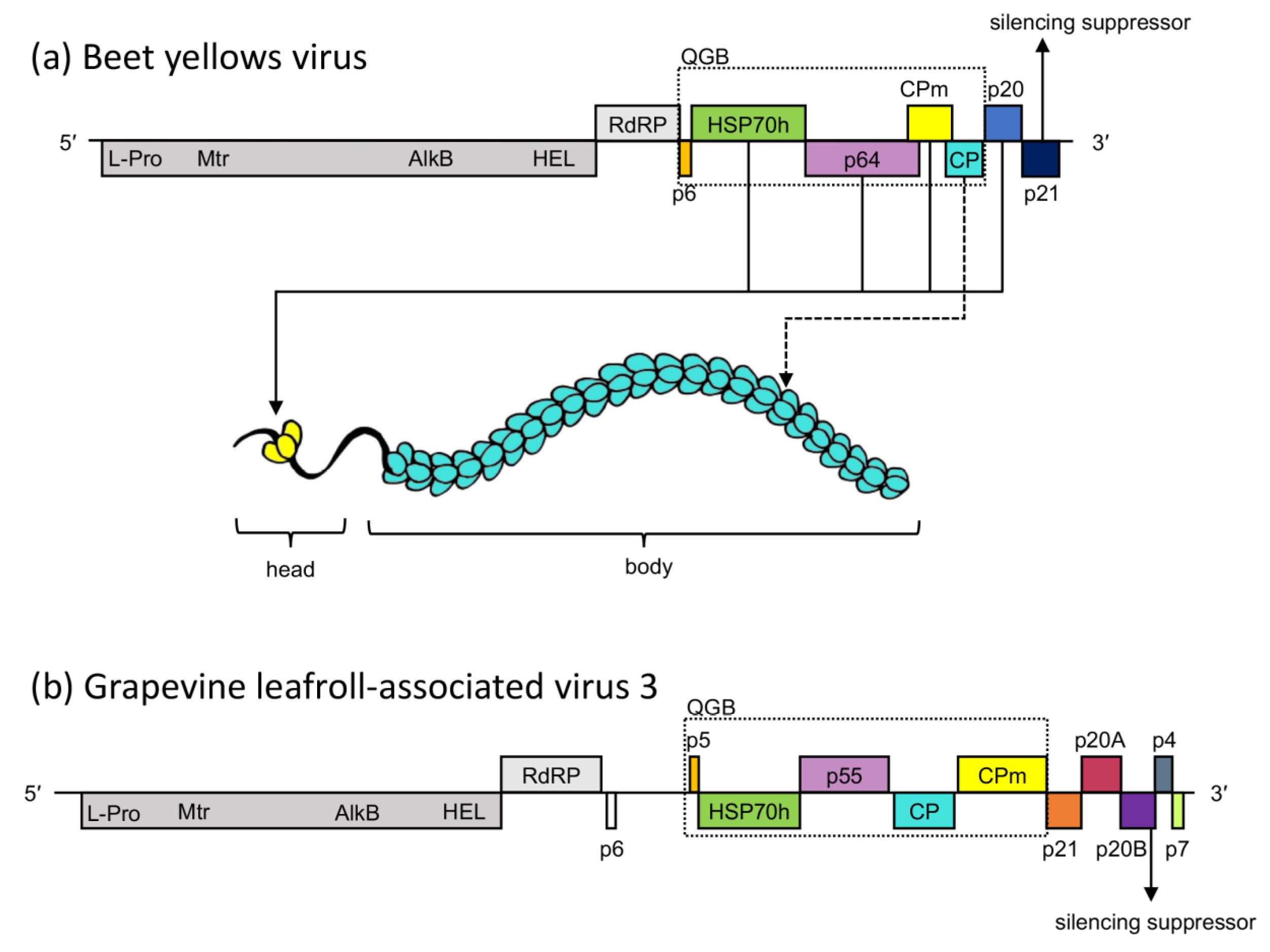
Identification of Interactions between Proteins Encoded by Grapevine Leafroll-Associated Virus 3
Abstract
1. Introduction
2. Materials and Methods
2.1. Source Material
2.2. Construction of Yeast Two-Hybrid Vectors
2.3. Yeast Transformation, Autoactivation and Toxicity Screening
2.4. SDS-PAGE and Western Blotting
2.5. Y2H Small-Scale Matings and Screening for Protein Interactions
2.6. Bimolecular Fluorescence Complementation Vectors
2.7. Bimolecular Fluorescence Complementation Assays in Nicotiana benthamiana
- The highest performing combination of orientations (Protein A and B)
- Protein A with A. thaliana CIPK24 in the orientation of Protein B
- Protein B with A. thaliana CIPK24 in the orientation of Protein A
- No YFP constructs, to serve as an internal reference for expression strength and a baseline of background fluorescence.
