild Duck and Poultry as One Measure of Infectivity and Adaptation to Poultry. Avian Dis. 2008, 52, 455–460. [Google Scholar] [CrossRef]
- Youk, S.S.; Lee, D.H.; Leyson, C.M.; Smith, D.; Criado, M.F.; DeJesus, E.; Swayne, D.E.; Pantin-Jackwood, M.J. Loss of Fitness of Mexican H7N3 Highly Pathogenic Avian Influenza Virus in Mallards after Circulating in Chickens. J. Virol. 2019, 93, e00543-19. [Google Scholar] [CrossRef] [PubMed]
- Pantin-Jackwood, M.J.; Stephens, C.B.; Bertran, K.; Swayne, D.E.; Spackman, E. The pathogenesis of H7N8 low and highly pathogenic avian influenza viruses from the United States 2016 outbreak in chickens, turkeys and mallards. PLoS ONE 2017, 12, e0177265. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J. Immunohistochemical Staining of Influenza Virus in Tissues. Methods Mol. Biol. 2020, 2123, 29–36. [Google Scholar] [CrossRef]
- Pedersen, J.C. Hemagglutination-inhibition assay for influenza virus subtype identification and the detection and quantitation of serum antibodies to influenza virus. Methods Mol. Biol. 2014, 1161, 11–25. [Google Scholar] [CrossRef]
- Jenson, T.A. Agar Gel Immunodiffusion Assay to Detect Antibodies to Type A Influenza Virus. Methods Mol. Biol. 2020, 2123, 165–175. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lee, C.W.; Suarez, D.L. Application of real-time RT-PCR for the quantitation and competitive replication study of H5 and H7 subtype avian influenza virus. J. Virol. Methods 2004, 119, 151–158. [Google Scholar] [CrossRef]
- Das, A.; Spackman, E.; Pantin-Jackwood, M.J.; Suarez, D.L. Removal of real-time reverse transcription polymerase chain reaction (RT-PCR) inhibitors associated with cloacal swab samples tissues for improved diagnosis of Avian influenza virus by, R.T.-P.C.R. J. Vet. Diagn. Investig. 2019, 21, 771–778. [Google Scholar] [CrossRef]
- Katoh, K.M.K.; Kuma Ki Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- The Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, E.; Costa-Hurtado, M.; Smith, D.; Lee, D.H.; Spackman, E.; Kapczynski, D.R.; Torchetti, M.K.; Killian, M.L.; Suarez, D.L.; Swayne, D.E.; et al. Changes in adaptation of H5N2 highly pathogenic avian influenza H5 clade 2.3.4.4 viruses in chickens and mallards. Virology 2016, 499, 52–64. [Google Scholar] [CrossRef]
- Bertran, K.; Swayne, D.E.; Pantin-Jackwood, M.J.; Kapczynski, D.R.; Spackman, E.; Suarez, D.L. Lack of chicken adaptation of newly emergent Eurasian H5N8 and reassortant H5N2 high pathogenicity avian influenza viruses in the U.S. is consistent with restricted poultry outbreaks in the Pacific flyway during 2014–2015. Virology 2016, 494, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Leyson, C.M.; Youk, S.; Ferreira, H.L.; Suarez, D.L.; Pantin-Jackwood, M. Multiple Gene Segments Are Associated with Enhanced Virulence of Clade 2.3.4.4 H5N8 Highly Pathogenic Avian Influenza Virus in Mallards. J. Virol. 2021, 95, e0095521. [Google Scholar] [CrossRef]
- Youk, S.S.; Leyson, C.M.; Seibert, B.A.; Jadhao, S.; Perez, D.R.; Suarez, D.L.; Pantin-Jackwood, M.J. Mutations in PB1, NP, HA, and NA Contribute to Increased Virus Fitness of H5N2 Highly Pathogenic Avian Influenza Virus Clade 2.3.4.4 in Chickens. J. Virol. 2021, 95, e01675-20. [Google Scholar] [CrossRef]
