Identification of Critical Amino Acids of Coxsackievirus A10 Associated with Cell Tropism and Viral RNA Release during Uncoating
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses and Antibodies
2.2. Sequencing of CV-A10 Strains
2.3. Construction and Generation of Recombinant CV-A10 Strains
2.4. Fifty Percent of Cell Culture Infective Dose (CCID50) Assay
2.5. Immunofluorescence Assay (IFA)
2.6. Viral Binding and Internalization Assays by qPCR
2.7. Viral Uncoating Assay
2.8. One-Step Growth Analysis for CV-A10 Strains
2.9. Small Interfering RNA (siRNA) Interference in HEK293 Cells
2.10. Western Blotting and Co-Immunoprecipitation
2.11. Statistical Analysis
3. Results
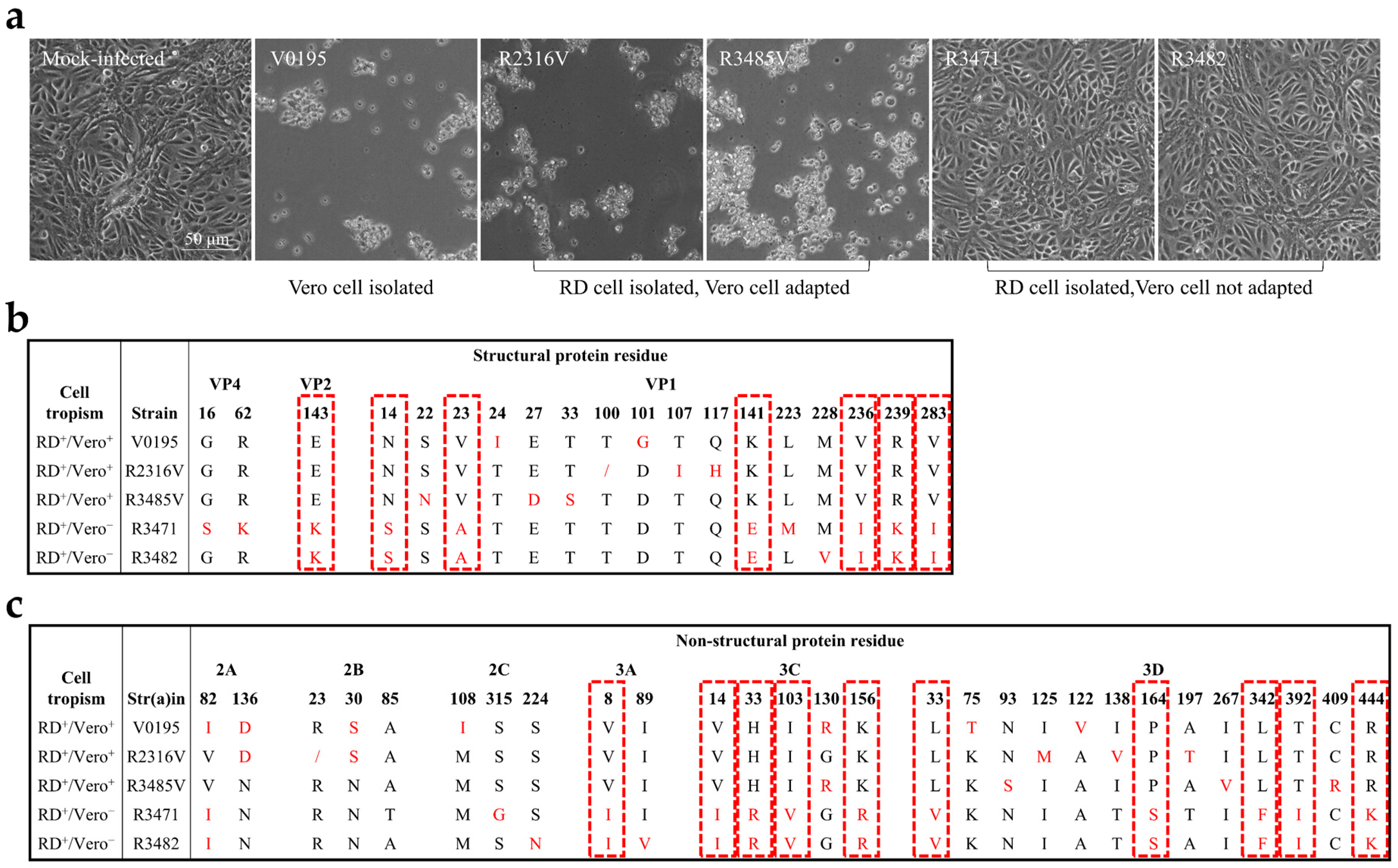
3.1. Cell Tropism of CV-A10 to Vero Cells
3.2. Identification of the Residues Related to CV-A10 Cell Tropism
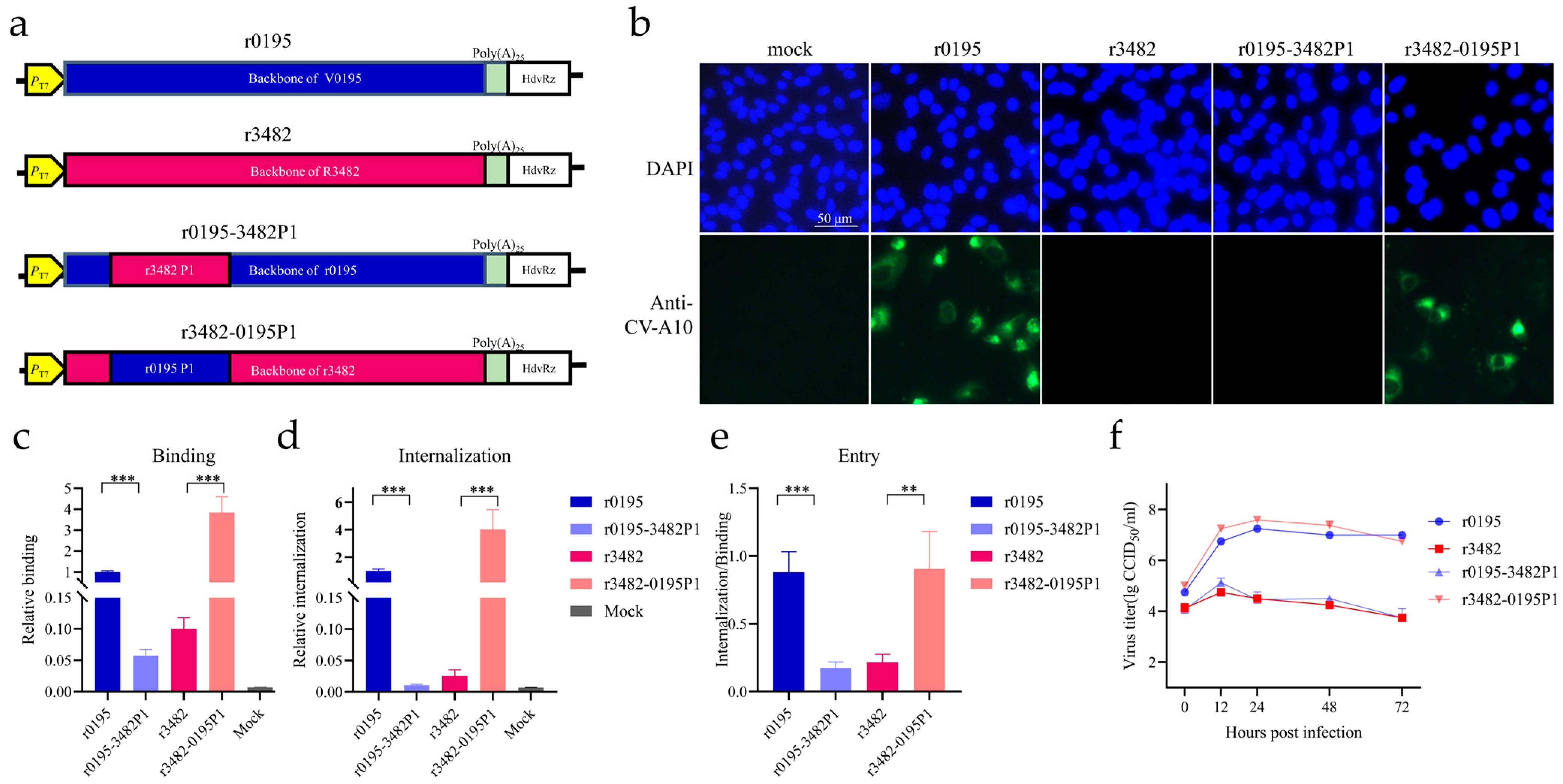
3.3. Cell Tropism of CV-A10 to Vero Cells Mainly Determined by the Structural Proteins
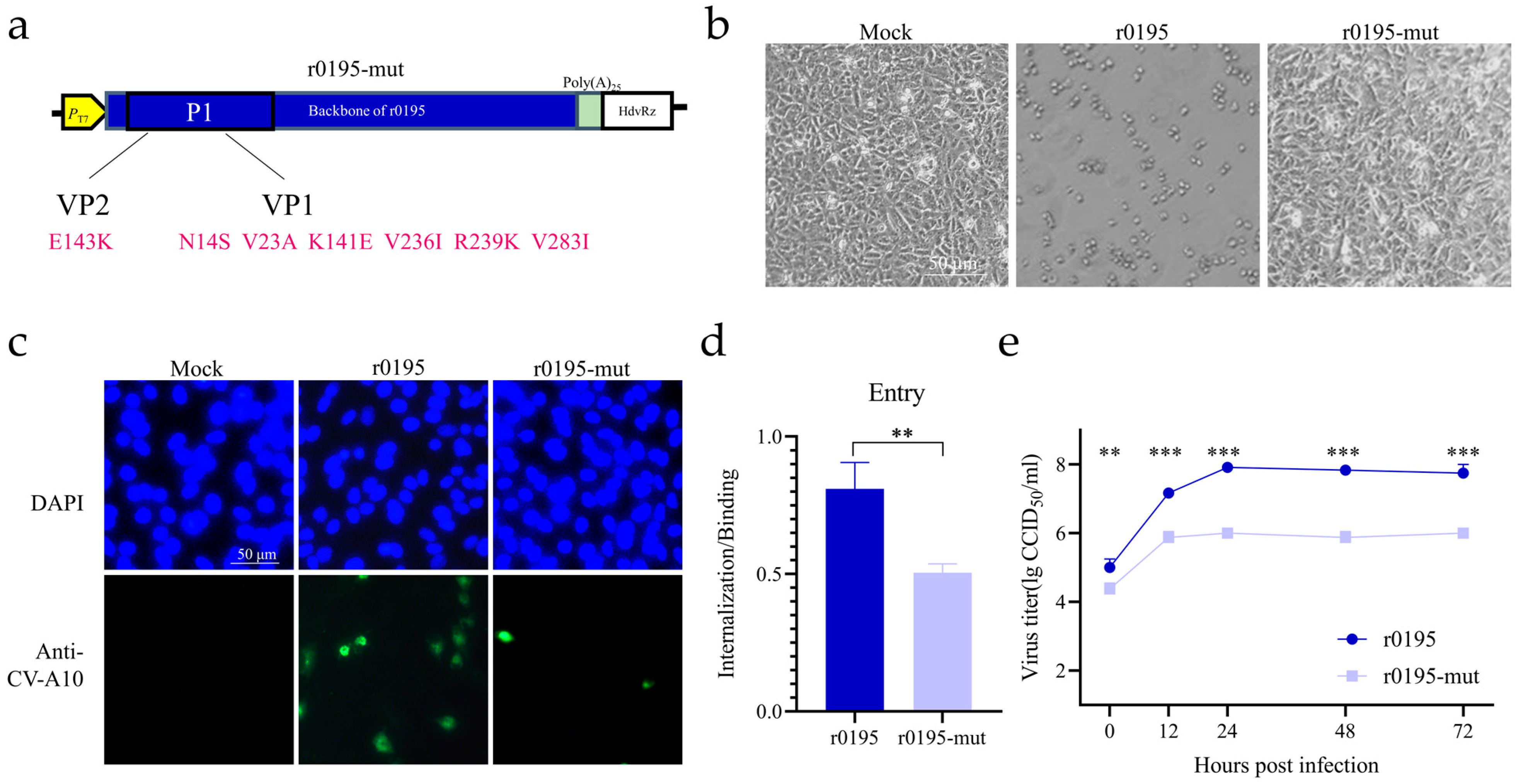
3.4. Seven Mutations on the P1 Region Reversing the Cell Tropism
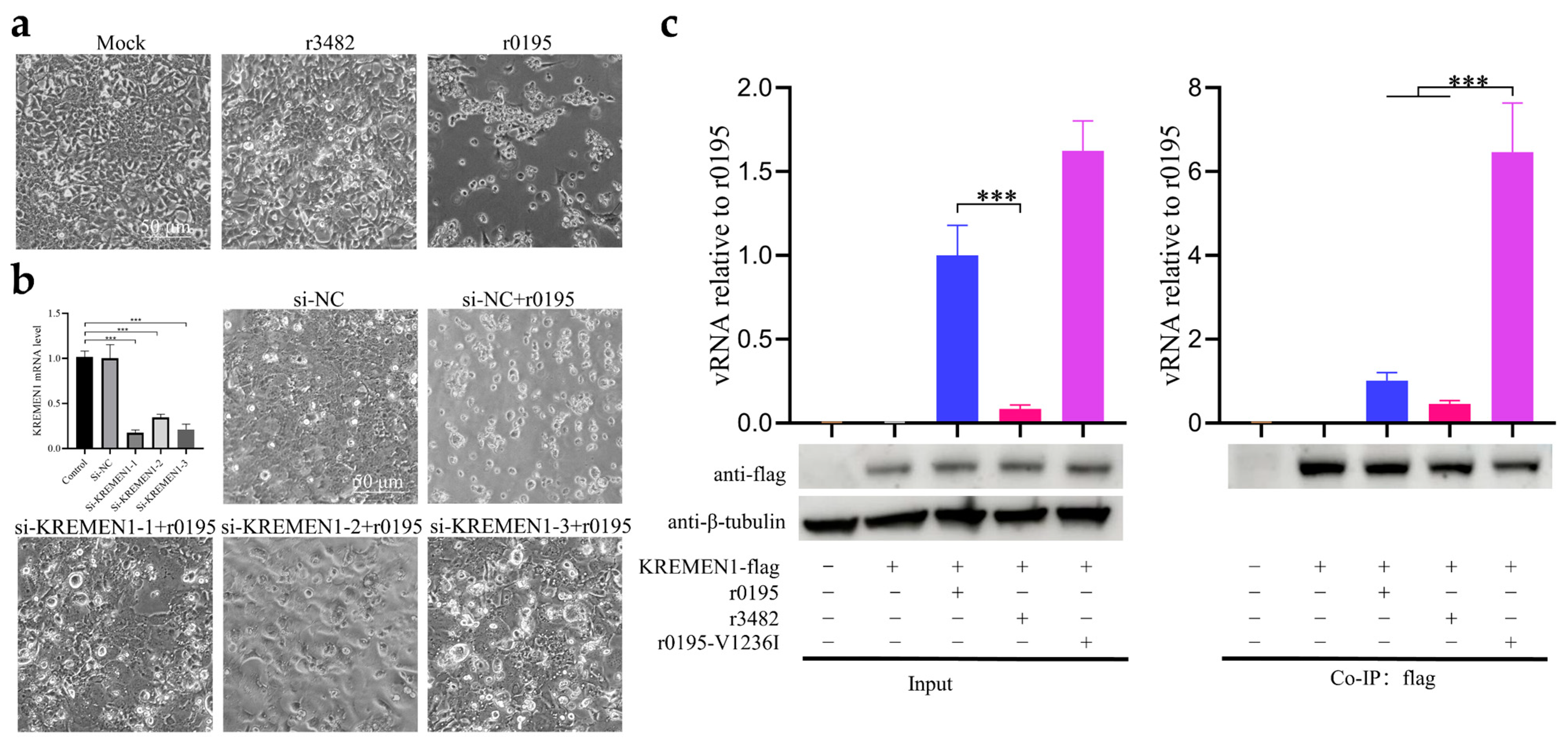
3.5. V1236 Is Critical for Viral Uncoating in Vero Cells
3.6. V1236I Mutation Reducing Viral RNA Release during KREMEN1-Dependent Uncoating
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ji, H.; Fan, H.; Lu, P.X.; Zhang, X.F.; Ai, J.; Shi, C.; Huo, X.; Bao, C.J.; Shan, J.; Jin, Y. Surveillance for severe hand, foot, and mouth disease from 2009 to 2015 in Jiangsu province: Epidemiology, etiology, and disease burden. BMC Infect. Dis. 2019, 19, 79. [Google Scholar] [CrossRef]
- Meng, X.D.; Tong, Y.; Wei, Z.N.; Wang, L.; Mai, J.Y.; Wu, Y.; Luo, Z.Y.; Li, S.; Li, M.; Wang, S.; et al. Epidemical and etiological study on hand, foot and mouth disease following EV-A71 vaccination in Xiangyang, China. Sci. Rep. 2020, 10, 20909. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Gao, F.; Mao, Q.; Sun, S.; Wu, X.; Liu, S.; Yang, X.; Liang, Z. Hand, foot, and mouth disease associated with coxsackievirus A10: More serious than it seems. Expert. Rev. Anti. Infect. Ther. 2019, 17, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, S.; Klemola, P.; Kaijalainen, S.; Paananen, A.; Simonen, M.L.; Vuorinen, T.; Roivainen, M. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J. Clin. Virol. 2010, 48, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; Wada, M.; Sato, H.; Yamaguchi, Y.; Tanji, H.; Kurokawa, K.; Kawanami, T.; Takahashi, T.; Kato, T. Neuromyelitis optica preceded by hyperCKemia and a possible association with coxsackie virus group A10 infection. Intern. Med. 2013, 52, 2665–2668. [Google Scholar] [CrossRef]
- Kumar, A.; Shukla, D.; Kumar, R.; Idris, M.Z.; Jauhari, P.; Srivastava, S.; Dhole, T.N. Molecular identification of enteroviruses associated with aseptic meningitis in children from India. Arch. Virol. 2013, 158, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Fuschino, M.E.; Lamson, D.M.; Rush, K.; Carbone, L.S.; Taff, M.L.; Hua, Z.; Landi, K.; George, K.S. Detection of coxsackievirus A10 in multiple tissues of a fatal infant sepsis case. J. Clin. Virol. 2012, 53, 259–261. [Google Scholar] [CrossRef]
- Davia, J.L.; Bel, P.H.; Ninet, V.Z.; Bracho, M.A.; Gonzalez-Candelas, F.; Salazar, A.; Gobernado, M.; Bosch, I.F. Onychomadesis outbreak in Valencia, Spain associated with hand, foot, and mouth disease caused by enteroviruses. Pediatr. Dermatol. 2011, 28, 1–5. [Google Scholar] [CrossRef]
- Chen, M.; He, S.; Yan, Q.; Xu, X.; Wu, W.; Ge, S.; Zhang, S.; Chen, M.; Xia, N. Severe hand, foot and mouth disease associated with coxsackievirus A10 infections in Xiamen, China in 2015. J. Clin. Virol. 2017, 93, 20–24. [Google Scholar] [CrossRef]
- Yi, E.J.; Kim, Y.I.; Song, J.H.; Ko, H.J.; Chang, S.Y. Intranasal immunization with curdlan induce Th17 responses and enhance protection against enterovirus 71. Vaccine 2023, 41, 2243–2252. [Google Scholar] [CrossRef]