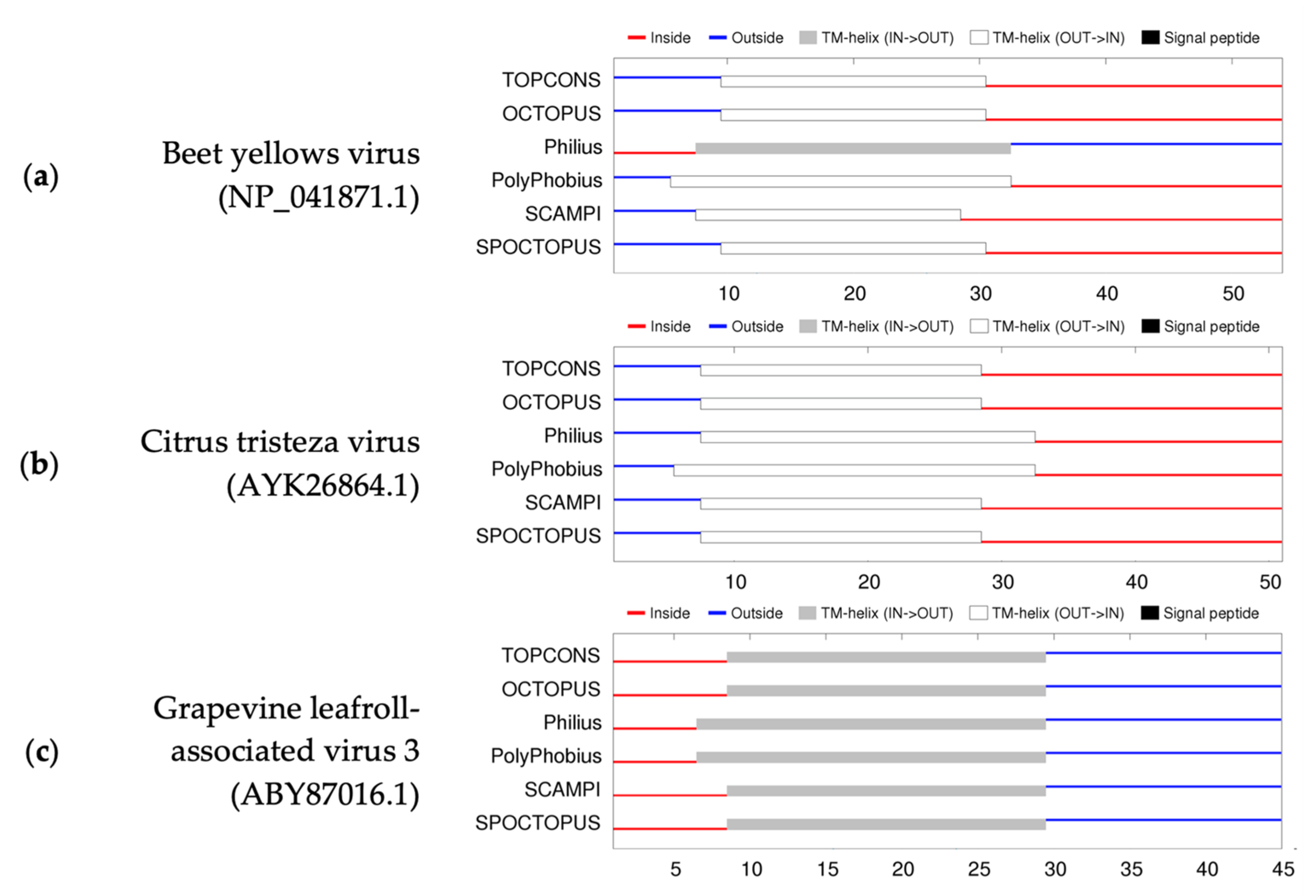
2.8. GLRaV-3 Transmembrane Protein Analyses
3. Results
3.1. Yeast Two-Hybrid Assays
- Y2HGold[pGBKT7::HSP70h] x Y187[pGADT7::HSP70h]
- Y2HGold[pGBKT7::CP] x Y187[pGADT7::HSP70h]
- Y2HGold[pGBKT7::CP] x Y187[pGADT7::p6]
- Y2HGold[pGBKT7::p20B] x Y187[pGADT7::p20B]
3.2. Bimolecular Fluorescence Complementation Assays
3.3. GLRaV-3 Transmembrane Protein Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martelli, G.P. A Brief Historical Account of the Family Closteroviridae. In Citrus Tristeza Virus: Methods and Protocols; Catara, A.F., Bar-Joseph, M., Licciardello, G., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 7–13. ISBN 9781493995585. [Google Scholar]
- Fuchs, M.; Bar-Joseph, M.; Candresse, T.; Maree, H.J.; Martelli, G.P.; Melzer, M.J.; Menzel, W.; Minafra, A.; Sabanadzovic, S.; Report Consortium, I. ICTV Virus Taxonomy Profile: Closteroviridae. J. Gen. Virol. 2020, 101, 364–365. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Boyko, V.P.; Agranovsky, A.A.; Koonin, E.V. Phylogeny of Capsid Proteins of Rod-Shaped and Filamentous RNA Plant Viruses: Two Families with Distinct Patterns of Sequence and Probably Structure Conservation. Virology 1991, 184, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Callaway, A.; Giesman-Cookmeyer, D.; Gillock, E.T.; Sit, T.L.; Lommel, S.A. The Multifunctional Capsid Proteins of Plant RNA Viruses. Annu. Rev. Phytopathol. 2001, 39, 419–460. [Google Scholar] [CrossRef] [PubMed]
- Agranovsky, A.A.; Lesemann, D.E.; Maiss, E.; Hull, R.; Atabekov, J.G. “Rattlesnake” Structure of a Filamentous Plant RNA Virus Built of Two Capsid Proteins. Proc. Natl. Acad. Sci. USA 1995, 92, 2470–2473. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier Academic Press: San Diego, CA, USA, 2011; ISBN 9780123846846. [Google Scholar]
- Alzhanova, D.V.; Napuli, A.J.; Creamer, R.; Dolja, V.V. Cell-to-Cell Movement and Assembly of a Plant Closterovirus: Roles for the Capsid Proteins and Hsp70 Homolog. EMBO J. 2001, 20, 6997–7007. [Google Scholar] [CrossRef] [PubMed]
- Napuli, A.J.; Alzhanova, D.V.; Doneanu, C.E.; Barofsky, D.F.; Koonin, E.V.; Dolja, V.V. The 64-Kilodalton Capsid Protein Homolog of Beet Yellows Virus Is Required for Assembly of Virion Tails. J. Virol. 2003, 77, 2377–2384. [Google Scholar] [CrossRef]
