Abstract
Vibrio alginolyticus is one of the major pathogens causing vibriosis to a variety of aquatic animals as well as bringing about severe food safety concerns. Nowadays, phage therapy has received increasing attention as an alternative to the antibiotics that have being limited for use in aquaculture industries. In this work, a potent bacteriophage, vB_ValM_PVA23 (PVA23), which efficiently infects pathogenic strains of V. alginolyticus, was isolated from sewage water and characterized by microbiological and genomic analyses. Based on the transmission electronic observation, the phage was characterized to be the Myoviridae family. It has a latent period of 10 min and a burst size of 203 PFUs/infected bacterium, and was stable over a broad pH range (5.0–11.0) and a wide temperature span (−80 °C to 60 °C), respectively. Genome sequencing results show that PVA23 has a 246,962-bp double-stranded DNA with a G + C content of 41.25%. The lab and plant shrimp farming trials demonstrated that phage preparation derived from PVA23 out-performed the chemical disinfectant iodine treatment in the prevention of V. alginolyticus propagation, and the phage application could rapidly yet significantly reduce the level of V. alginolyticus in the pond within 12 h, with negligible rebound observed. These results suggests that phage PVA23 has the potential to be used as an anti-V. alginolyticus agent in aquaculture industries.
1. Introduction
Aquatic products are one of the major sources of food protein for humans in the world. With the rapid thriving of aquatic farming worldwide, the diseases derived from fatal aquatic bacterial pathogens, such as Vibrio, are posed as a huge threat. Some strains of Vibrio produce multiple virulence factors, such as hemolysin, caseinase, lipase, gelatinase and phospholipase, which are detrimental to aquatic animal health [1,2]. Among them, V. alginolyticus is a major pathogenic species that is widely distributed in coastal waters worldwide. It is well known that V. alginolyticus causes vibriosis to aquatic animals like shrimp, fish and shellfish, leading to mass mortality of economic marine animals as well as severe economic losses [3,4]. V. alginolyticus also brings about food safety concerns by polluting the sea food chains and producing toxins that are dangerous to human beings [5,6].
Currently, application of chemical agents, such as chemical disinfectants and antibiotics, is still the routine approach to control Vibrio in aquatic farming practice. Except for disease prevention, chemical disinfectants are also used widely for clearing the pond water, bathing the seedling, disinfecting pond mud and so on. However, it was found that the chemical disinfectants could adversely affect the growth of the farming animals as well as the beneficial microbes due to their toxic nature and non-selective killing effects [7]. Furthermore, the antimicrobial activities of chemical disinfectants are concentration dependent, so the routine water change over and the exposure to continuous airing may lower their concentration and largely discount their activity. On the other hand, the long-term overuse of the chemical antibiotics has led to adverse consequences such as the accumulation of chemical residuals, the emergence of multidrug resistances of Vibrio, and the microbial changeover, which not only arouses food safety issues, but also makes aquatic farming unsustainable [8]. Therefore, demand of safer alternatives for Vibrio control is on the rise.
Bacteriophages are viruses that can infect and kill their specific host bacteria and regulate bacterial populations by inducing bacterial lysis [9]. The use of phage therapy can be traced back to a century ago, and later on it was widely accepted, thanks to a panel of advantages over traditional antibiotic therapies [9]. Phage therapy has been considered as a potential alternative to antibiotic therapy in treating bacterial infections, and it had been successfully used to control several pathogenic infections in humans, and veterinary and agricultural settings [10]. Recently, some investigators have explored phage therapy for aquatic farming application as well as for bio-preservation use in food industries [8,11,12]. A panel of investigations have confirmed that preparations of phage or phage cocktails could effectively kill a variety of pathogenic Vibrio isolated from the ponds that farm different aquatic animals, such as shrimp, fish, oyster and sea cucumber [13,14,15,16,17]. Phage therapy has also been confirmed to be active against some dangerous emerging pathogens like multidrug-resistant bacteria in fish farming [8]. Furthermore, phage therapy has also been documented for the inhibition of biofilms formulated by Vibrio [18]. In short, phage therapy is becoming a promising antibiotic alternative for use in aquaculture.
