RNA Editing-Dependent and -Independent Roles of Adenosine Deaminases Acting on RNA Proteins in Herpesvirus Infection—Hints on Another Layer of Complexity
Abstract
1. Introduction
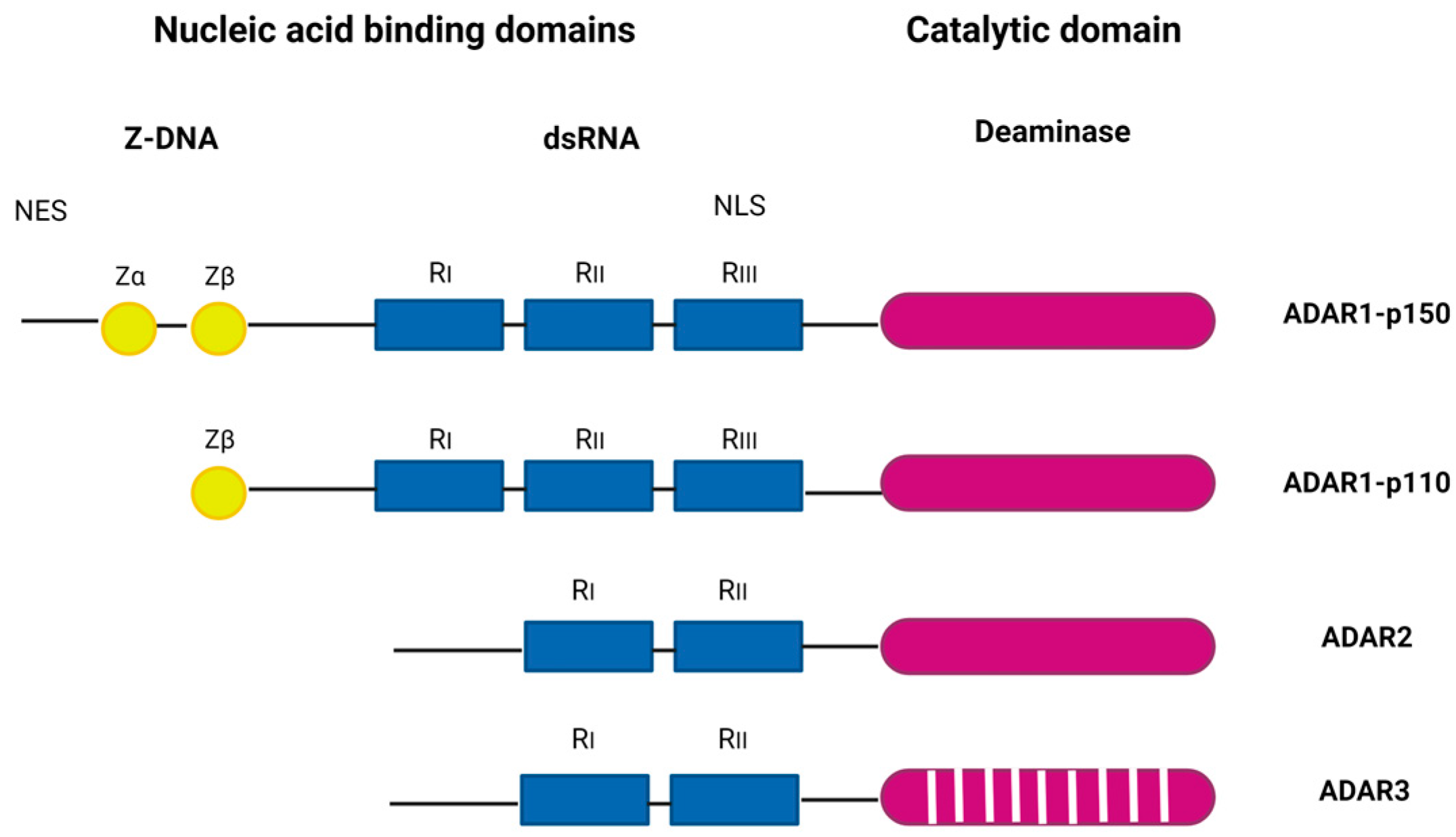
2. The ADAR Protein Family in Brief
3. Herpesviruses in Brief
The Orthoherpesviridae
4. ADAR and Herpesviruses
4.1. Evidence for ADAR-Mediated RNA Editing in Alphaherpesviruses
4.1.1. Herpes Simplex Virus 1 (HSV-1)—Latent miRNA Editing
4.1.2. Varicella Zoster Virus (VZV)—Edited Novel Viral Transcripts
4.1.3. Gallid Herpesvirus 2 (GaHV-2, or Marek’s Disease Virus 1 (MDV-1)
4.2. Evidence of ADAR-Mediated RNA Editing in Betaherpesviruses—Edited Host miRNA
4.3. Gammaherpesviruses: Editing-Dependent and Editing-Independent Roles of ADAR Proteins
4.3.1. Epstein–Barr Virus (EBV, HHV-4)—Editing Affects miRNA Biogenesis
4.3.2. Kaposi’s Sarcoma-Associated Herpesvirus (KSHV, HHV-8)
- KSHV—RNA Editing Phenomena
- KSHV—Editing-Independent Roles of ADAR
4.4. The Malacoherpesviridae–Herpesviruses of Mollusks
5. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Schlee, M.; Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016, 16, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Vukic, D.; Michalik, D.; O’Connell, M.A.; Keegan, L.P. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 2017, 136, 1265–1278. [Google Scholar] [CrossRef]
- Tomaselli, S.; Bonamassa, B.; Alisi, A.; Nobili, V.; Locatelli, F.; Gallo, A. ADAR enzyme and miRNA story: A nucleotide that can make the difference. Int. J. Mol. Sci. 2013, 14, 22796–22816. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, S.; Galeano, F.; Locatelli, F.; Gallo, A. ADARs and the Balance Game between Virus Infection and Innate Immune Cell Response. Curr. Issues Mol. Biol. 2015, 17, 37–51. [Google Scholar]
- Samuel, C.E. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. J. Biol. Chem. 2019, 294, 1710–1720. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014, 24, 365–376. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, Y.; Tan, B.C.; Kang, L.; Tian, Z.; Zhu, Y.; Zhang, W.; Liang, Y.; Hu, X.; Tan, X.; et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012, 30, 253–260. [Google Scholar] [CrossRef]
- Pfaller, C.K.; George, C.X.; Samuel, C.E. Adenosine Deaminases Acting on RNA (ADARs) and Viral Infections. Annu. Rev. Virol. 2021, 8, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Rosenberg, B.R.; Chung, H.; Rice, C.M. Identification of ADAR1 p150 and p110 Associated Edit Sites. Methods Mol. Biol. 2023, 2651, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Nachmani, D.; Zimmermann, A.; Oiknine Djian, E.; Weisblum, Y.; Livneh, Y.; Khanh Le, V.T.; Galun, E.; Horejsi, V.; Isakov, O.; Shomron, N.; et al. MicroRNA editing facilitates immune elimination of HCMV infected cells. PLoS Pathog. 2014, 10, e1003963. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef]