- Tilkunova, T.Y.; Zinovkin, R.A.; Agranovsky, A.A. RNA-Binding Properties of the Proteins of Beet Yellows Closterovirus. Mol. Biol. 2004, 38, 464–468. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Gowda, S.; Ayllón, M.A.; Dawson, W.O. Closterovirus Bipolar Virion: Evidence for Initiation of Assembly by Minor Coat Protein and Its Restriction to the Genomic RNA 5′ Region. Proc. Natl. Acad. Sci. USA 2004, 101, 799–804. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Hagiwara, Y.; Dolja, V.V. HSP70 Homolog Functions in Cell-to-Cell Movement of a Plant Virus. Proc. Natl. Acad. Sci. USA 1999, 96, 14771–14776. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Andreev, I.A.; Prokhnevsky, A.I.; Duncan, G.H.; Taliansky, M.E.; Dolja, V.V. Complex Molecular Architecture of Beet Yellows Virus Particles. Proc. Natl. Acad. Sci. USA 2004, 101, 5030–5035. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Pan, Y.-W.; Dolja, V.V. Movement Protein of a Closterovirus Is a Type III Integral Transmembrane Protein Localized to the Endoplasmic Reticulum. J. Virol. 2004, 78, 3704–3709. [Google Scholar] [CrossRef]
- Prokhnevsky, A.I.; Peremyslov, V.V.; Napuli, A.J.; Dolja, V.V. Interaction between Long-Distance Transport Factor and Hsp70-Related Movement Protein of Beet Yellows Virus. J. Virol. 2002, 76, 11003–11011. [Google Scholar] [CrossRef]
- Reed, J.C.; Kasschau, K.D.; Prokhnevsky, A.I.; Gopinath, K.; Pogue, G.P.; Carrington, J.C.; Dolja, V.V. Suppressor of RNA Silencing Encoded by Beet Yellows Virus. Virology 2003, 306, 203–209. [Google Scholar] [CrossRef]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.-X.; Falk, B.W.; Dawson, W.O.; Ding, S.-W. Three Distinct Suppressors of RNA Silencing Encoded by a 20-Kb Viral RNA Genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef]
- Chiba, M.; Reed, J.C.; Prokhnevsky, A.I.; Chapman, E.J.; Mawassi, M.; Koonin, E.V.; Carrington, J.C.; Dolja, V.V. Diverse Suppressors of RNA Silencing Enhance Agroinfection by a Viral Replicon. Virology 2006, 346, 7–14. [Google Scholar] [CrossRef]
- Gouveia, P.; Dandlen, S.; Costa, Â.; Marques, N.; Nolasco, G. Identification of an RNA Silencing Suppressor Encoded by Grapevine Leafroll-Associated Virus 3. Eur. J. Plant. Pathol. 2012, 133, 237–245. [Google Scholar] [CrossRef]
- Alzhanova, D.V.; Hagiwara, Y.; Peremyslov, V.V.; Dolja, V.V. Genetic Analysis of the Cell-to-Cell Movement of Beet Yellows Closterovirus. Virology 2000, 268, 192–200. [Google Scholar] [CrossRef]
- Tian, T.; Rubio, L.; Yeh, H.-H.; Crawford, B.; Falk, B.W.Y. Lettuce Infectious Yellows Virus: In Vitro Acquisition Analysis Using Partially Purified Virions and the Whitefly Bemisia Tabaci. J. Gen. Virol. 1999, 80, 1111–1117. [Google Scholar] [CrossRef]
- Hull, R. Architecture and Assembly of Virus Particles. In Plant Virology; Academic Press: Cambridge, CA, USA, 2013; pp. 69–143. ISBN 9780123848727. [Google Scholar]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine Leafroll-Associated Virus 3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef]