- Huang, K. Structural basis for neutralization of enterovirus. Curr. Opin. Virol. 2021, 51, 199–206. [Google Scholar] [CrossRef]
- Zhu, R.; Xu, L.; Zheng, Q.; Cui, Y.; Li, S.; He, M.; Yin, Z.; Liu, D.; Li, S.; Li, Z.; et al. Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10. Sci. Adv. 2018, 4, eaat7459. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Song, J.; Wang, J.; Li, Y.; Deng, H.; Li, M.; Gao, N.; Zhai, S.; Dang, S.; Zhang, X.; et al. Cost-effectiveness of a national enterovirus 71 vaccination program in China. PLoS Negl. Trop. Dis. 2017, 11, e0005899. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Chiu, C.H.; Lin, C.Y.; Chiu, N.C.; Chen, P.Y.; Le, T.T.V.; Le, D.N.; Duong, A.H.; Nguyen, V.L.; Huynh, T.N.; et al. Efficacy, safety, and immunogenicity of an inactivated, adjuvanted enterovirus 71 vaccine in infants and children: A multiregion, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet 2022, 399, 1708–1717. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, Y.; Ye, Y.; Jiang, F.; He, H.; Luo, L.; Chen, H.; Shi, L.; Mu, Q.; Chen, W.; et al. Immunogenicity and safety of an inactivated enterovirus 71 vaccine coadministered with trivalent split-virion inactivated influenza vaccine: A phase 4, multicenter, randomized, controlled trial in China. Front. Immunol. 2022, 13, 1080408. [Google Scholar] [CrossRef]
- Schmidt, N.J.; Lennette, E.H.; Ho, H.H. Comparative sensitivity of human fetal diploid kidney cell strains and monkey kidney cell cultures for isolation of certain human viruses. Am. J. Clin. Pathol. 1965, 43, 297–301. [Google Scholar] [CrossRef]
- St Geme, J.W., Jr. Behavior of coxsackie A6 virus in murine cells: Failure of newer techniques to detect multipication in vitro. J. Bacteriol. 1964, 87, 969–970. [Google Scholar] [CrossRef] [PubMed]