- Hood, J.L.; Morabito, M.V.; Martinez, C.R., 3rd; Gilbert, J.A.; Ferrick, E.A.; Ayers, G.D.; Chappell, J.D.; Dermody, T.S.; Emeson, R.B. Reovirus-mediated induction of ADAR1 (p150) minimally alters RNA editing patterns in discrete brain regions. Mol. Cell. Neurosci. 2014, 61, 97–109. [Google Scholar] [CrossRef]
- Wang, Q.; Miyakoda, M.; Yang, W.; Khillan, J.; Stachura, D.L.; Weiss, M.J.; Nishikura, K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004, 279, 4952–4961. [Google Scholar] [CrossRef]
- Rice, G.I.; Kasher, P.R.; Forte, G.M.; Mannion, N.M.; Greenwood, S.M.; Szynkiewicz, M.; Dickerson, J.E.; Bhaskar, S.S.; Zampini, M.; Briggs, T.A.; et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012, 44, 1243–1248. [Google Scholar] [CrossRef]
- Herbert, A. Mendelian disease caused by variants affecting recognition of Z-DNA and Z-RNA by the Zalpha domain of the double-stranded RNA editing enzyme ADAR. Eur. J. Hum. Genet. 2020, 28, 114–117. [Google Scholar] [CrossRef]
- Miyamura, Y.; Suzuki, T.; Kono, M.; Inagaki, K.; Ito, S.; Suzuki, N.; Tomita, Y. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am. J. Hum. Genet. 2003, 73, 693–699. [Google Scholar] [CrossRef]
- Baker, A.R.; Slack, F.J. ADAR1 and its implications in cancer development and treatment. Trends Genet. 2022, 38, 821–830. [Google Scholar] [CrossRef]
- Stok, J.E.; Oosenbrug, T.; Ter Haar, L.R.; Gravekamp, D.; Bromley, C.P.; Zelenay, S.; Reis e Sousa, C.; van der Veen, A.G. RNA sensing via the RIG-I-like receptor LGP2 is essential for the induction of a type I IFN response in ADAR1 deficiency. EMBO J. 2022, 41, e109760. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Deng, P.; Zhu, Z.; Zhu, J.; Wang, G.; Zhang, L.; Chen, A.F.; Wang, T.; Sarkar, S.N.; Billiar, T.R.; et al. Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J. Immunol. 2014, 193, 3436–3445. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Calis, J.J.A.; Wu, X.; Sun, T.; Yu, Y.; Sarbanes, S.L.; Dao Thi, V.L.; Shilvock, A.R.; Hoffmann, H.H.; Rosenberg, B.R.; et al. Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 2018, 172, 811–824.e814. [Google Scholar] [CrossRef] [PubMed]
- de Reuver, R.; Verdonck, S.; Dierick, E.; Nemegeer, J.; Hessmann, E.; Ahmad, S.; Jans, M.; Blancke, G.; Van Nieuwerburgh, F.; Botzki, A.; et al. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 2022, 607, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, N.W.; Ames, J.M.; Maurano, M.; Chu, L.H.; Somfleth, K.Y.; Gokhale, N.S.; Werner, M.; Snyder, J.M.; Lichauco, K.; Savan, R.; et al. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature 2022, 607, 769–775. [Google Scholar] [CrossRef]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar] [CrossRef]
- Hajji, K.; Sedmik, J.; Cherian, A.; Amoruso, D.; Keegan, L.P.; O’Connell, M.A. ADAR2 enzymes: Efficient site-specific RNA editors with gene therapy aspirations. RNA 2022, 28, 1281–1297. [Google Scholar] [CrossRef]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef]
