Enterococcus faecium Bacteriophage vB_EfaH_163, a New Member of the Herelleviridae Family, Reduces the Mortality Associated with an E. faecium vanR Clinical Isolate in a Galleria mellonella Animal Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample, Strains and Culture Conditions
2.2. Phage Titre Determination
2.3. Phage Isolation and Propagation
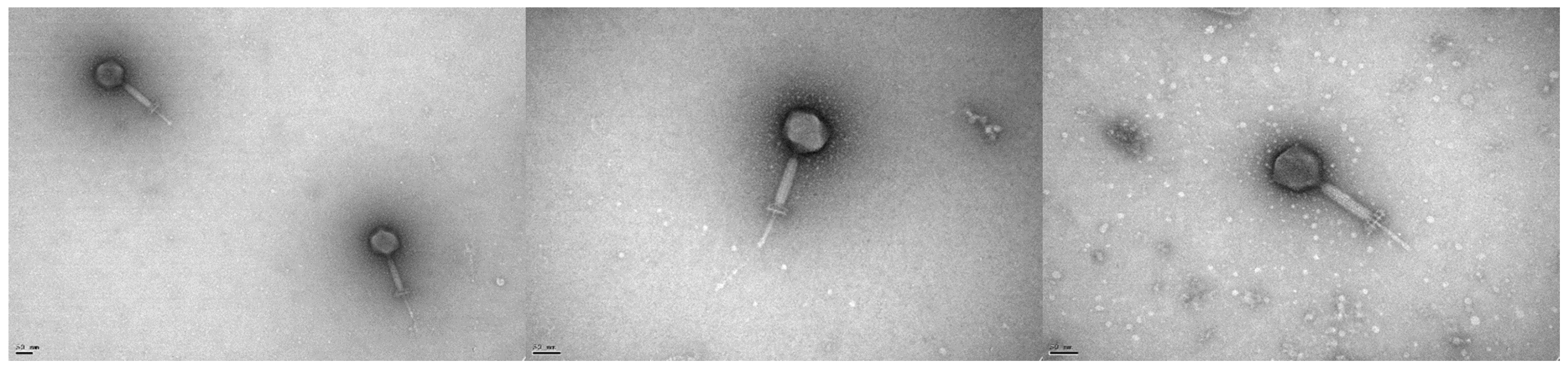
2.4. Electron Microscopy
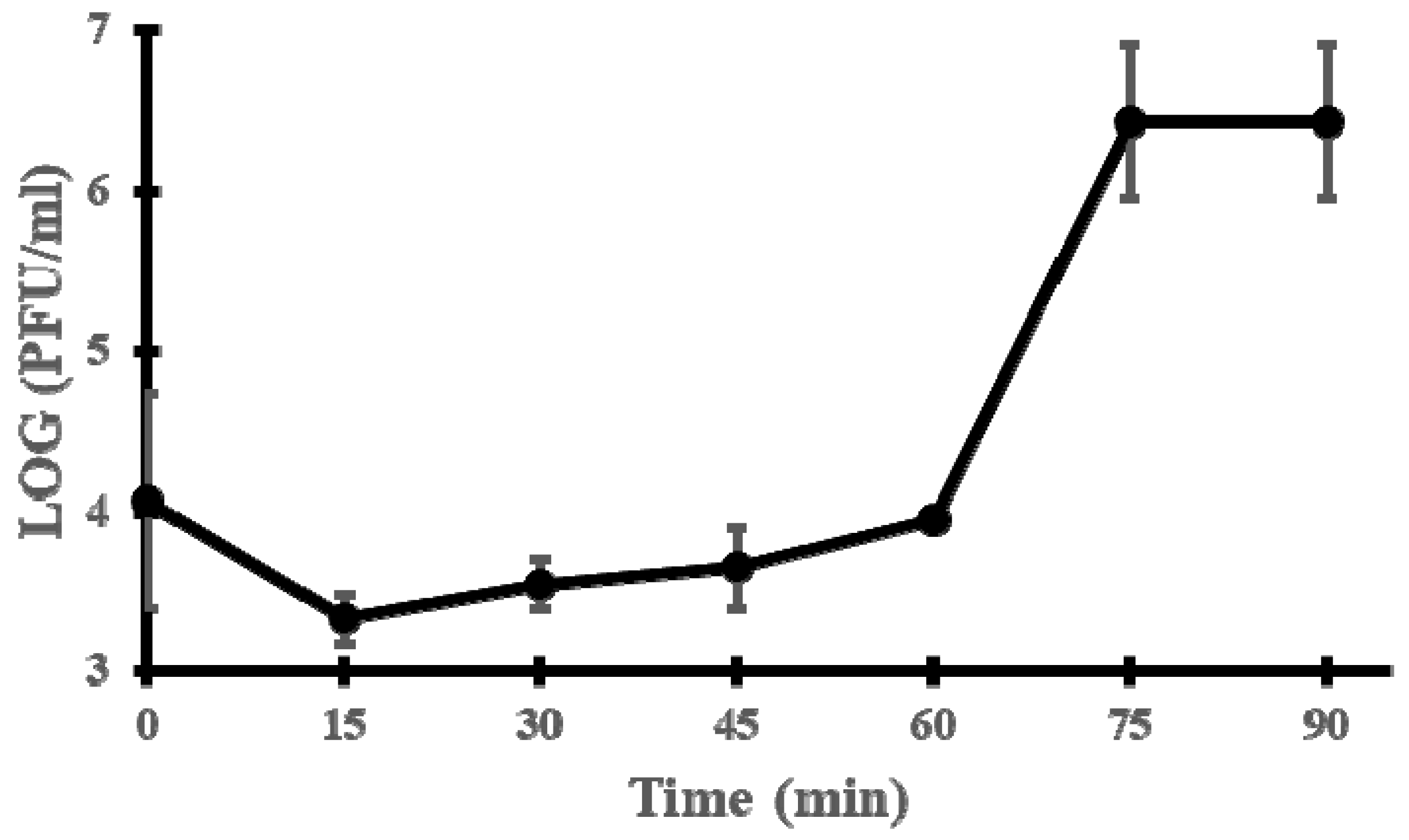
2.5. One-Step Growth Curve
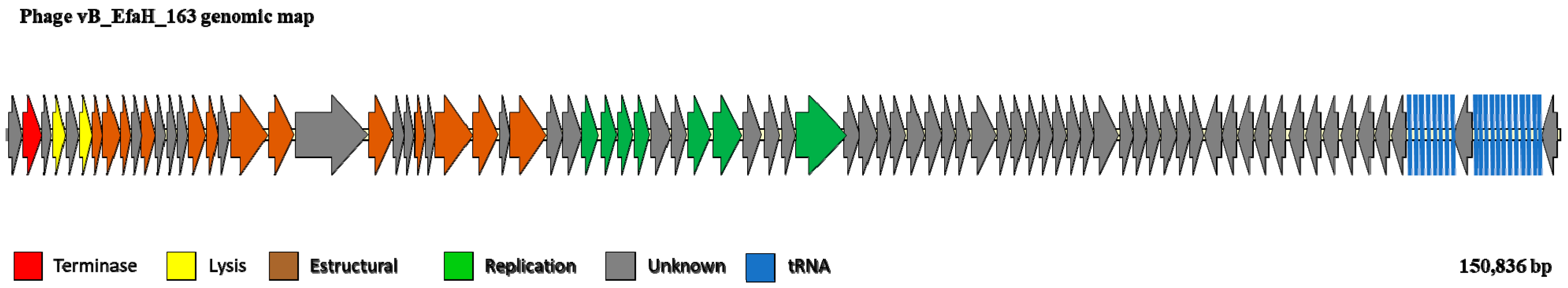
2.6. Phage Genome Sequencing and Analysis
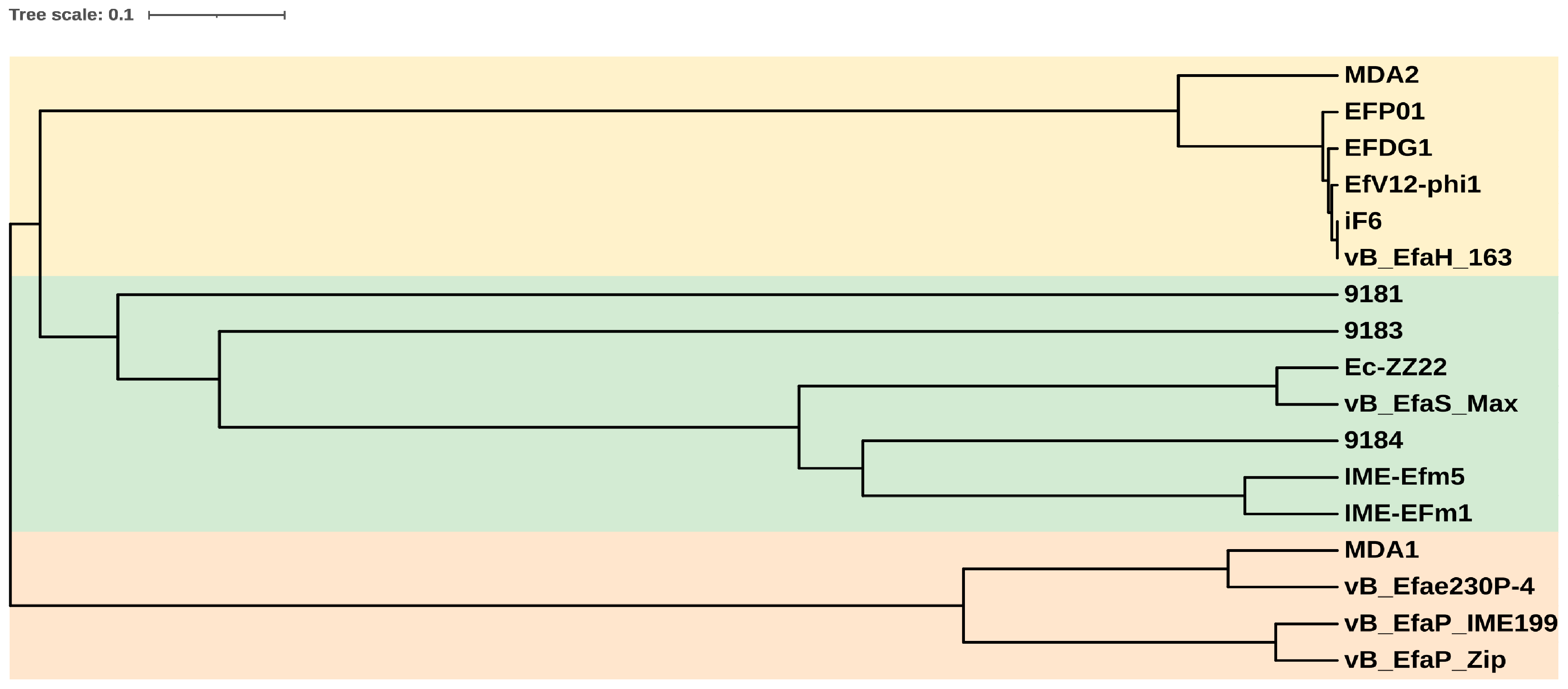
2.7. Phylogenetic Analysis
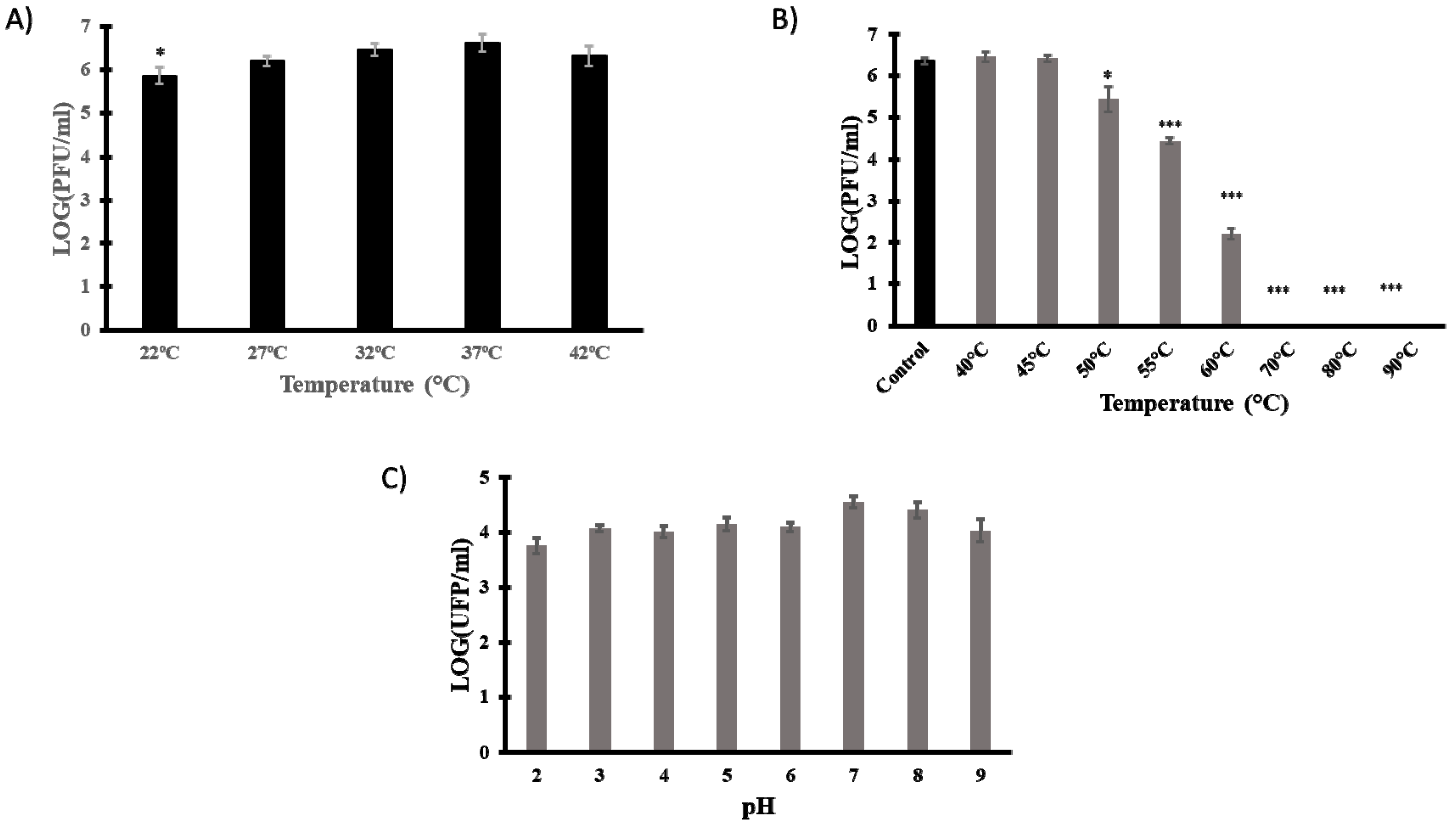
2.8. Technological Characterisation
2.9. Functional Characterisation
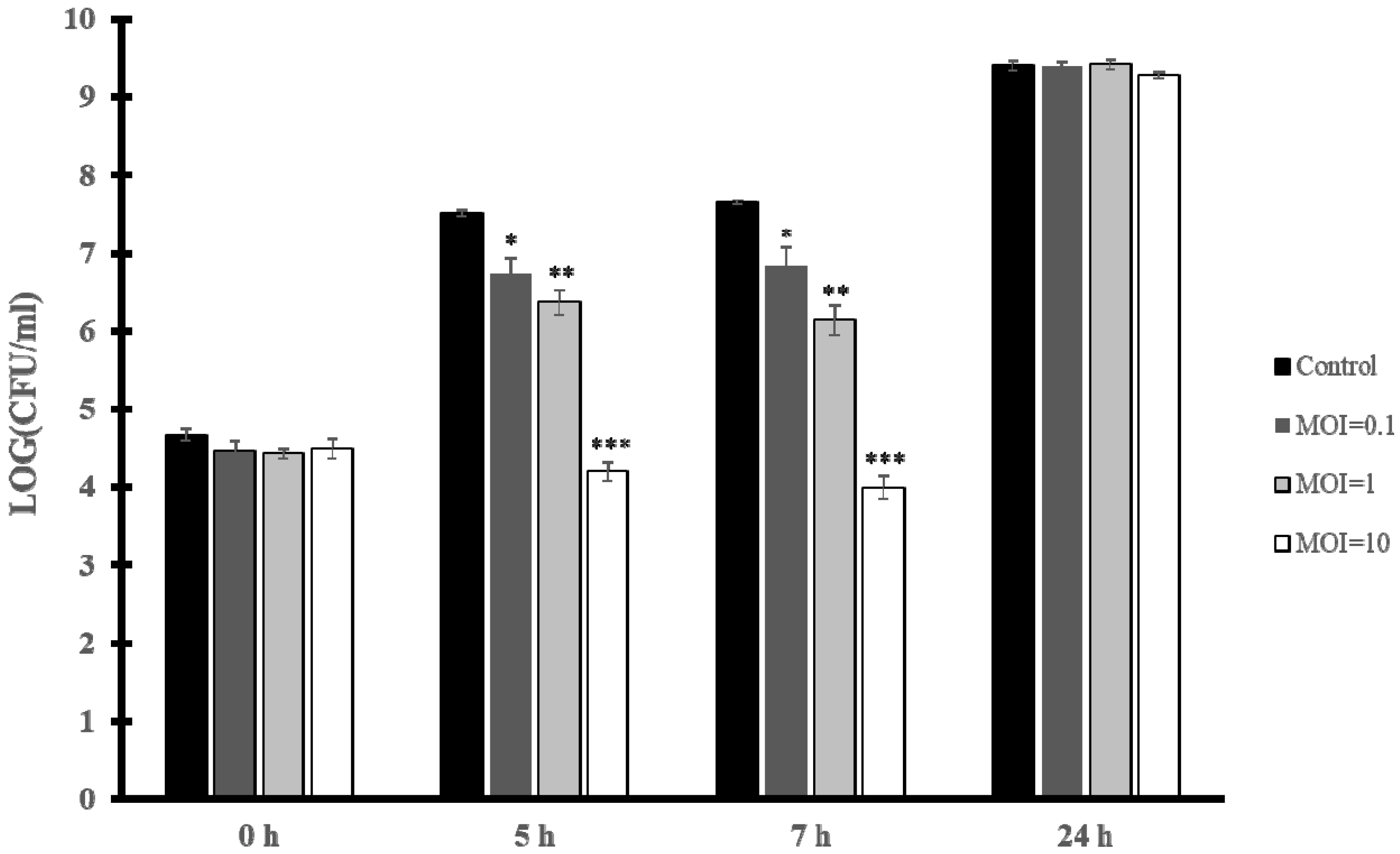
2.9.1. Biocontrol of the E. faecium VR-13 vanR Clinical Isolate by vB_EfaH_163 Infection in Broth
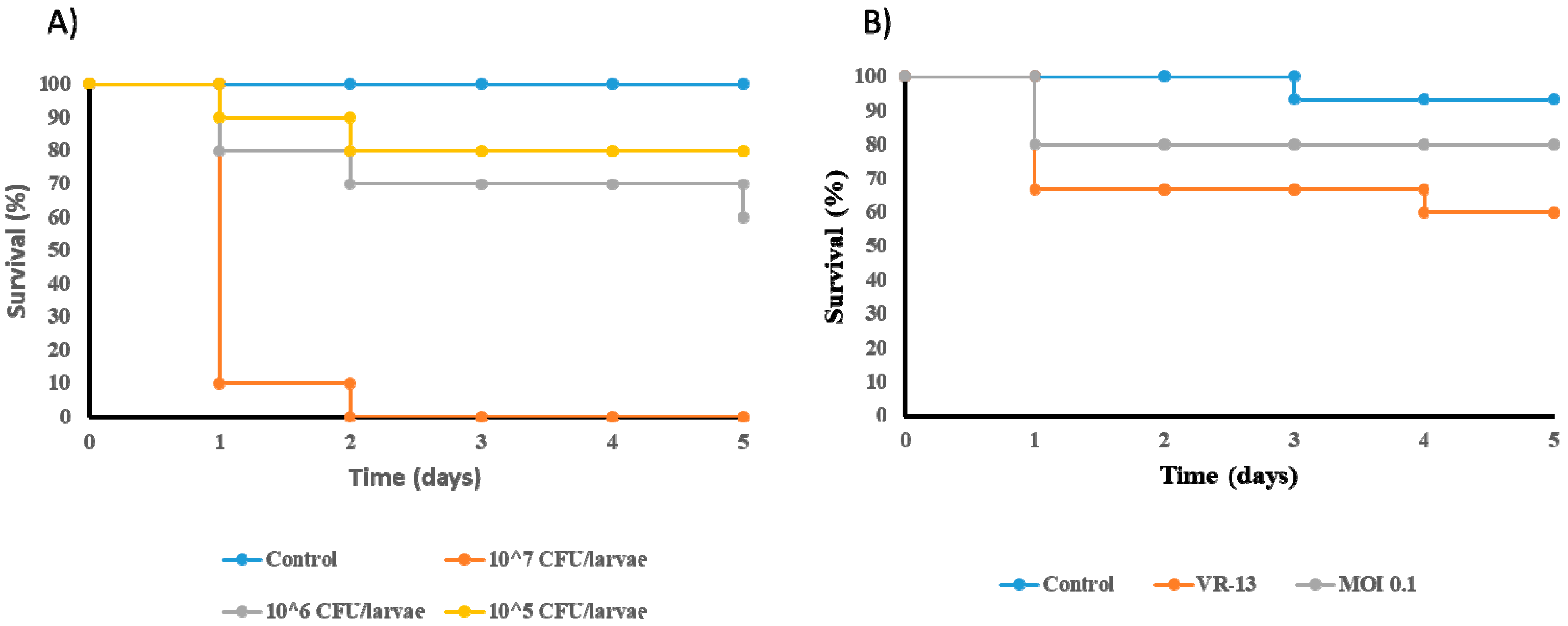
2.9.2. In Vivo Effectiveness of Phage Treatment in the Galleria mellonella Model
2.10. Statistical Analysis
3. Results
3.1. Phage vB_EfaH_163 Isolation
3.2. Microbiological Characterisation of Phage vB_EfaH_163
3.3. The vB_EfmH_163 Genome: Characterisation and Phylogenetic Analysis
3.4. Technological Characterisation
3.5. Biocontrol of E. faecium VR-13 by vB_EfaH_163
3.6. Reduction in the Mortality of Galleria mellonella Infected by E. faecium VR-13 due to Treatment with Phage vB_EfaH_163
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mestrovic, T.; Robles Aguilar, G.; Swetschinski, L.R.; Ikuta, K.S.; Gray, A.P.; Davis Weaver, N.; Han, C.; Wool, E.E.; Gershberg Hayoon, A.; Hay, S.I.; et al. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: A cross-country systematic analysis. Lancet Public Health 2022, 2667, e897–e913. [Google Scholar] [CrossRef]
- Shankar, P.R. Book review: Tackling drug-resistant infections globally. Arch. Pharm. Pract. 2016, 7, 110. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.R.; Ferreira, R.; Costa, A.R.; Oliveira, H.; Azeredo, J. Efficacy and safety assessment of two enterococci phages in an in vitro biofilm wound model. Sci. Rep. 2019, 9, 6643. [Google Scholar] [CrossRef] [PubMed]
- Dahl, A.; Bruun, N.E. Enterococcus faecalis infective endocarditis: Focus on clinical aspects. Expert Rev. Cardiovasc. Ther. 2013, 11, 1247–1257. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef]
- Stuart, C.; Schwartz, S.; Beeson, T.; Owatz, C. Enterococcus faecalis: Its role in root canal treatment failure and current concepts in retreatment. J. Endod. 2006, 32, 93–98. [Google Scholar] [CrossRef]
- Tebruegge, M.; Pantazidou, A.; Clifford, V.; Gonis, G.; Ritz, N.; Connell, T.; Curtis, N. The age-related risk of co-existing meningitis in children with urinary tract infection. PLoS ONE 2011, 6, e26576. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Sun, L.; Wang, X. Vancomycin-resistant Enterococcus faecium pneumonia in a uremic patient on hemodialysis: A case report and review of the literature. BMC Infect. Dis. 2020, 20, 167. [Google Scholar] [CrossRef]
