Characterization of SARS-CoV-2 Mutational Signatures from 1.5+ Million Raw Sequencing Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Variant Calling
2.2. Dataset
2.3. Mutational Signatures Analysis
2.4. Pango Analysis
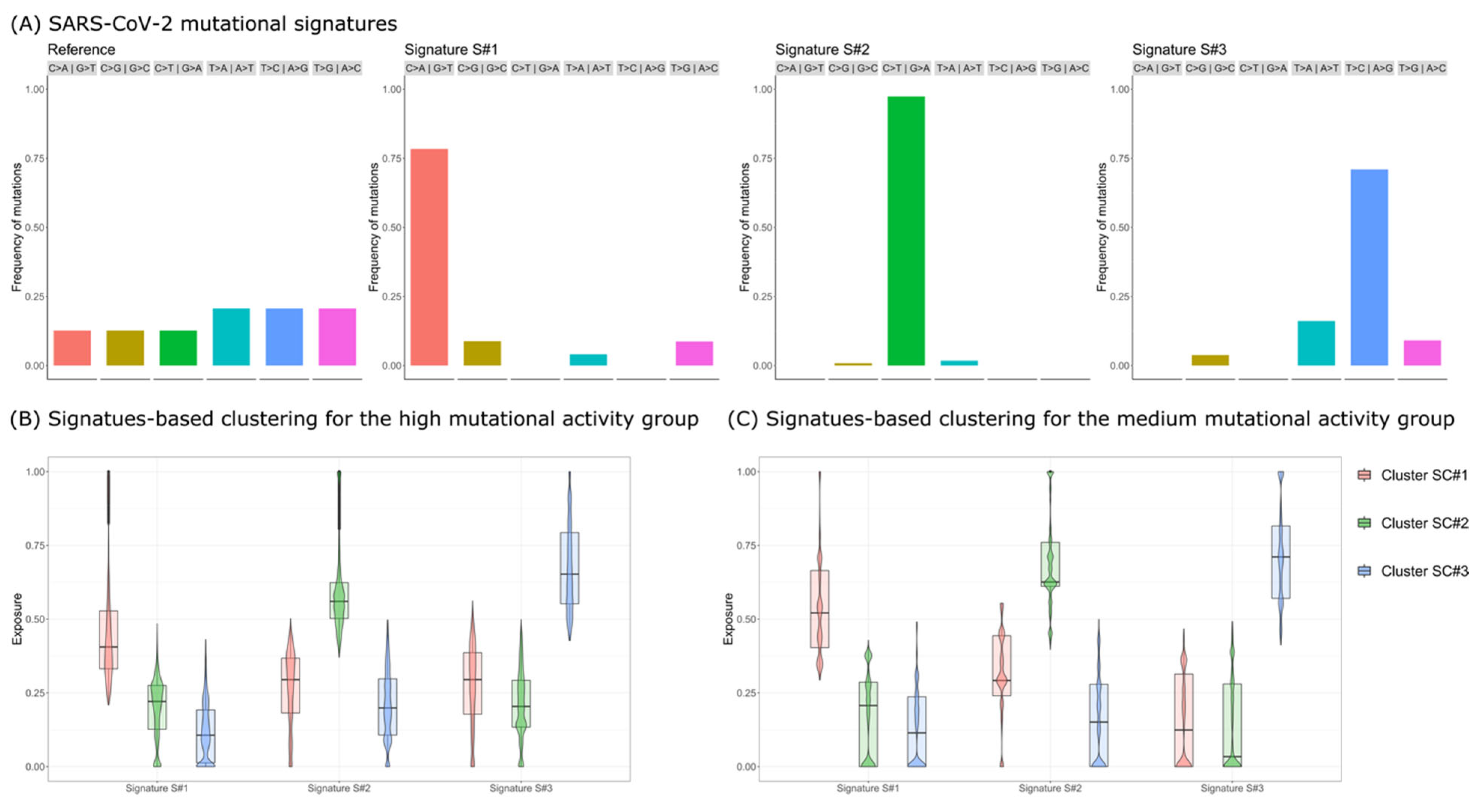
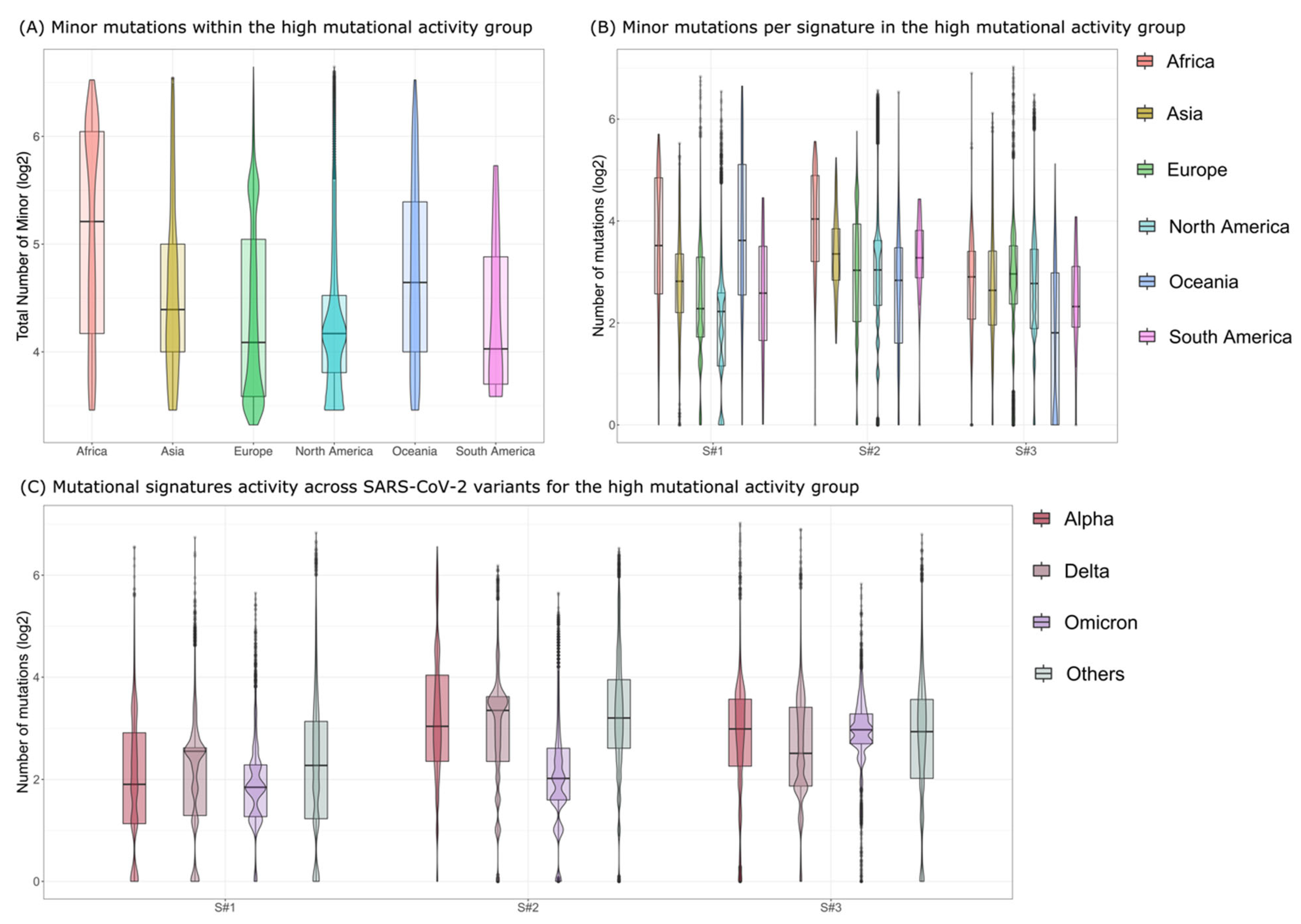
3. Results
- Signature S#1: mostly characterized by C>A|G>T mutations and previously associated with reactive oxygen species (ROS) activity [20];
- Signature S#2: mostly characterized by C>T|G>A mutations and previously associated with APOBEC activity [19];
- Signature S#3: mostly characterized by T>C|A>G mutations and previously associated with adenosine deaminase acting on RNA (ADAR) activity [20].
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Machingaidze, S.; Wiysonge, C.S. Understanding COVID-19 vaccine hesitancy. Nat. Med. 2021, 27, 1338–1339. [Google Scholar] [CrossRef] [PubMed]
- Solís Arce, J.S.; Warren, S.S.; Meriggi, N.F.; Scacco, A.; McMurry, N.; Voors, M.; Syunyaev, G.; Malik, A.A.; Aboutajdine, S.; Adeojo, O.; et al. COVID-19 vaccine acceptance and hesitancy in low-and middle-income countries. Nat. Med. 2021, 27, 1385–1394. [Google Scholar] [CrossRef]
- Patwary, M.M.; Alam, A.; Bardhan, M.; Disha, A.S.; Haque, Z.; Billah, S.M.; Kabir, P.; Browning, M.H.E.M.; Rahman, M.; Parsa, A.D.; et al. COVID-19 Vaccine Acceptance among Low- and Lower-Middle-Income Countries: A Rapid Systematic Review and Meta-Analysis. Vaccines 2022, 10, 427. [Google Scholar] [CrossRef]
- Africa CDC. Africa CDC—COVID-19 Daily Updates. Africa CDC. Available online: https://africacdc.org/covid-19/ (accessed on 6 October 2022).