- Chen, C.X.; Cho, D.S.; Wang, Q.; Lai, F.; Carter, K.C.; Nishikura, K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000, 6, 755–767. [Google Scholar] [CrossRef]
- Oakes, E.; Anderson, A.; Cohen-Gadol, A.; Hundley, H.A. Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma. J. Biol. Chem. 2017, 292, 4326–4335. [Google Scholar] [CrossRef]
- Raghava Kurup, R.; Oakes, E.K.; Manning, A.C.; Mukherjee, P.; Vadlamani, P.; Hundley, H.A. RNA binding by ADAR3 inhibits adenosine-to-inosine editing and promotes expression of immune response protein MAVS. J. Biol. Chem. 2022, 298, 102267. [Google Scholar] [CrossRef] [PubMed]
- Mladenova, D.; Barry, G.; Konen, L.M.; Pineda, S.S.; Guennewig, B.; Avesson, L.; Zinn, R.; Schonrock, N.; Bitar, M.; Jonkhout, N.; et al. Adar3 Is Involved in Learning and Memory in Mice. Front. Neurosci. 2018, 12, 243. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 2011, 411, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benko, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef]
- Pellett, E.P.; Roizman, B. Herpesviridae. In Fields of Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2017; Volume 2. [Google Scholar]
- Murthy, S.; Couacy-Hymann, E.; Metzger, S.; Nowak, K.; De Nys, H.; Boesch, C.; Wittig, R.; Jarvis, M.A.; Leendertz, F.H.; Ehlers, B. Absence of frequent herpesvirus transmission in a nonhuman primate predator-prey system in the wild. J. Virol. 2013, 87, 10651–10659. [Google Scholar] [CrossRef]
- Rosani, U.; Gaia, M.; Delmont, T.O.; Krupovic, M. Tracing the invertebrate herpesviruses in the global sequence datasets. Front. Mar. Sci. 2023, 10, 1159754. [Google Scholar] [CrossRef]
- Roizman, B.; Knipe, D.M.; Whitley, R.J. Herpes Simplex Viruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: New York, NY, USA, 2013; Volume 2. [Google Scholar]
- Cliffe, A.R.; Garber, D.A.; Knipe, D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009, 83, 8182–8190. [Google Scholar] [CrossRef]
- Chen, S.H.; Kramer, M.F.; Schaffer, P.A.; Coen, D.M. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 1997, 71, 5878–5884. [Google Scholar] [CrossRef]
- Perng, G.C.; Jones, C.; Ciacci-Zanella, J.; Stone, M.; Henderson, G.; Yukht, A.; Slanina, S.M.; Hofman, F.M.; Ghiasi, H.; Nesburn, A.B.; et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 2000, 287, 1500–1503. [Google Scholar] [CrossRef]
- Thompson, R.L.; Sawtell, N.M. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 2001, 75, 6660–6675. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Zhou, C.; Johnson, K.E.; Colgrove, R.C.; Coen, D.M.; Knipe, D.M. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 2005, 102, 16055–16059. [Google Scholar] [CrossRef] [PubMed]