- Chonmaitree, T.; Ford, C.; Sanders, C.; Lucia, H.L. Comparison of cell cultures for rapid isolation of enteroviruses. J. Clin. Microbiol. 1988, 26, 2576–2580. [Google Scholar] [CrossRef]
- Kiesslich, S.; Kamen, A.A. Vero cell upstream bioprocess development for the production of viral vectors and vaccines. Biotechnol. Adv. 2020, 44, 107608. [Google Scholar] [CrossRef]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef]
- Barrett, P.N.; Mundt, W.; Kistner, O.; Howard, M.K. Vero cell platform in vaccine production: Moving towards cell culture-based viral vaccines. Expert. Rev. Vaccines 2009, 8, 607–618. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Strating, J.; van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Koike, S. Cellular receptors for enterovirus A71. J. Biomed. Sci. 2020, 27, 23. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Sun, D.; Guo, J.; Elgner, F.; Wang, M.; Hildt, E.; Cheng, A. Multifunctionality of structural proteins in the enterovirus life cycle. Future Microbiol. 2019, 14, 1147–1157. [Google Scholar] [CrossRef]
- Staring, J.; van den Hengel, L.G.; Raaben, M.; Blomen, V.A.; Carette, J.E.; Brummelkamp, T.R. KREMEN1 is a host entry receptor for a major group of enteroviruses. Cell. Host. Microbe. 2018, 23, 636–643.e635. [Google Scholar] [CrossRef]
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV virus taxonomy profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Peng, R.; Song, H.; Tong, Z.; Qu, X.; Liu, S.; Zhao, X.; Chai, Y.; Wang, P.; Gao, G.F.; et al. Molecular basis of coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1. Proc. Natl. Acad. Sci. USA 2020, 117, 18711–18718. [Google Scholar] [CrossRef]
- Pascal, S.M.; Garimella, R.; Warden, M.S.; Ponniah, K. Structural biology of the enterovirus replication-linked 5′–cloverleaf RNA and associated virus proteins. Microbiol. Mol. Biol. Rev. 2020, 84, e00062-19. [Google Scholar] [CrossRef]