- Bogs, J.; Veits, J.; Gohrbandt, S.; Hundt, J.; Stech, O.; Breithaupt, A.; Teifke, J.P.; Mettenleiter, T.C.; Stech, J. Highly pathogenic H5N1 influenza viruses carry virulence determinants beyond the polybasic hemagglutinin cleavage site. PLoS ONE 2010, 5, e11826. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Sun, H.; Qu, Y.; Wang, C.; Gao, W.; Zhu, J.; Sun, Y.; Bi, Y.; Huang, Y.; Chang, K.C.; et al. M Gene Reassortment in H9N2 Influenza Virus Promotes Early Infection and Replication: Contribution to Rising Virus Prevalence in Chickens in China. J. Virol. 2017, 91, e02055-16. [Google Scholar] [CrossRef] [PubMed]
- Kajihara, M.; Sakoda, Y.; Soda, K.; Minari, K.; Okamatsu, M.; Takada, A.; Kida, H. The PB2, PA, HA, NP, and NS genes of a highly pathogenic avian influenza virus A/whooper swan/Mongolia/3/2005 (H5N1) are responsible for pathogenicity in ducks. Virol. J. 2013, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Verhagen, J.H.; Herfst, S.; Fouchier, R.A.M. How a virus travels the world. Science 2015, 347, 616–617. [Google Scholar] [CrossRef]
- Lee, D.-H.; Torchetti, M.K.; Winker, K.; Ip, H.S.; Song, C.-S.; Swayne, D.E. Intercontinental Spread of Asian-Origin H5N8 to North America through Beringia by Migratory Birds. J. Virol. 2015, 89, 6521–6524. [Google Scholar] [CrossRef]
- Lee, D.H.; Bahl, J.; Torchetti, M.K.; Killian, M.L.; Ip, H.S.; DeLiberto, T.J.; Swayne, D.E. Highly Pathogenic Avian Influenza Viruses and Generation of Novel Reassortants, United States, 2014–2015. Emerg. Infect. Dis. 2016, 22, 1283–1285. [Google Scholar] [CrossRef]
- Lee, D.H.; Torchetti, M.K.; Hicks, J.; Killian, M.L.; Bahl, J.; Pantin-Jackwood, M.; Swayne, D.E. Transmission Dynamics of Highly Pathogenic Avian Influenza Virus A(H5Nx) Clade 2.3.4.4, North America, 2014–2015. Emerg. Infect. Dis. 2018, 24, 1840–1848. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control; European Reference Laboratory; Adlhoch, C.; Fusaro, A.; Gonzales, J.L.; Kuiken, T.; Marangon, S.; Niqueux, E.; Staubach, C.; et al. Avian influenza overview September–December 2021. EFSA J. 2021, 19, e07108. [Google Scholar]
- Engelsma, M.; Heutink, R.; Harders, F.; Germeraad, E.A.; Beerens, N. Multiple Introductions of Reassorted Highly Pathogenic Avian Influenza H5Nx Viruses Clade 2.3.4.4b Causing Outbreaks in Wild Birds and Poultry in The Netherlands, 2020–2021. Microbiol. Spectr. 2022, 10, e0249921. [Google Scholar] [CrossRef]
- Verhagen, J.H.; Fouchier, R.A.M.; Lewis, N. Highly Pathogenic Avian Influenza Viruses at the Wild-Domestic Bird Interface in Europe: Future Directions for Research and Surveillance. Viruses 2021, 13, 212. [Google Scholar] [CrossRef]
- Caliendo, V.; Lewis, N.S.; Pohlmann, A.; Baillie, S.R.; Banyard, A.C.; Beer, M.; Brown, I.H.; Fouchier, R.A.M.; Hansen, R.D.E.; Lameris, T.K.; et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. Sci. Rep. 2022, 12, 11729. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.H.; Bertran, K.; Lee, D.H.; Criado, M.F.; Killmaster, L.; Pantin-Jackwood, M.J.; Swayne, D.E. Diverse infectivity, transmissibility, and pathobiology of clade 2.3.4.4 H5Nx highly pathogenic avian influenza viruses in chickens. Emerg. Microbes Infect. 2023, 12, 2218945. [Google Scholar] [CrossRef]
- Bertran, K.; Lee, D.H.; Pantin-Jackwood, M.J.; Spackman, E.; Balzli, C.; Suarez, D.L.; Swayne, D.E. Pathobiology of Clade 2.3.4.4 H5Nx High-Pathogenicity Avian Influenza Virus Infections in Minor Gallinaceous Poultry Supports Early Backyard Flock Introductions in the Western United States in 2014–2015. J. Virol. 2017, 91, e00960-17. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Patton, C.; Jones, J.C.; Jeevan, T.; Harrington, W.N.; Trifkovic, S.; Seiler, J.P.; Fabrizio, T.; Woodard, K.; Turner, J.C.; et al. Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America. Nat. Commun. 2023, 14, 3082. [Google Scholar] [CrossRef]