- Jurak, I.; Kramer, M.F.; Mellor, J.C.; van Lint, A.L.; Roth, F.P.; Knipe, D.M.; Coen, D.M. Numerous Conserved and Divergent MicroRNAs Expressed by Herpes Simplex Viruses 1 and 2. J. Virol. 2010, 84, 4659–4672. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Nagel, M.A.; Cohrs, R.J.; Gilden, D.H.; Cullen, B.R. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J. Virol. 2009, 83, 10677–10683. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Wang, K.; Tang, S.; Krause, P.R.; Mont, E.K.; Cohen, J.I.; Cullen, B.R. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 2010, 84, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Y.; Liu, X.J.; Chen, X.Q.; Zhou, X.S.; Du, T.; Roizman, B.; Zhou, G.Y. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc. Natl. Acad. Sci. USA 2016, 113, E894–E901. [Google Scholar] [CrossRef]
- Cokaric Brdovcak, M.; Zubkovic, A.; Ferencic, A.; Sosa, I.; Stemberga, V.; Cuculic, D.; Rokic, F.; Vugrek, O.; Hackenberg, M.; Jurak, I. Herpes simplex virus 1 miRNA sequence variations in latently infected human trigeminal ganglia. Virus Res. 2018, 256, 90–95. [Google Scholar] [CrossRef]
- Pan, D.; Pesola, J.M.; Li, G.; McCarron, S.; Coen, D.M. Mutations Inactivating Herpes Simplex Virus 1 MicroRNA miR-H2 Do Not Detectably Increase ICP0 Gene Expression in Infected Cultured Cells or Mouse Trigeminal Ganglia. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Flores, O.; Nakayama, S.; Whisnant, A.W.; Javanbakht, H.; Cullen, B.R.; Bloom, D.C. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J. Virol. 2013, 87, 6589–6603. [Google Scholar] [CrossRef]
- Barrozo, E.R.; Nakayama, S.; Singh, P.; Neumann, D.M.; Bloom, D.C. Herpes Simplex Virus 1 MicroRNA miR-H8 Is Dispensable for Latency and Reactivation In Vivo. J. Virol. 2021, 95, e02179-20. [Google Scholar] [CrossRef]
- Jurak, I.; Silverstein, L.B.; Sharma, M.; Coen, D.M. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J. Virol. 2012, 86, 10093–10102. [Google Scholar] [CrossRef] [PubMed]
- Zubkovic, A.; Gomes, C.; Parchure, A.; Cesarec, M.; Ferencic, A.; Rokic, F.; Jakovac, H.; Whitford, A.L.; Dochnal, S.A.; Cliffe, A.R.; et al. HSV-1 miRNAs are post-transcriptionally edited in latently infected human ganglia. J. Virol. 2023, e0073023. [Google Scholar] [CrossRef] [PubMed]
- Arvin, A.; Gilden, D. Varicela-Zoster Virus. In Fields of Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: New York, NY, USA, 2013; Volume 2. [Google Scholar]
- Depledge, D.P.; Sadaoka, T.; Ouwendijk, W.J.D. Molecular Aspects of Varicella-Zoster Virus Latency. Viruses 2018, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Prazsak, I.; Moldovan, N.; Balazs, Z.; Tombacz, D.; Megyeri, K.; Szucs, A.; Csabai, Z.; Boldogkoi, Z. Long-read sequencing uncovers a complex transcriptome topology in varicella zoster virus. BMC Genom. 2018, 19, 873. [Google Scholar] [CrossRef]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus Genome Integration into Telomeric Repeats of Host Cell Chromosomes. Annu. Rev. Virol. 2014, 1, 215–235. [Google Scholar] [CrossRef]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef]