- Chuang, Y.-C.; Lin, H.-Y.; Chen, P.-Y.; Lin, C.-Y.; Wang, J.-T.; Chang, S.-C. Daptomycin versus linezolid for the treatment of vancomycin-resistant enterococcal bacteraemia: Implications of daptomycin dose. Clin. Microbiol. Infect. 2016, 22, 890.e1–890.e7. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; Leon-Sampedro, R.; Del Campo, R.; Coque, T.M.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 185–227. [Google Scholar] [CrossRef] [PubMed]
- Palareti, G.; Legnani, C.; Cosmi, B.; Antonucci, E.; Erba, N.; Poli, D.; Testa, S.; Tosetto, A.; De Micheli, V.; Ghirarduzzi, A.; et al. Comparison between different D-Dimer cutoff values to assess the individual risk of recurrent venous thromboembolism: Analysis of results obtained in the DULCIS study. Int. J. Lab. Hematol. 2016, 38, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.W.; Kozak, R.A.; Eshaghi, A.; Avaness, M.; Salt, N.; Patel, S.N.; Simor, A.E.; Leis, J.A. Nosocomial outbreak of vanD -carrying vancomycin-resistant Enterococcus faecium. Infect. Control Hosp. Epidemiol. 2018, 39, 1266–1268. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.A.; Shore, A.C.; O’Connell, B.; Brennan, G.I.; Coleman, D.C. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: High prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 2020, 75, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Chatterjee, S.; Datta, S.; Prasad, R.; Dubey, D.; Prasad, R.K.; Vairale, M.G. Bacteriophages and its applications: An overview. Folia Microbiol. 2017, 62, 17–55. [Google Scholar] [CrossRef]
- Cisek, A.A.; Dąbrowska, I.; Gregorczyk, K.P.; Wyżewski, Z. Phage therapy in bacterial infections treatment: One hundred years after the discovery of bacteriophages. Curr. Microbiol. 2017, 74, 277–283. [Google Scholar] [CrossRef]
- Elbreki, M.; Ross, R.P.; Hill, C.; O’Mahony, J.; McAuliffe, O.; Coffey, A. Bacteriophages and their derivatives as biotherapeutic agents in disease prevention and treatment. J. Viruses 2014, 2014, 382539. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Upadrasta, A.; Bettio, P.H.A.; Clooney, A.G.; Draper, L.A.; Ross, R.P.; Hill, C. Evaluation of phage therapy in the context of Enterococcus faecalis and its associated diseases. Viruses 2019, 11, 366. [Google Scholar] [CrossRef]
- Fernández, L.; Gutiérrez, D.; García, P.; Rodríguez, A. The perfect bacteriophage for therapeutic applications—A quick guide. Antibiotics 2019, 8, 126. [Google Scholar] [CrossRef]