- Puranik, A.; Slomka, M.J.; Warren, C.J.; Thomas, S.S.; Mahmood, S.; Byrne, A.M.P.; Ramsay, A.M.; Skinner, P.; Watson, S.; Everett, H.E.; et al. Transmission dynamics between infected waterfowl and terrestrial poultry: Differences between the transmission and tropism of H5N8 highly pathogenic avian influenza virus (clade 2.3.4.4a) among ducks, chickens and turkeys. Virology 2020, 541, 113–123. [Google Scholar] [CrossRef]
- James, J.; Billington, E.; Warren, C.J.; De Sliva, D.; Di Genova, C.; Airey, M.; Meyer, S.M.; Lewis, T.; Peers-Dent, J.; Thomas, S.S.; et al. Clade 2.3.4.4b H5N1 high pathogenicity avian influenza virus (HPAIV) from the 2021/22 epizootic is highly duck adapted and poorly adapted to chickens. J. Gen. Virol. 2023, 104, 001852. [Google Scholar] [CrossRef]
- Cha, R.M.; Lee, Y.N.; Park, M.J.; Baek, Y.G.; Shin, J.I.; Jung, C.H.; Sagong, M.; Heo, G.B.; Kang, Y.M.; Lee, K.N.; et al. Genetic Characterization and Pathogenesis of H5N1 High Pathogenicity Avian Influenza Virus Isolated in South Korea during 2021–2022. Viruses 2023, 15, 1403. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.R.; Swayne, D.E.; Davison, S.; Casavant, S.; Gutierrez, C. Validation of egg yolk antibody testing as a method to determine influenza status in white leghorn hens. Avian Dis. 2003, 47, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Morales, A.C., Jr.; Hilt, D.A.; Williams, S.M.; Pantin-Jackwood, M.J.; Suarez, D.L.; Spackman, E.; Stallknecht, D.E.; Jackwood, M.W. Biologic characterization of H4, H6, and H9 type low pathogenicity avian influenza viruses from wild birds in chickens and turkeys. Avian Dis. 2009, 53, 552–562. [Google Scholar] [CrossRef]
- Spackman, E.; Gelb, J., Jr.; Preskenis, L.A.; Ladman, B.S.; Pope, C.R.; Pantin-Jackwood, M.J.; McKinley, E.T. The pathogenesis of low pathogenicity H7 avian influenza viruses in chickens, ducks and turkeys. Virol. J. 2010, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.P.; Pantin-Jackwood, M.; Suarez, D.L.; Saif, Y.M.; Lee, C.W. Pathobiological characterization of low-pathogenicity H5 avian influenza viruses of diverse origins in chickens, ducks and turkeys. Arch. Virol. 2010, 155, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.P.; Pantin-Jackwood, M.; Yassine, H.M.; Saif, Y.M.; Lee, C.W. The high susceptibility of turkeys to influenza viruses of different origins implies their importance as potential intermediate hosts. Avian Dis. 2010, 54, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Criado, M.F.; Leyson, C.M.; Youk, S.; DeBlois, S.; Olivier, T.; Killian, M.L.; Torchetti, M.L.; Parris, D.J.; Spackman, E.; Kapczynski, D.R.; et al. The pathobiology of H7N3 low and high pathogenicity avian influenza viruses from the United States outbreak in 2020 differs between turkeys and chickens. Viruses 2021, 13, 1851. [Google Scholar] [CrossRef] [PubMed]
- Veterinary Services, Surveillance, Preparedness, and Response Services, Animal and Plant Health Inspection Service, United States Department of Agriculture. 2016. Final Report for the 2014–2015 Outbreak of Highly Pathogenic Avian Influenza (HPAI) in the United States. Available online: https://www.aphis.usda.gov/animal_health/emergency_management/downloads/hpai/2015-hpai-final-report.pdf (accessed on 15 July 2023).