- Cantello, J.L.; Anderson, A.S.; Morgan, R.W. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek’s disease virus. J. Virol. 1994, 68, 6280–6290. [Google Scholar] [CrossRef]
- Figueroa, T.; Boumart, I.; Coupeau, D.; Rasschaert, D. Hyperediting by ADAR1 of a new herpesvirus lncRNA during the lytic phase of the oncogenic Marek’s disease virus. J. Gen. Virol. 2016, 97, 2973–2988. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Shenk, T.; Griffiths, P.D.; Pass, R.F. Cytomegaloviruses. In Fields of Virology, 6th ed.; Lippincott Williams & Wilklins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Goodrum, F. The complex biology of human cytomegalovirus latency. Adv. Virus Res. 2022, 112, 31–85. [Google Scholar] [CrossRef]
- Slavuljica, I.; Krmpotic, A.; Jonjic, S. Manipulation of NKG2D ligands by cytomegaloviruses: Impact on innate and adaptive immune response. Front. Immunol. 2011, 2, 85. [Google Scholar] [CrossRef]
- Nachmani, D.; Lankry, D.; Wolf, D.G.; Mandelboim, O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 2010, 11, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Dolken, L.; Perot, J.; Cognat, V.; Alioua, A.; John, M.; Soutschek, J.; Ruzsics, Z.; Koszinowski, U.; Voinnet, O.; Pfeffer, S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 2007, 81, 13771–13782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Longnecker, R.M.; Kieff, E.; Cohen, J.I. Epstein-Barr Virus. In Fields of Virology, 6th ed.; Lippincott Williams & Wilklins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Frappier, L. Epstein-Barr virus: Current questions and challenges. Tumour Virus Res. 2021, 12, 200218. [Google Scholar] [CrossRef] [PubMed]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. EBV Noncoding RNAs. Curr. Top. Microbiol. Immunol. 2015, 391, 181–217. [Google Scholar] [CrossRef]
- Iizasa, H.; Wulff, B.E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; et al. Editing of Epstein-Barr Virus-encoded BART6 MicroRNAs Controls Their Dicer Targeting and Consequently Affects Viral Latency. J. Biol. Chem. 2010, 285, 33358–33370. [Google Scholar] [CrossRef]
- Lei, T.; Yuen, K.S.; Tsao, S.W.; Chen, H.; Kok, K.H.; Jin, D.Y. Perturbation of biogenesis and targeting of Epstein-Barr virus-encoded miR-BART3 microRNA by adenosine-to-inosine editing. J. Gen. Virol. 2013, 94, 2739–2744. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Corcoran, D.L.; Gottwein, E.; Frank, C.L.; Kang, D.; Hafner, M.; Nusbaum, J.D.; Feederle, R.; Delecluse, H.J.; Luftig, M.A.; et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012, 8, e1002484. [Google Scholar] [CrossRef]
- Cao, S.; Moss, W.; O’Grady, T.; Concha, M.; Strong, M.J.; Wang, X.; Yu, Y.; Baddoo, M.; Zhang, K.; Fewell, C.; et al. New Noncoding Lytic Transcripts Derived from the Epstein-Barr Virus Latency Origin of Replication, oriP, Are Hyperedited, Bind the Paraspeckle Protein, NONO/p54nrb, and Support Viral Lytic Transcription. J. Virol. 2015, 89, 7120–7132. [Google Scholar] [CrossRef]