- Saldarelli, P.; Giampetruzzi, A.; Maree, H.; Al Rwahnih, M. High-Throughput Sequencing: Advantages Beyond Virus Identification. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer: Cham, Switzerland, 2017; pp. 625–642. ISBN 9783319577043. [Google Scholar]
- Bester, R.; Burger, J.T.; Maree, H.J. Transcriptome Analysis Reveals Differentially Expressed Small RNAs and Genes Associated with Grapevine Leafroll-Associated Virus 3 Infections. Physiol. Mol. Plant Pathol. 2017, 100, 220–236. [Google Scholar] [CrossRef]
- Bester, R.; Burger, J.T.; Maree, H.J. Differential Expression of MiRNAs and Associated Gene Targets in Grapevine Leafroll-Associated Virus 3-Infected Plants. Arch. Virol. 2017, 162, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.J.; Zheng, Y.; Jagadeeswaran, G.; Sunkar, R.; Naidu, R.A. High-Throughput Sequence Analysis of Small RNAs in Grapevine (Vitis vinifera L.) Affected by Grapevine Leafroll Disease. Mol. Plant Pathol. 2012, 13, 1060–1076. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Hanner, R.H.; Meng, B. Transcriptomic Analyses of Grapevine Leafroll-Associated Virus 3 Infection in Leaves and Berries of ‘Cabernet Franc’. Viruses 2022, 14, 1831. [Google Scholar] [CrossRef] [PubMed]
- Prator, C.A.; Chooi, K.M.; Jones, D.; Davy, M.W.; MacDiarmid, R.M.; Almeida, R.P.P. Comparison of Two Different Host Plant Genera Responding to Grapevine Leafroll-Associated Virus 3 Infection. Sci. Rep. 2020, 10, 8505. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, A.B.; Bester, R.; Olmos, A.; Maree, H.J. Bioinformatic Tools and Genome Analysis of Citrus Tristeza Virus. In Citrus Tristeza Virus: Methods and Protocols; Catara, A.F., Bar-Joseph, M., Licciardello, G., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 163–178. ISBN 9781493995585. [Google Scholar]
- Bester, R.; Pepler, P.T.; Burger, J.T.; Maree, H.J. Relative Quantitation Goes Viral: An RT-QPCR Assay for a Grapevine Virus. J. Virol. Methods 2014, 210, 67–75. [Google Scholar] [CrossRef]
- Kushnirov, V.V. Rapid and Reliable Protein Extraction from Yeast. Yeast 2000, 16, 857–860. [Google Scholar] [CrossRef]
- Brunelle, J.L.; Green, R. One-Dimensional SDS-Polyacrylamide Gel Electrophoresis (1D SDS-PAGE). Methods Enzymol. 2014, 541, 151–159. [Google Scholar] [CrossRef]
- Waadt, R.; Schmidt, L.K.; Lohse, M.; Hashimoto, K.; Bock, R.; Kudla, J. Multicolor Bimolecular Fluorescence Complementation Reveals Simultaneous Formation of Alternative CBL/CIPK Complexes in Planta. TPJ 2008, 56, 505–516. [Google Scholar] [CrossRef]
- Waadt, R.; Kudla, J. In Planta Visualization of Protein Interactions Using Bimolecular Fluorescence Complementation (BiFC). Cold Spring Harb. Protoc. 2008, 2008, pdb.prot4995. [Google Scholar] [CrossRef]