In this study, a phage PVA23 was isolated from local sewage samples for its high efficiency in lysing potential against V. alginolyticus. The phage was characterized genomically and phenotypically, and its application against V. alginolyticus was evaluated in pond trials in parallel with disinfectants commonly used in farming practice.
2. Materials and Methods
2.1. Isolation and Identification of V. alginolyticus Strains
V. alginolyticus strains used in this study were isolated from both diseased fishes and shrimps; the wastewater samples were collected from aquaculture ponds in different regions of China using the methods as reported [19]. Fish and shrimp samples were initially disinfected with 0.1% (w/v) benzalkonium chloride and then rinsed with sterile 0.85% (w/v) saline. Under aseptic conditions, animal tissues were ground, homogenized in a glass homogenizer with 5 mL of sterile saline, then the juice was pipetted promptly onto thiosulphate citrate bile salts sucrose agar culture medium (TCBS, Hope Biotechnology Co., Ltd. of Qingdao, Qingdao, China) plates. The plates were incubated at 37 °C for 24 h. Morphologically uniform yellow colonies with a diameter of about 2–4 mm were picked and thread onto new TCBSs to obtain pure cultures. The species of these purified strains were identified by a 16S rRNA PCR using the following universal primers: 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1492R: 5′-GGCTACCTTGTTACGACTT-3′. The PCR products obtained were sequenced and compared with the reference 16S rRNA sequence of V. alginolyticus in the GenBank. All identified isolates were incubated at 37 °C with shaking at 170 rpm in 2216E liquid medium (Hope Biotechnology Co., Ltd. of Qingdao, Qingdao, China) and stored at −80 °C with 20% glycerol.
2.2. Detection of Virulence Genes of the V. alginolyticus Isolates
A panel of common virulence genes of V. alginolyticus, including collagenase gene (Col), alkaline serine protease gene (AspA), helmolysin genes (tlh, tdh and trh), flagellar filament protein gene (flaA), outer membrane protein genes (ompW and ompK), ferric uptake system gene (fur), gyrase B subunit (gryB) and virulence-regulating genes (toxR and toxS), were detected by PCR assay [2,20] (Table 1). The PCR amplification was performed under the following conditions: initial denaturation at 94 °C for 4 min, followed by 35 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1.5 min; and a final extension at 72 °C for 8 min. Amplified PCR products were visualized on a 1% agarose gel stained with ethidium bromide, running at 90 V for 25 min. The presence of virulence genes was confirmed by gel electrophoresis showing the corresponding target band by the gel imaging system after repeating the PCR three times.

Table 1.
Primers used in this study for amplification of virulent genes of V. alginolyticus.
2.3. Isolation, Purification and Propagation of Bacteriophages
Isolation of phages was conducted using the conventional double-layer agar method [13]. The V. alginolyticus that isolated with the most virulent genes from the above experiment were then used as hosts to isolate their specific phages. The sewage sample was collected from a large-scale aquatic market in Qingdao, China. First, sewage water (20 mL) was centrifuged at 8000× g for 10 min, and filtered through a 0.22 µm membrane to get rid of bacteria. Next, 5 mL of the filtrate and 5 mL of V. alginolyticus culture (107 CFU/mL) were added to 50 mL of sterilized 2216E medium. The mixture was incubated at 37 °C with orbital shaking for 6 h to enrich possible phages, then it was centrifuged at 8000× g for 10 min and filtered through a 0.22 µm membrane to remove the remaining bacteria. 100 µL of the filtrate, mixed with 100 μL of V. alginolyticus culture, was poured into 5 mL of molten soft 2216E upper agar (2216E liquid medium with 0.7% agar) and then poured onto the surface of a prepared and cooled 2216E lower plate (2216E liquid medium with 1.5% agar). The phage plaque’s appearance was observed after the overnight incubation at 37 °C.