- Damania, B.A.; Ceserman, E. Kaposi’s Sarcoma-Associated Herpesvirus. In Fields of Virology, 6th ed.; Lippincott Williams & Wilklins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Dissinger, N.J.; Damania, B. Recent advances in understanding Kaposi’s sarcoma-associated herpesvirus. F1000Research 2016, 5, 740. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Wang, H.; Herndier, B.; Ganem, D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 1996, 93, 6641–6646. [Google Scholar] [CrossRef] [PubMed]
- Sadler, R.; Wu, L.; Forghani, B.; Renne, R.; Zhong, W.; Herndier, B.; Ganem, D. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1999, 73, 5722–5730. [Google Scholar] [CrossRef] [PubMed]
- Forte, E.; Raja, A.N.; Shamulailatpam, P.; Manzano, M.; Schipma, M.J.; Casey, J.L.; Gottwein, E. MicroRNA-mediated transformation by the Kaposi’s sarcoma-associated herpesvirus Kaposin locus. J. Virol. 2015, 89, 2333–2341. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grasser, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef]
- Gottwein, E.; Cai, X.; Cullen, B.R. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 2006, 80, 5321–5326. [Google Scholar] [CrossRef]
- Gandy, S.Z.; Linnstaedt, S.D.; Muralidhar, S.; Cashman, K.A.; Rosenthal, L.J.; Casey, J.L. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol. 2007, 81, 13544–13551. [Google Scholar] [CrossRef]
- Rajendren, S.; Ye, X.; Dunker, W.; Richardson, A.; Karijolich, J. The cellular and KSHV A-to-I RNA editome in primary effusion lymphoma and its role in the viral lifecycle. Nat. Commun. 2023, 14, 1367. [Google Scholar] [CrossRef]
- Liu, Q.; Rao, Y.; Tian, M.; Zhang, S.; Feng, P. Modulation of Innate Immune Signaling Pathways by Herpesviruses. Viruses 2019, 11, 572. [Google Scholar] [CrossRef]
- O’Connor, C.M.; Sen, G.C. Innate Immune Responses to Herpesvirus Infection. Cells 2021, 10, 2122. [Google Scholar] [CrossRef]
- Zhang, H.; Ni, G.; Damania, B. ADAR1 Facilitates KSHV Lytic Reactivation by Modulating the RLR-Dependent Signaling Pathway. Cell Rep. 2020, 31, 107564. [Google Scholar] [CrossRef] [PubMed]
- Mushegian, A.; Karin, E.L.; Pupko, T. Sequence analysis of malacoherpesvirus proteins: Pan-herpesvirus capsid module and replication enzymes with an ancient connection to “Megavirales”. Virology 2018, 513, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Rosani, U.; Bai, C.M.; Maso, L.; Shapiro, M.; Abbadi, M.; Domeneghetti, S.; Wang, C.M.; Cendron, L.; MacCarthy, T.; Venier, P. A-to-I editing of Malacoherpesviridae RNAs supports the antiviral role of ADAR1 in mollusks. BMC Evol. Biol. 2019, 19, 149. [Google Scholar] [CrossRef] [PubMed]