- Khalifa, L.; Shlezinger, M.; Beyth, S.; Houri-Haddad, Y.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Phage therapy against Enterococcus faecalis in dental root canals. J. Oral Microbiol. 2016, 8, 32157. [Google Scholar] [CrossRef]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.-A.; Beyth, N.; Hazan, R. Targeting Enterococcus faecalis biofilms with phage therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Gelman, D.; Shlezinger, M.; Dessal, A.L.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Defeating antibiotic- and phage-resistant Enterococcus faecalis using a phage cocktail in vitro and in a clot model. Front. Microbiol. 2018, 9, 326. [Google Scholar] [CrossRef] [PubMed]
- del Rio, B.; Sanchez-Llana, E.; Redruello, B.; Magadan, A.H.; Fernandez, M.; Martin, M.C.; Ladero, V.; Alvarez, M.A. Enterococcus faecalis bacteriophage 156 Is an effective biotechnological tool for reducing the presence of tyramine and putrescine in an experimental cheese model. Front. Microbiol. 2019, 10, 566. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Gomez-Sordo, C.; Sanchez-Llana, E.; del Rio, B.; Redruello, B.; Fernandez, M.; Martin, M.C.; Alvarez, M.A. Q69 (an E. faecalis-Infecting bacteriophage) as a biocontrol agent for reducing tyramine in dairy products. Front. Microbiol. 2016, 7, 445. [Google Scholar] [CrossRef] [PubMed]
- Biswas, B.; Adhya, S.; Washart, P.; Paul, B.; Trostel, A.N.; Powell, B.; Carlton, R.; Merril, C.R. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 2002, 70, 1664. [Google Scholar] [CrossRef]
- Gelman, D.; Beyth, S.; Lerer, V.; Adler, K.; Poradosu-Cohen, R.; Coppenhagen-Glazer, S.; Hazan, R. Combined bacteriophages and antibiotics as an efficient therapy against VRE Enterococcus faecalis in a mouse model. Res. Microbiol. 2018, 169, 531–539. [Google Scholar] [CrossRef]
- Letkiewicz, S.; Międzybrodzki, R.; Fortuna, W.; Weber-Dąbrowska, B.; Górski, A. Eradication of Enterococcus faecalis by phage therapy in chronic bacterial prostatitis—Case report. Folia Microbiol. 2009, 54, 457–461. [Google Scholar] [CrossRef]
- Ladero, V.; Fernandez, M.; Calles-Enriquez, M.; Sanchez-Llana, E.; Canedo, E.; Martin, M.C.; Alvarez, M.A. Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 2012, 30, 132–138. [Google Scholar] [CrossRef]
- Ladero, V.; Fernandez, M.; Alvarez, M.A. Isolation and identification of tyramine-producing enterococci from human fecal samples. Can. J. Microbiol. 2009, 55, 215–218. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Banerjei, L.; Myers, G.S.; Nelson, K.E.; Seshadri, R.; Read, T.D.; Fouts, D.E.; Eisen, J.A.; Gill, S.R.; Heidelberg, J.F.; et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2003, 299, 2071–2074. [Google Scholar] [CrossRef]
- Hevia, A.; Delgado, S.; Margolles, A.; Sánchez, B. Application of density gradient for the isolation of the fecal microbial stool component and the potential use thereof. Sci. Rep. 2015, 5, 16807. [Google Scholar] [CrossRef] [PubMed]