A single plaque was picked into 1 mL of sterilized PBS buffer (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4·12H2O, 0.24 g KH2PO4, 0.11 g CaCl2, adding water to 1000 mL and adjusting pH to 7.2 with Na2HPO4·12H2O or KH2PO4) and bathing in water at 40 °C for 30 min to harvest phages. Then, the phage-containing PBS buffer was centrifuged at 12,000× g for 5 min and filtered through a 0.22 µm membrane. This filtrate was diluted at a 10-fold gradient with PBS buffer to a suitable multiple which could obtain single plaques on the double-agar plate. The single-plaque isolation process by the double-layer agar method was repeated four more times until plaques of uniform size, shape and clarity were obtained.
The lysate of the pure single plaque in 100 µL PBS was mixed with an equal amount of fresh culture of the host strain, and then was added into 5 mL of 2216E tube and incubated at 37 °C with shaking at 170 rpm to get a phage proliferation solution. Cell debris of the solution was removed by centrifugation and filtration. The bacteriophage titer of the filtrate was determined using the double-layer agar method and expressed as plaque forming units (PFU) per mL filtrate. The proliferation solution was stored at 4 °C for further experiments.
2.4. Screening of Lytic Bacteriophages
The screening of lytic bacteriophages was based on the lytic capability and the host range against the Vibrio strains. Here, the host ranges of the isolated phages were determined by the method of standard spot tests assay [22]. All V. alginolyticus isolates from above experiment and the reference strain V. alginolyticus ATCC 17749 (purchased from Guangdong Microbial Culture Collection Center) were tested. Briefly, 100 µL of each bacterium (107 CFU/mL) was added to 5 mL of molten soft 2216E upper agar and then poured onto the surface of a prepared and cooled 2216E lower plate. 2 µL of PVA23 stock (109 PFU/mL) was spotted onto the lawn and incubated at 37 °C for 12 h. The bacterial hosts of a tested phage were recorded if a lysis circle could be observed in the plate. The experiments were conducted in triplicate.
2.5. Determination of Host Ranges of Phage PVA23
The phage PVA23 was selected for further experiments. The host range of PVA23 was determined by two methods of standard spot tests assay and efficiency of plating (EOP) assay [23]. All V. alginolyticus isolates from above experiment and the reference strain V. alginolyticus ATCC 17749 (purchased from Guangdong Microbial Culture Collection Center) were tested. The spot tests of PVA23 have been done in the above section of screening of lytic bacteriophage. For the EOP assay, phage host ranges were identified by the EOP, which was measured by comparing titers between the test strains and V. alginolyticus VA15 as a reference. The EOP of bacteriophage is generally close to 1, with EOP ≥ 0.5 meaning high production, 0.1 ≤ EOP < 0.5 meaning medium production, 0.001 ≤ EOP < 0.1 belonging to the low production, and EOP ≤ 0.001 meaning no production [24]. The experiment was conducted in triplicate.
In addition, other species of Vibrio and non-Vibrio commonly found in pond water and saved in our laboratory, were also tested to further determine the host range of phage PVA23 using the method of standard spot tests assay. The experiment was conducted in triplicate.
2.6. Transmission Electron Microscopy of Phage PVA23
The morphology of PVA23 was characterized by using transmission electron microscopy [13]. Phage particles were precipitated with 10% polyethylene glycol 8000 (PEG 8000) at 4 °C overnight, centrifuged at 10,000× g for 15 min, and subsequently suspended in PBS buffer. One drop of the concentrated phage particles was spotted onto the surface of carbon grids for 15 min and then negatively stained with 2% phosphotungstic acid for 10 min. The phage was then observed using a Hitachi H-7700 biological transmission electron microscope (Hitachi High-Technologies Corporation, Tokyo, Japan) at an accelerating voltage of 80 kV. It was classified and identified on the basis of International Committee on Taxonomy of Viruses (ICTV) guidelines.