- Cameron, C.; Oh, H.; Moustafa, I. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future. Microbiol. 2010, 5, 867–881. [Google Scholar] [CrossRef]
- Yang, X.; Xu, C.Z.; Chen, J.H.; Meng, S.L.; Wang, Z.J.; Shen, S.; Chen, X.Q. Epidemiological investigation of hand, foot and mouth diseases following vaccination of EV-A71 in 2019. Chin. J. Biol. 2021, 34, 843–848. (In Chinese) [Google Scholar] [CrossRef]
- An, H.H.; Li, M.; Liu, R.L.; Wu, J.; Meng, S.L.; Guo, J.; Wang, Z.J.; Qian, S.S.; Shen, S. Humoral and cellular immunogenicity and efficacy of a coxsackievirus A10 vaccine in mice. Emerg. Microbes. Infect. 2023, 12, e2147022. [Google Scholar] [CrossRef]
- Yan, J.; Wang, M.; Wang, M.; Dun, Y.; Zhu, L.; Yi, Z.; Zhang, S. Involvement of VCP/UFD1/Nucleolin in the viral entry of Enterovirus A species. Virus. Res. 2020, 283, 197974. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Leung, C.; Tahan, S.; Wang, D. Entry by multiple picornaviruses is dependent on a pathway that includes TNK2, WASL, and NCK1. eLife 2019, 8, e50276. [Google Scholar] [CrossRef]
- Nguyen, Y.; Jesudhasan, P.R.; Aguilera, E.R.; Pfeiffer, J.K. Identification and characterization of a poliovirus capsid mutant with enhanced thermal stability. J. Virol. 2019, 93, e01510–e01518. [Google Scholar] [CrossRef] [PubMed]
- Kuss, S.K.; Etheredge, C.A.; Pfeiffer, J.K. Multipl host barriers restrict poliovirus trafficking in mice. PLoS Pathog. 2008, 4, e1000082. [Google Scholar] [CrossRef] [PubMed]
- Prim, N.; Rodriguez, G.; Margall, N.; Del Cuerpo, M.; Trallero, G.; Rabella, N. Combining cell lines to optimize isolation of human enterovirus from clinical specimens: Report of 25 years of experience. J. Med. Virol. 2013, 85, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Lipson, S.M.; Walderman, R.; Costello, P.; Szabo, K. Sensitivity of rhabdomyosarcoma and guinea pig embryo cell cultures to field isolates of difficult-to-cultivate group A coxsackieviruses. J. Clin. Microbiol. 1988, 26, 1298–1303. [Google Scholar] [CrossRef]