- Blaurock, C.; Pfaff, F.; Scheibner, D.; Hoffmann, B.; Fusaro, A.; Monne, I.; Mettenleiter, T.C.; Breithaupt, A.; Abdelwhab, E.M. Evidence for Different Virulence Determinants and Host Response after Infection of Turkeys and Chickens with Highly Pathogenic H7N1 Avian Influenza Virus. J. Virol. 2022, 96, e0099422. [Google Scholar] [CrossRef]
- Bertran, K.; Lee, D.H.; Criado, M.F.; Smith, D.; Swayne, D.E.; Pantin-Jackwood, M.J. Pathobiology of Tennessee 2017 H7N9 low and high pathogenicity avian influenza viruses in commercial broiler breeders and specific pathogen free layer chickens. Vet. Res. 2018, 49, 82. [Google Scholar] [CrossRef] [PubMed]
- Bertran, K.; Lee, D.H.; Balzli, C.; Pantin-Jackwood, M.J.; Spackman, E.; Swayne, D.E. Age is not a determinant factor in susceptibility of broilers to H5N2 clade 2.3.4.4 high pathogenicity avian influenza virus. Vet. Res. 2016, 47, 116. [Google Scholar] [CrossRef]
- Lean, F.Z.X.; Nunez, A.; Banyard, A.C.; Reid, S.M.; Brown, I.H.; Hansen, R.D.E. Gross pathology associated with highly pathogenic avian influenza H5N8 and H5N1 in naturally infected birds in the UK (2020–2021). Vet. Rec. 2022, 190, e731. [Google Scholar] [CrossRef]
- Gaide, N.; Lucas, M.N.; Delpont, M.; Croville, G.; Bouwman, K.M.; Papanikolaou, A.; van der Woude, R.; Gagarinov, I.A.; Boons, G.J.; De Vries, R.P.; et al. Pathobiology of highly pathogenic H5 avian influenza viruses in naturally infected Galliformes and Anseriformes in France during winter 2015–2016. Vet. Res. 2022, 53, 11. [Google Scholar] [CrossRef]

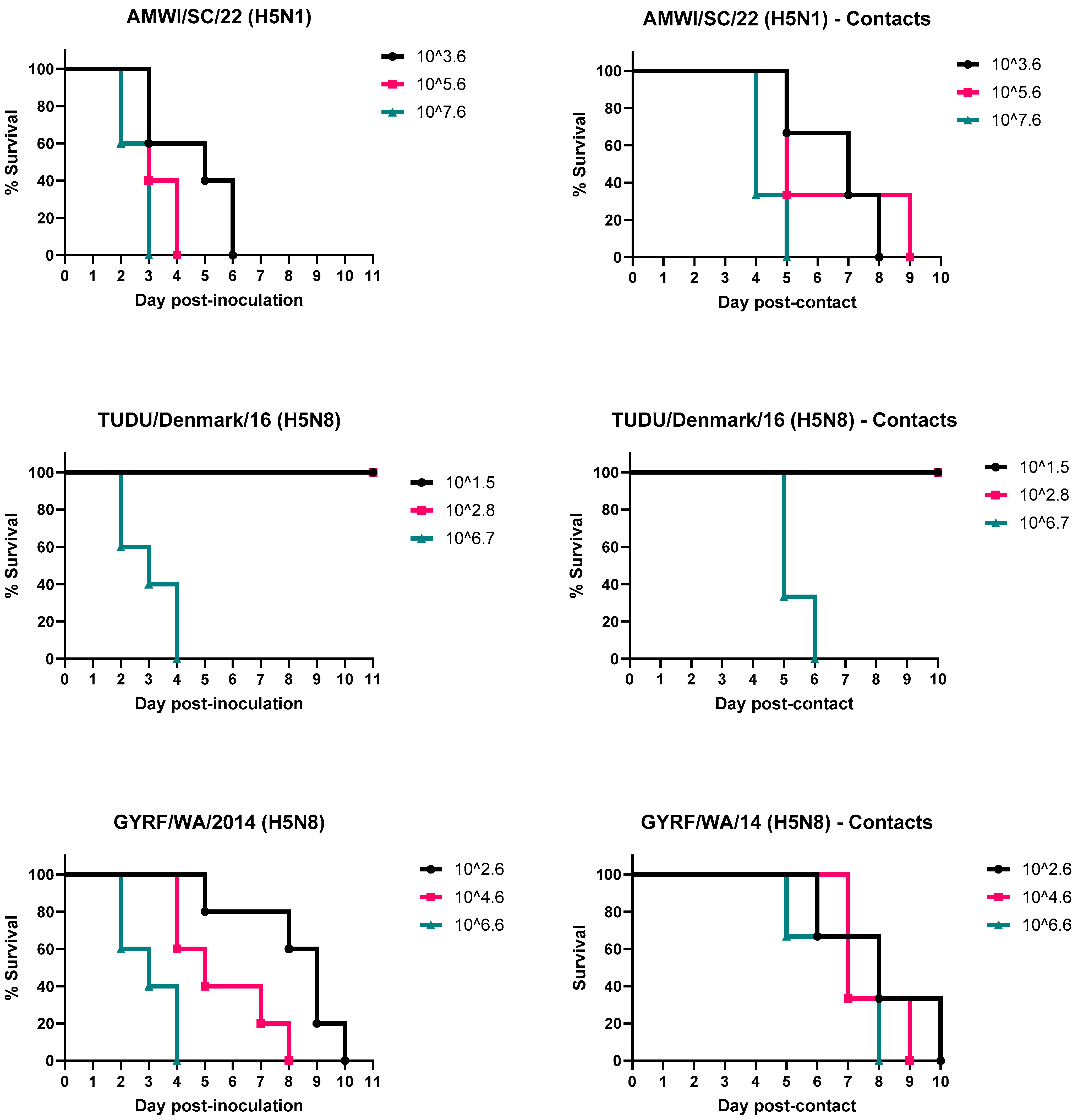

| Bird Species | Virus | Dose (log10 EID50) | Inoculated | Contact-Exposed | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # Birds Shedding Virus/Total 1 | # Dead Birds/ Total (MDT) 2 | # Birds HI Positive/ Total 3 | # Birds Infected/ Total 4 | BID50 (log10) 5 | # Birds Shedding Virus/Total | # Dead Birds/ Total (MDT) | # Birds HI Positive/Total | # Birds Infected/ Total | |||
| Chickens | AMWI/SC/21 | 3.6 | 5/5 | 3/5 (2.7) | 0/2 | 3/5 | 2.6 * | 3/3 | 0/3 | 0/3 | 0/3 |