- Binetti, A.G.; del Rio, B.; Martin, M.C.; Alvarez, M.A. Detection and characterization of Streptococcus thermophilus bacteriophages by use of the antireceptor gene sequence. Appl. Environ. Microbiol. 2005, 71, 6096–6103. [Google Scholar] [CrossRef] [PubMed]
- del Rio, B.; Sánchez-Llana, E.; Martínez, N.; Fernández, M.; Ladero, V.; Alvarez, M.A. Isolation and characterization of Enterococcus faecalis-infecting bacteriophages from different cheese types. Front. Microbiol. 2021, 11, 592172. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Rajandas, H.; Parimannan, S.; Manickam, R.; Marimuthu, K.; Petersen, B.; Clokie, M.R.J.; Millard, A.; Sicheritz-Pontén, T. PhageLeads: Rapid assessment of phage therapeutic suitability using an ensemble machine learning approach. Viruses 2022, 14, 342. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Br. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Wandro, S.; Oliver, A.; Gallagher, T.; Weihe, C.; England, W.; Martiny, J.B.H.; Whiteson, K. Predictable molecular adaptation of coevolving Enterococcus faecium and lytic phage EfV12-phi1. Front. Microbiol. 2019, 9, 3192. [Google Scholar] [CrossRef] [PubMed]
- El Haddad, L.; Angelidakis, G.; Clark, J.R.; Mendoza, J.F.; Terwilliger, A.L.; Chaftari, C.P.; Duna, M.; Yusuf, S.T.; Harb, C.P.; Stibich, M.; et al. Genomic and functional characterization of vancomycin-resistant enterococci-specific bacteriophages in the Galleria mellonella wax moth larvae model. Pharmaceutics 2022, 14, 1591. [Google Scholar] [CrossRef]
- Canfield, G.S.; Chatterjee, A.; Espinosa, J.; Mangalea, M.R.; Sheriff, E.K.; Keidan, M.; McBride, S.W.; McCollister, B.D.; Hang, H.C.; Duerkop, B.A. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 2021, 65, e00143-21. [Google Scholar] [CrossRef]
- Li, J.; Shi, H.; Zhao, C.; Hao, Y.; He, Y.; Sun, Y. Complete genome sequence of the siphoviral bacteriophage ec-zz2, which is capable of lysing Enterococcus faecium. Genome Announc. 2016, 4, e01167-16. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, W.; Lv, Y.; Zheng, W.; Mi, Z.; Pei, G.; An, X.; Xu, X.; Han, C.; Liu, J.; et al. Characterization and complete genome sequence analysis of novel bacteriophage IME-EFm1 infecting Enterococcus faecium. J. Gen. Virol. 2014, 95, 2565–2575. [Google Scholar] [CrossRef]
- Gong, P.; Cheng, M.; Li, X.; Jiang, H.; Yu, C.; Kahaer, N.; Li, J.; Zhang, L.; Xia, F.; Hu, L.; et al. Characterization of Enterococcus faecium bacteriophage IME-EFm5 and its endolysin LysEFm5. Virology 2016, 492, 11–20. [Google Scholar] [CrossRef]
- Xing, S.; Zhang, X.; Sun, Q.; Wang, J.; Mi, Z.; Pei, G.; Huang, Y.; An, X.; Fu, K.; Zhou, L.; et al. Complete genome sequence of a novel, virulent Ahjdlikevirus bacteriophage that infects Enterococcus faecium. Arch. Virol. 2017, 162, 3843–3847. [Google Scholar] [CrossRef]
- Manohar, P.; Nachimuthu, R.; Lopes, B.S. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol. 2018, 18, 97. [Google Scholar] [CrossRef]