- Rosani, U.; Bortoletto, E.; Montagnani, C.; Venier, P. ADAR-Editing during Ostreid Herpesvirus 1 Infection in Crassostrea gigas: Facts and Limitations. mSphere 2022, 7, e0001122. [Google Scholar] [CrossRef]
- Bai, C.M.; Rosani, U.; Zhang, X.; Xin, L.S.; Bortoletto, E.; Wegner, K.M.; Wang, C.M. Viral Decoys: The Only Two Herpesviruses Infecting Invertebrates Evolved Different Transcriptional Strategies to Deflect Post-Transcriptional Editing. Viruses 2021, 13, 1971. [Google Scholar] [CrossRef]
- Cheng, A.Z.; Moraes, S.N.; Shaban, N.M.; Fanunza, E.; Bierle, C.J.; Southern, P.J.; Bresnahan, W.A.; Rice, S.A.; Harris, R.S. APOBECs and Herpesviruses. Viruses 2021, 13, 390. [Google Scholar] [CrossRef]
- Shen, H.Q.; An, O.; Ren, X.; Song, Y.Y.; Tang, Z.J.; Ke, X.Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Molias, F.B.; et al. ADARs act as potent regulators of circular transcriptome in cancer. Nat. Commun. 2022, 13, 1508. [Google Scholar] [CrossRef]
- Kapoor, U.; Licht, K.; Amman, F.; Jakobi, T.; Martin, D.; Dieterich, C.; Jantsch, M.F. ADAR-deficiency perturbs the global splicing landscape in mouse tissues. Genome Res. 2020, 30, 1107–1118. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef]
- Haque, S.; Harries, L.W. Circular RNAs (circRNAs) in Health and Disease. Genes 2017, 8, 353. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, S.; Yang, N.; Zou, Y.; Zheng, D.; Xiao, T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017, 404, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, T.; Oh, D.; Dremel, S.; Mahesh, G.; Koparde, V.N.; Duncan, G.; Andresson, T.; Ziegelbauer, J.M. A virus-induced circular RNA maintains latent infection of Kaposi’s sarcoma herpesvirus. Proc. Natl. Acad. Sci. USA 2023, 120, e2212864120. [Google Scholar] [CrossRef] [PubMed]
- Garaigorta, U.; Chisari, F.V. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 2009, 6, 513–522. [Google Scholar] [CrossRef]
- Lei, M.; Liu, Y.; Samuel, C.E. Adenovirus VAI RNA antagonizes the RNA-editing activity of the ADAR adenosine deaminase. Virology 1998, 245, 188–196. [Google Scholar] [CrossRef] [PubMed]
- de Chassey, B.; Aublin-Gex, A.; Ruggieri, A.; Meyniel-Schicklin, L.; Pradezynski, F.; Davoust, N.; Chantier, T.; Tafforeau, L.; Mangeot, P.E.; Ciancia, C.; et al. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 2013, 9, e1003440. [Google Scholar] [CrossRef]
- Liu, Y.; Wolff, K.C.; Jacobs, B.L.; Samuel, C.E. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology 2001, 289, 378–387. [Google Scholar] [CrossRef]
| Virus Taxonomy | Virus | ADAR Activity | Ref. | ||
|---|---|---|---|---|---|
| Herpesvirales | Ortoherpesviridae | Alphaherpesvirinae | HSV-1 (HHV-1) | ADAR1 expression levels maintained during productive infection. Editing of HSV-1 miR-H2-3p in latency and to lesser extent in productive infection. Function: increased targeting repertoire of miR-H2-3p. | [13,49,54] |
| VZV (HHV-3) | Dynamics of ADAR expression levels: unknown. Editing of lncRNA NTO3 (antisense to ORF63). Function: unknown. | [57] | |||