| 5.6 | 5/5 | 5/5 (2) | na | 5/5 | 3/3 | 0/3 | 0/3 | 0/3 | |||
| 7.6 | 5/5 | 5/5 (1) | na | 5/5 | 3/3 | 0/3 | 0/3 | 0/3 | |||
| TUDU/ Denmark/16 | 1.5 | 0/5 | 0/5 | 0/5 | 0/5 | 4.2 | 0/3 | 0/3 | 0/3 | 0/3 | |
| 2.8 | 1/5 | 1/5 (3) | 0/4 | 1/5 | 0/3 | 0/3 | 0/3 | 0/3 | |||
| 6.7 | 5/5 | 5/5 (2) | na | 5/5 | 3/3 | 1/3 (4) | 0/2 | 1/3 | |||
| GYRF/WA/14 | 2.6 | 0/5 | 0/5 | 0/5 | 0/5 | 3.9 | 0/3 | 0/3 | 0/3 | 0/3 | |
| 4.6 | 5/5 | 4/5 (3) | 0/1 | 4/5 | 0/3 | 0/3 | 0/3 | 0/3 | |||
| 6.6 | 5/5 | 5/5 (2.6) | na | 5/5 | 3/3 | 1/3 (4) | 0/2 | 1/3 | |||
| Turkeys | AMWI/SC/21 | 3.6 | 4/5 | 5/5 (4.6) | na | 5/5 | 2.2 * | 3/3 | 3/3 (5.7) | na | 3/3 |
| 5.6 | 5/5 | 5/5 (3.4) | na | 5/5 | 3/3 | 3/3 (5.3) | na | 3/3 | |||
| 7.6 | 5/5 | 5/5 (2.6) | na | 5/5 | 3/3 | 3/3 (3.3) | na | 3/3 | |||
| TUDU/Denmark/16 | 1.5 | 0/5 | 0/5 | 0/5 | 0/5 | 4.7 | 0/3 | 0/3 | 0/3 | 0/3 | |
| 2.8 | 0/5 | 0/5 | 0/5 | 0/5 | 0/3 | 0/3 | 0/3 | 0/3 | |||
| 6.7 | 5/5 | 5/5 (3) | na | 5/5 | 3/3 | 3/3 (4.3) | na | 3/3 | |||
| GYRF/WA/14 | 2.6 | 5/5 | 5/5 (8.2) | na | 5/5 | 0.9 * | 3/3 | 3/3 (7) | na | 3/3 | |
| 4.6 | 5/5 | 5/5 (5.6) | na | 5/5 | 3/3 | 3/3 (6.7) | na | 3/3 | |||
| 6.6 | 5/5 | 5/5 (3) | na | 5/5 | 3/3 | 3/3 (5.7) | na | 3/3 | |||
| Tissue | Chickens | Turkeys | Viral Antigen-Stained Cell Types | ||||
|---|---|---|---|---|---|---|---|
| AW/SC/21 | TUDU/ Denmark/16 | GF/WA/ 14 | AW/SC/ 21 | TUDU/ Denmark /16 | GF/WA/ 14 | ||
| Nasal turbinate | +++ | +++/+++ | +++/+++ | na/+++ | +++/+++ | +/++ | Nasal epithelial cells, nasal gland epithelial cells, mononuclear cells, vascular endothelial cells |
| Trachea | ++ | +/na | +/+ | ++/++ | ++/++ | +/+ | Epithelial cells, mononuclear cells, vascular endothelial cells |
| Lung | +++ | +++/+++ | +++/++ | ++/+++ | +++/++ | +/++ | Pneumocytes, vascular endothelial cells, mononuclear scattered in alveolar septa |
| Comb | +++ | +++/+++ | +++/+++ | na/+++ | na/na | na/na | Vascular endothelial cells, mononuclear cells, feather pulp |