- Jeon, J.; Park, J.-H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Lagatolla, C.; Milic, J.; Imperi, F.; Cervoni, M.; Bressan, R.; Luzzati, R.; Di Bella, S. Synergistic activity of fosfomycin and chloramphenicol against vancomycin-resistant Enterococcus faecium (VREfm) isolates from bloodstream infections. Diagn. Microbiol. Infect. Dis. 2021, 99, 115241. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, H.; Morita, M. Phage DNA packaging. Genes Cells 2003, 2, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Barylski, J.; Kropinski, A.M.; Alikhan, N.-F.; Adriaenssens, E.M. ICTV report consortium ICTV virus taxonomy profile: Herelleviridae. J. Gen. Virol. 2020, 101, 362. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Tolstoy, I.; Kropinski, A.M. Phage annotation guide: Guidelines for assembly and high-quality annotation. PHAGE 2021, 2, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.; Brüssow, H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef]
- Kot, W.; Neve, H.; Heller, K.J.; Vogensen, F.K. Bacteriophages of leuconostoc, oenococcus, and weissella. Front. Microbiol. 2014, 5, 186. [Google Scholar] [CrossRef]
- Uchiyama, J.; Rashel, M.; Maeda, Y.; Takemura, I.; Sugihara, S.; Akechi, K.; Muraoka, A.; Wakiguchi, H.; Matsuzaki, S. Isolation and characterization of a novel Enterococcus faecalis bacteriophage φEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 2008, 278, 200–206. [Google Scholar] [CrossRef]
- Yin, Y.; Fischer, D. Identification and investigation of ORFans in the viral world. BMC Genom. 2008, 9, 24. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Gutiérrez, D.; Donovan, D.M.; Martínez, B.; Rodríguez, A.; García, P. Phage lytic proteins: Biotechnological applications beyond clinical antimicrobials. Crit. Rev. Biotechnol. 2015, 36, 542–552. [Google Scholar] [CrossRef]
- Wang, R.; Xing, S.; Zhao, F.; Li, P.; Mi, Z.; Shi, T.; Liu, H.; Tong, Y. Characterization and genome analysis of novel phage vB_EfaP_IME195 infecting Enterococcus faecalis. Virus Genes 2018, 54, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Casey, E.; van Sinderen, D.; Mahony, J. In vitro characteristics of phages to guide ‘Real Life’ phage therapy suitability. Viruses 2018, 10, 163. [Google Scholar] [CrossRef]
- Ladero, V.; Calles-Enríquez, M.; Fernández, M.; Alvarez, M.A. Toxicological effects of dietary biogenic amines. Curr. Nutr. Food Sci. 2010, 6, 145–156. [Google Scholar] [CrossRef]
- McLaughlin, M.; Malczynski, M.; Qi, C.; Barajas, G.; Radetski, J.; Zembower, T.; Scheetz, M.H. Virulence of Vancomycin-resistant Enterococcus faecium according to linezolid resistance and clinical outbreak status. Antimicrob. Agents Chemother. 2013, 57, 3923–3927. [Google Scholar] [CrossRef]