- Petersen, E.; Ntoumi, F.; Hui, D.S.; Abubakar, A.; Kramer, L.D.; Obiero, C.; Tambyah, P.A.; Blumberg, L.; Yapi, R.; Al-Abri, S.; et al. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B. 1.1. 529)-highlights Africa’s research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int. J. Infect. Dis. 2022, 114, 268–272. [Google Scholar] [CrossRef]
- Araf, Y.; Akter, F.; Tang, Y.D.; Fatemi, R.; Parvez MS, A.; Zheng, C.; Hossain, M.G. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022, 94, 1825–1832. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Njenga, M.K.; Dawa, J.; Nanyingi, M.; Gachohi, J.; Ngere, I.; Letko, M.; Otieno, C.F.; Gunn, B.M.; Osoro, E. Why is There Low Morbidity and Mortality of COVID-19 in Africa? Am. J. Trop. Med. Hyg. 2020, 103, 564–569. [Google Scholar] [CrossRef]
- Olson, M.E.; Harris, R.S.; Harki, D.A. APOBEC Enzymes as Targets for Virus and Cancer Therapy. Cell Chem. Biol. 2017, 25, 36–49. [Google Scholar] [CrossRef]
- Willems, L.; Gillet, N. APOBEC3 Interference during Replication of Viral Genomes. Viruses 2015, 7, 2999–3018. [Google Scholar] [CrossRef]
- Harris, R.S. Enhancing immunity to HIV through APOBEC. Nat. Biotechnol. 2008, 26, 1089–1090. [Google Scholar] [CrossRef]
- Guylaine, H.; Mansky, L.M.; Harris, R.S. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006, 8, 148–157. [Google Scholar]
- Matume, N.D.; Tebit, D.M.; Gray, L.R.; Turner, S.D.; Rekosh, D.; Bessong, P.O.; Hammarskjöld, M.-L. Characterization of APOBEC3 variation in a population of HIV-1 infected individuals in northern South Africa. BMC Med. Genet. 2019, 20, 21. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Jeremy, R.; Simmonds, P. Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution. Virology 2021, 556, 62–72. [Google Scholar]
- Ramazzotti, D.; Angaroni, F.; Maspero, D.; Mauri, M.; D’Aliberti, D.; Fontana, D.; Antoniotti, M.; Elli, E.M.; Graudenzi, A.; Piazza, R. Large-scale analysis of SARS-CoV-2 synonymous mutations reveals the adaptation to the human codon usage during the virus evolution. Virus Evol. 2022, 8, veac026. [Google Scholar] [CrossRef]
- Graudenzi, A.; Maspero, D.; Angaroni, F.; Piazza, R.; Ramazzotti, D. Mutational signatures and heterogeneous host response revealed via large-scale characterization of SARS-CoV-2 genomic diversity. iScience 2021, 24, 102116. [Google Scholar] [CrossRef]
- Ramazzotti, D.; Angaroni, F.; Maspero, D.; Gambacorti-Passerini, C.; Antoniotti, M.; Graudenzi, A.; Piazza, R. VERSO: A comprehensive framework for the inference of robust phylogenies and the quantification of intra-host genomic diversity of viral samples. Patterns 2021, 2, 100212. [Google Scholar] [CrossRef]
- Maspero, D.; Angaroni, F.; Porro, D.; Piazza, R.; Graudenzi, A.; Ramazzotti, D. VirMutSig: Discovery and assignment of viral mutational signatures from sequencing data. STAR Protoc. 2021, 2, 100911. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Mella, L.; Lal, A.; Angaroni, F.; Maspero, D.; Piazza, R.; Sidow, A.; Antoniotti, M.; Graudenzi, A.; Ramazzotti, D. SparseSignatures: An R package using LASSO-regularized non-negative matrix factorization to identify mutational signatures from human tumor samples. STAR Protoc. 2022, 3, 101513. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Liu, K.; Tibshirani, R.; Sidow, A.; Ramazzotti, D. De novo mutational signature discovery in tumor genomes using SparseSignatures. PLOS Comput. Biol. 2021, 17, e1009119. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aroldi, A.; Angaroni, F.; D’Aliberti, D.; Spinelli, S.; Crespiatico, I.; Crippa, V.; Piazza, R.; Graudenzi, A.; Ramazzotti, D. Characterization of SARS-CoV-2 Mutational Signatures from 1.5+ Million Raw Sequencing Samples. Viruses 2023, 15, 7. https://doi.org/10.3390/v15010007
Aroldi A, Angaroni F, D’Aliberti D, Spinelli S, Crespiatico I, Crippa V, Piazza R, Graudenzi A, Ramazzotti D. Characterization of SARS-CoV-2 Mutational Signatures from 1.5+ Million Raw Sequencing Samples. Viruses. 2023; 15(1):7. https://doi.org/10.3390/v15010007
Chicago/Turabian StyleAroldi, Andrea, Fabrizio Angaroni, Deborah D’Aliberti, Silvia Spinelli, Ilaria Crespiatico, Valentina Crippa, Rocco Piazza, Alex Graudenzi, and Daniele Ramazzotti. 2023. "Characterization of SARS-CoV-2 Mutational Signatures from 1.5+ Million Raw Sequencing Samples" Viruses 15, no. 1: 7. https://doi.org/10.3390/v15010007
APA StyleAroldi, A., Angaroni, F., D’Aliberti, D., Spinelli, S., Crespiatico, I., Crippa, V., Piazza, R., Graudenzi, A., & Ramazzotti, D. (2023). Characterization of SARS-CoV-2 Mutational Signatures from 1.5+ Million Raw Sequencing Samples. Viruses, 15(1), 7. https://doi.org/10.3390/v15010007