| Eyelid | +++ | +++/+++ | +++/+++ | ++/na | +/na | na/na | Vascular endothelial cells, interstitium, epithelial cells |
| Heart | +++ | +++/+++ | +++/+++ | ++/+++ | +++/++ | +/na | Myocardiocytes |
| Brain | +++ | +++/+++ | +++/+++ | +++/+++ | +/+ | +/++ | Neurons, Purkinje cells, glial cells, vascular endothelial cells |
| Proventriculus | + | +++/+++ | ++/+ | +/+ | +/+ | +/++ | Epithelial cells of the mucosa and gland, mononuclear cells infiltrating mucosa and submucosa |
| Duodenum | - | ++/++ | +/- | +/+ | na/+ | -/+ | Villi enterocytes and mononuclear cells infiltrating the mucosa and submucosa |
| Cecal tonsils | -/na | +++/+++ | ++/+ | +/na | +++/++ | -/+ | Mononuclear cells infiltrating in mucosa and submucosa |
| Pancreas | +++ | +++/+++ | ++/+ | ++/+++ | ++/++ | +/+ | Acinar cells, mononuclear cells, vascular endothelial cells |
| Liver | ++ | +++/++ | na/++ | ++/++ | +++/+++ | -/+ | Kupffer cells, hepatocytes, mononuclear cells |
| Kidney | ++ | +++/+++ | ++/+ | +++/++ | +++/+ | -/na | Tubular epithelial cells, interstitial mononuclear cells, vascular endothelial cells |
| Adrenal gland | na | +++/+++ | na/++ | na/na | na/+ | -/++ | Corticotropic cells and infiltrating mononuclear cells |
| Spleen | +++ | +++/+++ | +++/+++ | +++/+++ | +++/+++ | +/++ | Mononuclear cells, vascular endothelial cells |
| Thymus | ++ | +++/+++ | ++/+ | +/++ | ++/++ | -/+ | Thymic epithelium in medullar area, mononuclear cells |
| Cloacal bursa | + | +++/+++ | +/+ | +++/+ | +++/na | -/+++ | Mononuclear cells |
| Skeletal muscle | ++ | +/+++ | -/++ | ++/++ | +/+ | -/- | Mononuclear cells |
| Ovaries/Testis | ++ | na/+++ | na/na | na/+++ | ++/++ | -/- | Tegument/interstitial tissue, endothelial cells, infiltrating mononuclear cells |
| Bird Species | Virus | Bird ID | Muscle 1 | Lung | Spleen | Heart | Brain |
|---|---|---|---|---|---|---|---|
| Chickens | AMWI/SC/21 | CK-1 | 6.0 | 6.2 | 6.6 | 7.4 | 7.4 |
| TUDU/Denmark/16 | CK-1 | 8.5 | 8.2 | 8.6 | 10.1 | 9.0 | |
| CK-2 | 8.5 | 8.1 | 8.2 | 9.6 | 8.8 | ||
| GYRF/WA/14 | CK-1 | 7.8 | 7.4 | 7.8 | 9.0 | 8.5 | |
| CK-2 | 7.8 | 6.9 | 7.4 | 8.3 | 8.1 | ||