- Chandler-Bostock, R.; Mata, C.P.; Bingham, R.J.; Dykeman, E.C.; Meng, B.; Tuthill, T.J.; Rowlands, D.J.; Ranson, N.A.; Twarock, R.; Stockley, P.G. Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts. PLoS Pathog. 2020, 16, e1009146. [Google Scholar] [CrossRef]
- Royston, L.; Essaidi-Laziosi, M.; Perez-Rodriguez, F.J.; Piuz, I.; Geiser, J.; Krause, K.H.; Huang, S.; Constant, S.; Kaiser, L.; Garcin, D.; et al. Viral chimeras decrypt the role of enterovirus capsid proteins in viral tropism, acid sensitivity and optimal growth temperature. PLoS Pathog. 2018, 14, e1006962. [Google Scholar] [CrossRef]
- Lin, J.Y.; Shih, S.R. Cell and tissue tropism of enterovirus 71 and other enteroviruses infections. J. Biomed. Sci. 2014, 21, 18. [Google Scholar] [CrossRef]
- Ross, C.; Knox, C.; Tastan Bishop, O. Interacting motif networks located in hotspots associated with RNA release are conserved in enterovirus capsids. FEBS Lett. 2017, 591, 1687–1701. [Google Scholar] [CrossRef]
- Adeyemi, O.O.; Nicol, C.; Stonehouse, N.J.; Rowlands, D.J. Increasing type 1 poliovirus capsid stability by thermal selection. J. Virol. 2017, 91, e01586-16. [Google Scholar] [CrossRef]
- Xu, L.; He, D.; Yang, L.; Li, Z.; Ye, X.; Yu, H.; Zhao, H.; Li, S.; Yuan, L.; Qian, H.; et al. A broadly cross-protective vaccine presenting the neighboring epitopes within the VP1 GH loop and VP2 EF loop of enterovirus 71. Sci. Rep. 2015, 5, 12973. [Google Scholar] [CrossRef]
- Zhou, D.; Zhao, Y.; Kotecha, A.; Fry, E.E.; Kelly, J.T.; Wang, X.; Rao, Z.; Rowlands, D.J.; Ren, J.; Stuart, D.I. Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. Nat. Microbiol. 2019, 4, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Yeo, H.; Chong, C.W.H.; Chen, E.W.; Lim, Z.Q.; Ng, Q.Y.; Yan, B.; Chu, J.J.H.; Chow, V.T.K.; Alonso, S. A single amino acid substitution in structural protein VP2 abrogates the neurotropism of enterovirus A-71 in mice. Front. Microbiol. 2022, 13, 821976. [Google Scholar] [CrossRef] [PubMed]