- Horstman, A.; Nougalli Tonaco, I.A.; Boutilier, K.; Immink, R.G.H. A Cautionary Note on the Use of Split-YFP/BiFC in Plant Protein-Protein Interaction Studies. Int. J. Mol. Sci. 2014, 15, 9628–9643. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Peterson, P. F2PY: A Tool for Connecting Fortran and Python Programs. IJCSE 2009, 4, 296. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proceedings of the Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 92–96. [Google Scholar]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS Web Server for Consensus Prediction of Membrane Protein Topology and Signal Peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef]
- Ling, K.-S.; Zhu, H.-Y.; Gonsalves, D. Complete Nucleotide Sequence and Genome Organization of Grapevine Leafroll-Associated Virus 3, Type Member of the Genus Ampelovirus. J. Gen. Virol. 2004, 85, 2099–2102. [Google Scholar] [CrossRef]
- Zamyatnin, A.A.; Solovyev, A.G.; Bozhkov, P.V.; Valkonen, J.P.T.; Morozov, S.Y.; Savenkov, E.I. Assessment of the Integral Membrane Protein Topology in Living Cells. Plant J. 2006, 46, 145–154. [Google Scholar] [CrossRef]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and Functional Genomics of Closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Gowda, S.; Mawassi, M.; Albiach-Marti, M.; Ayllón, M.; Robertson, C.; Garnsey, S.; Dawson, W. Closterovirus Encoded HSP70 Homolog and P61 in Addition to Both Coat Proteins Function in Efficient Virion Assembly. Virology 2001, 278, 253–265. [Google Scholar] [CrossRef]
- Lentze, N.; Auerbach, D. Membrane-Based Yeast Two-Hybrid System to Detect Protein Interactions. Curr. Protoc. Protein Sci. 2008, 52, 19.17.1–19.17.28. [Google Scholar] [CrossRef]
- Frand, A.R.; Cuozzo, J.W.; Kaiser, C.A. Pathways for Protein Disulphide Bond Formation. Trends Cell Biol. 2000, 10, 203–210. [Google Scholar] [CrossRef]
- Li, E.; Wimley, W.C.; Hristova, K. Transmembrane Helix Dimerization: Beyond the Search for Sequence Motifs. Biochim. Biophys. Acta 2012, 1818, 183–193. [Google Scholar] [CrossRef]
- Goddard, A.; Oates, J.; Watts, A. Membrane Proteins: Structure and Organization. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1478–1481. ISBN 9783642167126. [Google Scholar]
- Mayer, M.P. Recruitment of Hsp70 Chaperones: A Crucial Part of Viral Survival Strategies. In Reviews of Physiology, Biochemistry and Pharmacology; Reviews of Physiology, Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–46. ISBN 9783540273639. [Google Scholar]