2.7. Determination of Optimal Multiplicity of Infection (MOI) and One-Step Growth Curve of Phage PVA23
In order to determine the optimal MOI of PVA23, 100 µL of serial dilutions of stocked solution (109 PFU/mL) and 100 µL of the host V. alginolyticus VA15 culture in logarithmic phase (107 CFU/mL) were added to 5 mL of 2216E broth to generate the MOI at 0.000001, 0.00001, 0.0001, 0.001, 0.01, 0.1, 1, 10 and 100, respectively. The mix was incubated at 37 °C with orbital shaking for 12 h. Then, the titers of the phage at different MOIs were determined by the double-layer plate method. The one with the highest titer was the optimal MOI. The experiment was conducted in triplicate.
In this work, the one-step growth analysis of PVA23 was performed by previous descriptions [25] with some modifications. First, PVA23 stock (109 PFU/mL) was added to the host V. alginolyticus VA15 culture in logarithmic phase (107 CFU/mL) at a MOI of 1. After incubation at 37°C for 10 min, the broth was centrifuged at 12,000× g for 30 s to remove unabsorbed free phage. Next, the pellets were washed with 2216E for twice at 37 °C, and the suspension was transferred to 20 mL of 2216E followed by immediate incubation at 37 °C with shaking. This moment was defined as t = 0 s, and 200-μL sample was collected every 10 min up to 180 min. The titer of phage particles was conducted using the double-layer agar method. All experiments were performed in triplicate. The burst size of PVA23 was calculated as the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells.
2.8. Determination of Thermal and pH Stability of Phage PVA23
To determine the thermostability of PVA23, stocked solutions (109 PFU/mL) were treated at different temperatures of 40 °C, 50 °C, 60 °C, 70 °C, 75 °C and 80 °C, respectively. Aliquots of 200-μL were taken at 20, 40, and 60 min during the incubation at each temperature, respectively. Then, the double-layer agar method was used to calculate phage titer against V. alginolyticus VA15. The experiment was conducted in triplicate.
To evaluate pH stability of PVA23, aliquots of 1 mL stocked solution (109 PFU/mL) was treated at 37 °C for 2 h with 10 mL PBS buffers with varied pH values (2–13). Then the double-layer agar method was used to calculate the phage titer. The experiment was conducted in triplicate.
2.9. Determination of Chloroform and Ultraviolet (UV) Sensitivity of Phage PVA23
To determine the chloroform sensitivity of the phage, 5 mL of PVA23 stock (109 PFU/mL) was mixed with chloroform to get final concentration at 1%, 2%, 3%, 4% and 5%, respectively, and incubated at 37 °C. The mix was incubated at 37 °C for 30 min by shaking. Then, 200-μL aliquots was taken for phage titer measuring. The phage stock solution without chloroform treatment was used as the control. The experiment was conducted in triplicate.
To determine the impact of Ultraviolet (UV) irradiation, 20 mL of PVA23 stock (109 PFU/mL) was poured into individual 8 cm sterile glass plate and was allowed to be exposed to UV irradiation (20 W, 30 cm) for varied times, respectively. Aliquots (100 µL) of each was withdrawn every 3 min within 30 min. A phage particle not subjected to UV irradiation was used as the control. The experiment was conducted in triplicate. Phage titer was measured to check the ultraviolet (UV) sensitivity of PVA23.
2.10. Assay of Lysis Activity In Vitro of Phage PVA23
The V. alginolyticus VA15 culture (in logarithmic phase,107 CFU/mL) was mixed with a 10-fold serial dilution of the phage stock (109 PFU/mL) to make MOIs at 0.01, 0.1, 1 and 10, respectively. The mix was incubated at 37 °C for 300 min with orbital shaking. Host culture untouched with the phage was served as the control. Aliquots were collected at 10-min intervals and the optical density (OD600) was measured by a UV-vis spectrophotometer. The experiment was conducted in triplicate.