- Skinner, K.; Sandoe, J.A.T.; Rajendran, R.; Ramage, G.; Lang, S. Efficacy of rifampicin combination therapy for the treatment of enterococcal infections assessed in vivo using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 2017, 49, 507–511. [Google Scholar] [CrossRef]
- Tkhilaishvili, T.; Wang, L.; Tavanti, A.; Trampuz, A.; Di Luca, M. Antibacterial efficacy of two commercially available bacteriophage formulations, staphylococcal bacteriophage and pyo bacteriophage, against methicillin-resistant Staphylococcus aureus: Prevention and eradication of biofilm formation and control of a systemic infection of Galleria mellonella larvae. Front. Microbiol. 2020, 11, 110. [Google Scholar] [CrossRef] [PubMed]
| Specie | Strain | Origin | vB_EfmH_163 Infection | Reference |
|---|---|---|---|---|
| E. faecium | LMA2 | Camel milk | + | MicroMol |
| E. faecium | LMA3 | Camel milk | + | MicroMol |
| E. faecium | LMA4 | Camel milk | + | MicroMol |
| E. faecium | LMA5 | Camel milk | + | MicroMol |
| E. faecium | LMA6 | Camel milk | + | MicroMol |
| E. faecium | LMA8 | Camel milk | + | MicroMol |
| E. faecium | LMA9 | Camel milk | − | MicroMol |
| E. faecium | LMA10 | Camel milk | + | MicroMol |
| E. faecium | LGMY-2 | Camel milk | + | MicroMol |
| E. faecium | LGMY-5 | Camel milk | + | MicroMol |
| E. faecium | LGMY-1 | Cow milk | + | MicroMol |
| E. faecium | LMGY-10 | Cow milk | − | MicroMol |
| E. faecium | LGMY-6 | Sheep milk | + | MicroMol |
| E. faecium | LGMY-11 | Goat milk | + | MicroMol |
| E. faecium | C39 | Cheese | − | [28] |
| E. faecium | AM | Cheese | − | [28] |
| E. faecium | 103 | Cheese | − | [28] |
| E. faecium | LGMY-12 | Date | + | MicroMol |
| E. faecium | LMGY-13 | Date | + | LGM |
| E. faecium | LGM11397 | Meat | − | LGM |
| E. faecium | LGM14205 | Meat | − | LGM |
| E. faecium | LGM20641 | Meat | − | LGM |
| E. faecium | HF11 | Human | − | [29] |
| E. faecium | HF14 | Human | + | [29] |
| E. faecium | HF24 | Human | + | [29] |
| E. faecium | HF52 | Human | − | [29] |
| E. faecium | HF56 | Human | + | [29] |
| E. faecium | VR-1 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-2 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-3 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-4 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-5 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-6 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-7 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-8 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-9 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-10 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-11 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-12 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-13 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-14 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-15 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-16 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-17 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-18 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-19 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-20 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-20b | Clinical | + | Bierzo Hospital |