- Agranovsky, A.A.; Koonin, E.V.; Boyko, V.P.; Maiss, E.; Frötschl, R.; Lunina, N.A.; Atabekov, J.G. Beet Yellows Closterovirus: Complete Genome Structure and Identification of a Leader Papain-like Thiol Protease. Virology 1994, 198, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-Dimensional Structure of the ATPase Fragment of a 70K Heat-Shock Cognate Protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural Analysis of Substrate Binding by the Molecular Chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.V.; Haskell, D.W.; Guy, C.L. Differential Influence of ATP on Native Spinach 70-Kilodalton Heat-Shock Cognates. Plant Physiol. 1994, 104, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Takakuwa, J.E.; Nitika; Knighton, L.E.; Truman, A.W. Oligomerization of Hsp70: Current Perspectives on Regulation and Function. Front. Mol. Biosci. 2019, 6, 81. [Google Scholar] [CrossRef]
- Napuli, A.J.; Falk, B.W.; Dolja, V.V. Interaction between HSP70 Homolog and Filamentous Virions of the Beet Yellows Virus. Virology 2000, 274, 232–239. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Ling, K.S.; Goszczynski, D.E.; McFerson, J.R.; Gonsalves, D. Nucleotide Sequence and Genome Organization of Grapevine Leafroll-Associated Virus-2 Are Similar to Beet Yellows Virus, the Closterovirus Type Member. J. Gen. Virol. 1998, 79, 1289–1298. [Google Scholar] [CrossRef]
- Alzhanova, D.V.; Prokhnevsky, A.I.; Peremyslov, V.V.; Dolja, V.V. Virion Tails of Beet Yellows Virus: Coordinated Assembly by Three Structural Proteins. Virology 2007, 359, 220–226. [Google Scholar] [CrossRef]
- Ayllón, M.A.; Gowda, S.; Satyanarayana, T.; Dawson, W.O. Cis-Acting Elements at Opposite Ends of the Citrus Tristeza Virus Genome Differ in Initiation and Termination of Subgenomic RNAs. Virology 2004, 322, 41–50. [Google Scholar] [CrossRef]
- Borroto-Fernández, E.G.; Torres-Acosta, J.A.; Laimer, M. RT-PCR Detection and Protein-Protein Interaction of Viral Components of Pineapple Mealybug Wilt-Associated Virus 2 in Cuba. JPP 2007, 89, 435–439. [Google Scholar]
- Gowda, S.; Satyanarayana, T.; Ayllón, M.A.; Albiach-Martí, M.R.; Mawassi, M.; Rabindran, S.; Garnsey, S.M.; Dawson, W.O. Characterization of the Cis-Acting Elements Controlling Subgenomic MRNAs of Citrus Tristeza Virus: Production of Positive- and Negative-Stranded 3′-Terminal and Positive-Stranded 5′-Terminal RNAs. Virology 2001, 286, 134–151. [Google Scholar] [CrossRef]
- Stewart, L.R.; Medina, V.; Tian, T.; Turina, M.; Falk, B.W.; Ng, J.C.K. A Mutation in the Lettuce Infectious Yellows Virus Minor Coat Protein Disrupts Whitefly Transmission but Not In Planta Systemic Movement. J. Virol. 2010, 84, 9. [Google Scholar] [CrossRef]
- Bukau, B.; Horwich, A.L. The Hsp70 and Hsp60 Chaperone Machines. Cell 1998, 92, 351–366. [Google Scholar] [CrossRef]
- Pelham, H.R. Speculations on the Functions of the Major Heat Shock and Glucose-Regulated Proteins. Cell 1986, 46, 959–961. [Google Scholar] [CrossRef]