- Tseligka, E.D.; Sobo, K.; Stoppini, L.; Cagno, V.; Abdul, F.; Piuz, I.; Meylan, P.; Huang, S.; Constant, S.; Tapparel, C. A VP1 mutation acquired during an enterovirus 71 disseminated infection confers heparan sulfate binding ability and modulates ex vivo tropism. PLoS Pathog. 2018, 14, e1007190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, D.; Ni, T.; Karia, D.; Kotecha, A.; Wang, X.; Rao, Z.; Jones, E.Y.; Fry, E.E.; Ren, J.; et al. Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of coxsackie virus A10. Nat. Commun. 2020, 11, 38. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, Y.; Fan, J.; Zhu, B.; Cao, L.; Gao, Q.; Zhang, Y.; Liu, H.; Rao, Z.; Wang, X. Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating. Nat. Commun. 2018, 9, 4985. [Google Scholar] [CrossRef]
- Strauss, M.; Levy, H.C.; Bostina, M.; Filman, D.J.; Hogle, J.M. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 2013, 87, 3903–3914. [Google Scholar] [CrossRef]
- Laitinen, O.H.; Svedin, E.; Kapell, S.; Nurminen, A.; Hytonen, V.P.; Flodstrom-Tullberg, M. Enteroviral proteases: Structure, host interactions and pathogenicity. Rev. Med. Virol. 2016, 26, 251–267. [Google Scholar] [CrossRef]
- Toyoda, H.; Nicklin, M.J.; Murray, M.G.; Anderson, C.W.; Dunn, J.J.; Studier, F.W.; Wimmer, E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 1986, 45, 761–770. [Google Scholar] [CrossRef]
- Leong, L.E.; Walker, P.A.; Porter, A.G. Human rhinovirus-14 protease 3C (3Cpro) binds specifically to the 5′–noncoding region of the viral RNA. Evidence that 3Cpro has different domains for the RNA binding and proteolytic activities. J. Biol. Chem. 1993, 268, 25735–25739. [Google Scholar] [CrossRef]
- Harris, K.S.; Xiang, W.; Alexander, L.; Lane, W.S.; Paul, A.V.; Wimmer, E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994, 269, 27004–27014. [Google Scholar] [CrossRef] [PubMed]