| E. faecium | VR-22 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-23 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-24 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-25 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-26 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-27 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-28 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-29 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-30 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-31 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-32 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-33 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-34 | Clinical | + | Bierzo Hospital |
| E. faecium | VR-35 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-36 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-37 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-38 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-39 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-40 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-41 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-42 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-43 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-44 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-45 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-46 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-47 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-48 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-49 | Clinical | − | Bierzo Hospital |
| E. faecium | VR-50 | Clinical | − | Bierzo Hospital |
| E. faecalis | CECT481T | Type strain | − | CECT |
| E. faecalis | 18a | Cheese | − | [28] |
| E. faecalis | 23a | Cheese | − | [28] |
| E. faecalis | V63 | Cheese | − | [28] |
| E. faecalis | 63c | Cheese | − | [28] |
| E. faecalis | 52c | Cheese | − | [28] |
| E. faecalis | HFS56 | Human | + | [29] |
| E. faecalis | HFS57 | Human | − | [29] |
| E. faecalis | VR-5 | Clinical | − | Bierzo Hospital |
| E. faecalis | VR-11 | Clinical | − | Bierzo Hospital |
| E. faecalis | optra5 | Clinical | + | Bierzo Hospital |
| E. faecalis | V583 | Clinical | − | [30] |
| Phage | Accession Number | Host | Family | Genome Size | Origin | Reference |
|---|---|---|---|---|---|---|
| vB_EfmH_163 | CAJDKA010000002.1 | E. faecium E. faecalis | Herelleviridae | 150,836 | Human faecal samples | This work |