- Ellis, R.J.; Hartl, F.U. Principles of Protein Folding in the Cellular Environment. Curr. Opin. Struct. Biol. 1999, 9, 102–110. [Google Scholar] [CrossRef]
- Bester, R.; Maree, H.J.; Burger, J.T. Complete Nucleotide Sequence of a New Strain of Grapevine Leafroll-Associated Virus 3 in South Africa. Arch. Virol. 2012, 157, 1815–1819. [Google Scholar] [CrossRef]
- Seah, Y.; Sharma, A.M.; Zhang, S.; Almeida, R.P.; Duffy, S. A Divergent Variant of Grapevine Leafroll-Associated Virus 3 Is Present in California. Virol. J. 2012, 9, 235. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Suppressors of RNA Silencing Encoded by Plant Viruses and Their Role in Viral Infections. FEBS Lett. 2005, 579, 5958–5964. [Google Scholar] [CrossRef]
- Ye, K.; Patel, D.J. RNA Silencing Suppressor P21 of Beet Yellows Virus Forms an RNA Binding Octameric Ring Structure. Structure 2005, 13, 1375–1384. [Google Scholar] [CrossRef]
- Gowda, S.; Satyanarayana, T.; Davis, C.L.; Navas-Castillo, J.; Albiach-Martí, M.R.; Mawassi, M.; Valkov, N.; Bar-Joseph, M.; Moreno, P.; Dawson, W.O. The P20 Gene Product of Citrus Tristeza Virus Accumulates in the Amorphous Inclusion Bodies. Virology 2000, 274, 246–254. [Google Scholar] [CrossRef]

| Protein A | Protein B | Y2H Detection | Optimal Combination of BiFC Plasmid Orientations | Average MIdiff |
|---|---|---|---|---|
| HSP70h | HSP70h | Yes | YC-HSP70h + YN-HSP70h | 51.78 |
| HSP70h | CP | Yes | YC-HSP70h + CP-YN | 34.63 |
| p20B | p20B | Yes | YC-p20B + YN-p20B | 92.7 * |
| CP | p6 | Yes | No YFP reconstitution observed | N/A |
| CP | CP | No | CP-YC + YN-CP | 26.99 |
| p5 | p5 | No | YC-p5 + YN-p5 | 113.86 |
| HSP70h | p20B | No | YC-p20B + YN-HSP70h | 45.55 * |
| CP | p20B | No | YC-p20B + YN-CP | 55.66 * |
| CPm | p20B | No | YC-p20B + YN-CPm | 27.24 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostert, I.; Bester, R.; Burger, J.T.; Maree, H.J. Identification of Interactions between Proteins Encoded by Grapevine Leafroll-Associated Virus 3. Viruses 2023, 15, 208. https://doi.org/10.3390/v15010208
Mostert I, Bester R, Burger JT, Maree HJ. Identification of Interactions between Proteins Encoded by Grapevine Leafroll-Associated Virus 3. Viruses. 2023; 15(1):208. https://doi.org/10.3390/v15010208
Chicago/Turabian StyleMostert, Ilani, Rachelle Bester, Johan T. Burger, and Hans J. Maree. 2023. "Identification of Interactions between Proteins Encoded by Grapevine Leafroll-Associated Virus 3" Viruses 15, no. 1: 208. https://doi.org/10.3390/v15010208
APA StyleMostert, I., Bester, R., Burger, J. T., & Maree, H. J. (2023). Identification of Interactions between Proteins Encoded by Grapevine Leafroll-Associated Virus 3. Viruses, 15(1), 208. https://doi.org/10.3390/v15010208