| GaHV-2 | Dynamics of ADAR expression levels: unknown. Editing of ERL lncRNA. Function: unknown. | [61] | |||
| Betaherpesvirinae | HCMV (HHV-5) | ADAR1 p110 is upregulated in productive infection. Editing of host miR-376a. Function: edited miRNA gains specificity to downregulates HLA-E and abolishes targeting of MICB (ligand of activating NKG2D receptor), facilitating elimination of HCMV infected cells. | [13] | ||
| Gammaherpesvirinae | EBV (HHV-4) | Dynamics of ADAR expression levels: unknown. Editing of pri-BHRF1-1, pri-miR-BART3,-BART6, -BART8, -BART11, and -BART16. Editing of vlncRNA oriPtL and oriPtR. Functions: affected Drosha processing of pri-miR-BART6 and -BART3 resulting in lower levels of miRNAs, and loss of posttranscriptional regulation of their targets (Dicer). miR-BART3 seed sequence editing abolished Dicer targeting. Functions of edited oriPtL and oriPtR: unknown. | [71,72,75] | ||
| KSHV (HHV-8) | ADAR1 expression levels maintained from latent to lytic infection. * ADAR1 (all forms) increased during reactivation. * Editing of K12 transcript, LANA, RTA, etc., and pri-miR-K12-10, pri-miR-K12-4 Functions: Editing eliminates K12 transforming activity and reduces pri-miR-K12-4 processing by Drosha. Increased repertoire of miR-K12-4 targets. ADAR1 prevents activation of RIG-I signaling and enables efficient virus reactivation. | [14,81,83,84,87] | |||
| Malacoherpesviridae | OsHV-1 HaHV-1 | ADAR1 upregulated in productively infected host. Editing: increased global editing of viral and host transcripts during infection. Function: unknown. | [89,90,91] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanišević, V.; Žilić, L.; Čunko, M.; Fadiga, H.; Munitić, I.; Jurak, I. RNA Editing-Dependent and -Independent Roles of Adenosine Deaminases Acting on RNA Proteins in Herpesvirus Infection—Hints on Another Layer of Complexity. Viruses 2023, 15, 2007. https://doi.org/10.3390/v15102007
Ivanišević V, Žilić L, Čunko M, Fadiga H, Munitić I, Jurak I. RNA Editing-Dependent and -Independent Roles of Adenosine Deaminases Acting on RNA Proteins in Herpesvirus Infection—Hints on Another Layer of Complexity. Viruses. 2023; 15(10):2007. https://doi.org/10.3390/v15102007
Chicago/Turabian StyleIvanišević, Vlatka, Lidia Žilić, Marina Čunko, Hana Fadiga, Ivana Munitić, and Igor Jurak. 2023. "RNA Editing-Dependent and -Independent Roles of Adenosine Deaminases Acting on RNA Proteins in Herpesvirus Infection—Hints on Another Layer of Complexity" Viruses 15, no. 10: 2007. https://doi.org/10.3390/v15102007
APA StyleIvanišević, V., Žilić, L., Čunko, M., Fadiga, H., Munitić, I., & Jurak, I. (2023). RNA Editing-Dependent and -Independent Roles of Adenosine Deaminases Acting on RNA Proteins in Herpesvirus Infection—Hints on Another Layer of Complexity. Viruses, 15(10), 2007. https://doi.org/10.3390/v15102007