2.11. Assay of Bacterial Phage-Insensitive Mutation Frequency (BIMF) of Phage PVA23
To determine bacterial phage-insensitive mutation frequency (BIMF), PVA23 was used to lyse the host V. alginolyticus VA15 for resistance observation by following the method previously described [26,27]. The fresh cultures of VA15 in logarithmic phase (107 CFU/mL, 100 µL) was used to co-culture with PVA23 stock (109 PFU/mL, 100 µL) at 37 °C for 10 min, the host bacterium without phage was used as the control. After serial dilution, the mixture was coated on TCBS plates and incubated at 37 °C for 18 h, and the number of phage-resistant colonies in the plate was recorded (the number of assumed insensitive colonies). Then, all the resistant colonies were picked for 2216E broth, and tolerance to PVA23 was confirmed by the spot assay. The number of colonies without lysis circle was recorded (the number of determined insensitive colonies). BIMF is the ratio of the number of determined insensitive colonies to the total number of colonies. The experiment was conducted in triplicate.
2.12. DNA Extraction, Genome Sequencing and Assembly of Phage PVA23
First, the concentrated phage PVA23 particles were treated with DNase I and RNase A (New England BioLab, England) followed by incubating at 37 °C for 30 min to remove any exogenous host genomic DNA and RNA. Then, phage PVA23 genomic DNA was extracted using the λ Bacteriophage Genomic DNA Rapid Extracted Kit (Yuanye Biotechnology Co., Ltd. of Shanghai, Shanghai, China) following the manufacturer’s instructions. The purified phage PVA23 DNA was performed on an Illumina NovaSeq sequencing platform (Weilai Biotechnology Co., Ltd. of Qingdao, Qingdao, China). The filtered reads were assembled using SOAPdenovo by the default parameters. The complete genome of phage PVA23 was finished and then manually inspected.
2.13. Genome Analysis and Phylogenetic Analysis of Phage PVA23
Prediction of all open reading frames (ORFs) of the genome sequence was performed using GeneMarkS [28]. Annotation and functional analysis were conducted using BLASTP algorithm against the non-redundant (nr) protein database of the National Center for Biotechnology Information (NCBI) [29]. The genome circle map was drawn by CGView server. The genome sequences of [20] other closely related homologous phages were downloaded from NCBI database, and similarities between the genomic sequences were determined using the average nucleotide identity MUMmer (ANIm) [30]. The putative tRNA-encoding genes were searched using tRNAscan-SE [31]. The genes for virulence and antibiotic resistance were detected by the VFDB database and the ARDB database [32], respectively.
To determine the taxonomy of PVA23, the phylogenetic trees based on the nucleotide sequences of the major capsid protein and the large terminase subunit were constructed using the ClustalW program in MEGA 7.0 software with the neighbor-joining method and 1000 bootstrap replications [33]. Comparative genomic analysis of PVA23 with the highest similar reference phages was performed using Easyfig 2.2 [34].