- Andino, R.; Rieckhof, G.E.; Achacoso, P.L.; Baltimore, D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′–end of viral RNA. EMBO J. 1993, 12, 3587–3598. [Google Scholar] [CrossRef]
- Xiang, W.; Harris, K.S.; Alexander, L.; Wimmer, E. Interaction between the 5′–terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 1995, 69, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.B.; Ghosh, S.K.; Roberts, A.W.; Sharma, S.D.; Yoder, J.D.; Arnold, J.J.; Gohara, D.W.; Barton, D.J.; Paul, A.V.; Cameron, C.E. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J. Biol. Chem 2002, 277, 31551–31562. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, J.; Liu, R.-L.; Yang, Z.-H.; Du, Y.-X.; Qian, S.-S.; Meng, S.-L.; Guo, J.; Zhang, B.; Shen, S. Identification of Critical Amino Acids of Coxsackievirus A10 Associated with Cell Tropism and Viral RNA Release during Uncoating. Viruses 2023, 15, 2114. https://doi.org/10.3390/v15102114
Pei J, Liu R-L, Yang Z-H, Du Y-X, Qian S-S, Meng S-L, Guo J, Zhang B, Shen S. Identification of Critical Amino Acids of Coxsackievirus A10 Associated with Cell Tropism and Viral RNA Release during Uncoating. Viruses. 2023; 15(10):2114. https://doi.org/10.3390/v15102114
Chicago/Turabian StylePei, Jie, Rui-Lun Liu, Zhi-Hui Yang, Ya-Xin Du, Sha-Sha Qian, Sheng-Li Meng, Jing Guo, Bo Zhang, and Shuo Shen. 2023. "Identification of Critical Amino Acids of Coxsackievirus A10 Associated with Cell Tropism and Viral RNA Release during Uncoating" Viruses 15, no. 10: 2114. https://doi.org/10.3390/v15102114
APA StylePei, J., Liu, R.-L., Yang, Z.-H., Du, Y.-X., Qian, S.-S., Meng, S.-L., Guo, J., Zhang, B., & Shen, S. (2023). Identification of Critical Amino Acids of Coxsackievirus A10 Associated with Cell Tropism and Viral RNA Release during Uncoating. Viruses, 15(10), 2114. https://doi.org/10.3390/v15102114