| EFDG1 | NC_029009 | E. faecium E faecalis | Herelleviridae | 147,589 | Sewage effluents | [21] |
| EfV12-phi1 | MH880817 | E. faecium | Herelleviridae | 152,770 | Sewage | [43] |
| EFP01 | NC_047796.1 | E. faecium | Herelleviridae | 155,053 | Sewage | - |
| iF6 | MT909815.1 | E. faecium | Herelleviridae | 156,592 | - | - |
| MDA2 | MW633168.1 | E. faecium | Herelleviridae | 140,226 | - | [44] |
| 9183 | MT939241.1 | E. faecium | Siphoviridae | 806,301 | Wastewater | [45] |
| 9181 | MT939240.1 | E. faecium | Siphoviridae | 71,854 | Wastewater | [45] |
| vB_EfaS_Max | MK360024 | E. faecium E. faecalis | Siphoviridae | 40,975 | Raw sewage from wastewater | [4] |
| 9184 | MT939242.1 | E. faecium | Siphoviridae | 44,108 | Wastewater | [45] |
| Ec-ZZ2 | NC_031260 | E. faecium | Siphoviridae | 41,170 | Sewage | [46] |
| IME-EFm1 | NC_024356 | E. faecium | Siphoviridae | 42,597 | Hospital sewage | [47] |
| IME-EFm5 | NC_028826 | E. faecium | Siphoviridae | 42,265 | Hospital sewage | [48] |
| vB_EfaP_Zip | MK360025 | E. faecium E. faecalis | Podoviridae | 18,742 | Raw sewage from wastewater | [4] |
| vB_Efae230p-4 | NC_025467 | E. faecium | Podoviridae | 17,972 | - | - |
| vB_EfaP_IME199 | KT945995 | E. faecium | Podoviridae | 18,838 | Sewage | [49] |
| MDA1 | MW623430.1 | E. faecium | Podoviridae | 18,058 | - | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pradal, I.; Casado, A.; del Rio, B.; Rodriguez-Lucas, C.; Fernandez, M.; Alvarez, M.A.; Ladero, V. Enterococcus faecium Bacteriophage vB_EfaH_163, a New Member of the Herelleviridae Family, Reduces the Mortality Associated with an E. faecium vanR Clinical Isolate in a Galleria mellonella Animal Model. Viruses 2023, 15, 179. https://doi.org/10.3390/v15010179
Pradal I, Casado A, del Rio B, Rodriguez-Lucas C, Fernandez M, Alvarez MA, Ladero V. Enterococcus faecium Bacteriophage vB_EfaH_163, a New Member of the Herelleviridae Family, Reduces the Mortality Associated with an E. faecium vanR Clinical Isolate in a Galleria mellonella Animal Model. Viruses. 2023; 15(1):179. https://doi.org/10.3390/v15010179
Chicago/Turabian StylePradal, Inés, Angel Casado, Beatriz del Rio, Carlos Rodriguez-Lucas, Maria Fernandez, Miguel A. Alvarez, and Victor Ladero. 2023. "Enterococcus faecium Bacteriophage vB_EfaH_163, a New Member of the Herelleviridae Family, Reduces the Mortality Associated with an E. faecium vanR Clinical Isolate in a Galleria mellonella Animal Model" Viruses 15, no. 1: 179. https://doi.org/10.3390/v15010179
APA StylePradal, I., Casado, A., del Rio, B., Rodriguez-Lucas, C., Fernandez, M., Alvarez, M. A., & Ladero, V. (2023). Enterococcus faecium Bacteriophage vB_EfaH_163, a New Member of the Herelleviridae Family, Reduces the Mortality Associated with an E. faecium vanR Clinical Isolate in a Galleria mellonella Animal Model. Viruses, 15(1), 179. https://doi.org/10.3390/v15010179