| Turkeys | AMWI/SC/21 | TK-1 | 6.7 | 6.9 | 7.2 | 7.2 | 7.5 |
| TK-2 | 6.3 | 7.8 | 7.5 | 7.7 | 8.0 | ||
| TUDU/Denmark/16 | TK-1 | 6.4 | 7.5 | 8.7 | 7.6 | 6.9 | |
| TK-2 | 7.0 | 7.8 | 8.9 | 2.9 | 7.4 | ||
| GYRF/WA/14 | TK-1 | 5.0 | 5.3 | 5.0 | 5.7 | 5.2 | |
| TK-2 | 6.1 | 5.9 | 5.9 | 6.8 | 6.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantin-Jackwood, M.J.; Spackman, E.; Leyson, C.; Youk, S.; Lee, S.A.; Moon, L.M.; Torchetti, M.K.; Killian, M.L.; Lenoch, J.B.; Kapczynski, D.R.; et al. Pathogenicity in Chickens and Turkeys of a 2021 United States H5N1 Highly Pathogenic Avian Influenza Clade 2.3.4.4b Wild Bird Virus Compared to Two Previous H5N8 Clade 2.3.4.4 Viruses. Viruses 2023, 15, 2273. https://doi.org/10.3390/v15112273
Pantin-Jackwood MJ, Spackman E, Leyson C, Youk S, Lee SA, Moon LM, Torchetti MK, Killian ML, Lenoch JB, Kapczynski DR, et al. Pathogenicity in Chickens and Turkeys of a 2021 United States H5N1 Highly Pathogenic Avian Influenza Clade 2.3.4.4b Wild Bird Virus Compared to Two Previous H5N8 Clade 2.3.4.4 Viruses. Viruses. 2023; 15(11):2273. https://doi.org/10.3390/v15112273
Chicago/Turabian StylePantin-Jackwood, Mary J., Erica Spackman, Christina Leyson, Sungsu Youk, Scott A. Lee, Linda M. Moon, Mia K. Torchetti, Mary L. Killian, Julianna B. Lenoch, Darrell R. Kapczynski, and et al. 2023. "Pathogenicity in Chickens and Turkeys of a 2021 United States H5N1 Highly Pathogenic Avian Influenza Clade 2.3.4.4b Wild Bird Virus Compared to Two Previous H5N8 Clade 2.3.4.4 Viruses" Viruses 15, no. 11: 2273. https://doi.org/10.3390/v15112273
APA StylePantin-Jackwood, M. J., Spackman, E., Leyson, C., Youk, S., Lee, S. A., Moon, L. M., Torchetti, M. K., Killian, M. L., Lenoch, J. B., Kapczynski, D. R., Swayne, D. E., & Suarez, D. L. (2023). Pathogenicity in Chickens and Turkeys of a 2021 United States H5N1 Highly Pathogenic Avian Influenza Clade 2.3.4.4b Wild Bird Virus Compared to Two Previous H5N8 Clade 2.3.4.4 Viruses. Viruses, 15(11), 2273. https://doi.org/10.3390/v15112273