2.14. Application Evaluation of Phage PVA23 for Vibrio Control in Laboratory Shrimp Culture Trials
To validate the performance of the phage in controlling Vibrio in practical applications, 500 healthy 60-day-old shrimps (Lenaeus vannamei; weight = 10 ± 0.5 g), cultured in 20 L glass tanks under appropriate conditions with aquatic water temperature maintaining at 25 ± 1 °C during the study, were equally divided into five groups. Each group contained 100 shrimp and was then divided equally into two parallel subgroups as replicates (50 shrimp per subgroup). The water used in the glass tanks of all tested groups was seawater, and it was sterilized before the experiment. The V. alginolyticus VA15 was used to challenge shrimp in the experiment. To prepare the cells of V. alginolyticus VA15, the culture in logarithmic phase was spined down and washed with sterile saline three times and then resuspended to make a stock at a concentration of 2 × 109 CFU/mL by measuring the optical density (OD600) and coating TCBS plates. A certain volume of VA15 suspension was added to each test group to reach the final concentration to 2 × 106 CFU/mL in the water. After 1 h, group 2 was added with an iodic disinfectant (found in market) at dose of 5 mg/L, and group 3, group 4 and group 5 receiving varied amounts of PVA23 stock (2 × 1010 CFU/mL) to obtain MOI at 0.1, 0.01 and 0.001, respectively. Group 1 was the blank control and received no treatment. The water samples of test groups were taken every 2 h and the Vibrio content was measured by coating TCBS plates, and the experiment was conducted in triplicate. The cumulative mortality of shrimps was recorded daily during the experiment.
2.15. Application Evaluation of Phage PVA23 for Vibrio Control in Shrimp Farming Plant
The experiment was made in a shrimp farming plant located in Rizhao, China, which was farming for shrimps (P. vannamei, average weight = 8 g ± 0.6g), kept in 30-m3 pond under appropriate conditions (water temperature 26 °C ± 1 °C, salinity 25‰). The pond water in this plant was changed by 20% every day. To control the propagation of pathogenic Vibrio, the pond was regularly added with agents, such as povidone-iodine, complex iodine, chlorine dioxide and probiotic products, etc., which were stopped at least 72 h before the phage tests.
The experiments contained four groups, and the Vibrio contents of the ponds in each group were detected by coating TCBS plates before the start. Group 1 was the blank control receiving no control agent. Group 2 was supplemented with an iodic disinfectant (found in market) at a dose of 5 mg/L. Group 3 and group 4 were supplemented with PVA23 stock with MOI at 0.01 and 0.001, respectively. Each group of the tests repeated twice, with pond water sampled every 4 h and the Vibrio content checked.
2.16. Nucleotide Sequence Accession Number and Phage Preservation
Phage PVA23 was preserved in the China General Microbiological Culture Collection Center (Beijing, China) with CGMCC number of No.24119. The complete genome sequence of PVA8 have been submitted to the NCBI GenBank database under accession number of ON190025.
2.17. Statistical Analyses
Data were analyzed by one-way analysis of variance (ANOVA) using Graph Pad Prism 8.0. Significant differences were determined using Tukey's Multiple Comparison Test. Statistical significance was considered at the p < 0.05 level, and the results were expressed as mean ± standard deviation (SD).
3. Results
3.1. Profile of Virulent Genes of V. alginolyticus Isolates
In this work, in total, 40 V. alginolyticus strains were successfully isolated and identified. In order to investigate the pathogenic features of the isolates of V. alginolyticus, virulence-associated genes, collagenase gene (Col), alkaline serine protease gene (AspA), helmolysin genes (tlh, tdh and trh), flagellar filament protein gene (flaA), outer membrane protein genes (ompK and ompW), ferric uptake system gene (fur), gyrase B subunit (gyrB) and virulence-regulating genes (toxR and toxS) were checked by PCR assay. As shown in Figure 1, AspA, fur and gyrB were the most prevalent genes with the detection frequencies of 100% (40/40), followed by tlh (92.5%, 37/40), Col (90%, 36/40), ompW and FlaA (85%, 34/40), ompK (80%, 32/40), toxR (62.5%, 25/40), trh (37.5%, 15/40), tdh (27.5%, 11/40), and toxS (17.5%, 7/40), respectively. All the V. alginolyticus isolates caried virulent genes, and the majority carried multi-virulent genes. Among them, two isolates, V. alginolyticus VA15 and VA17, carried all the twelve virulent genes, thus they were selected as the presentative strains for later phage lysis testing as well as hosts for the phage screening.
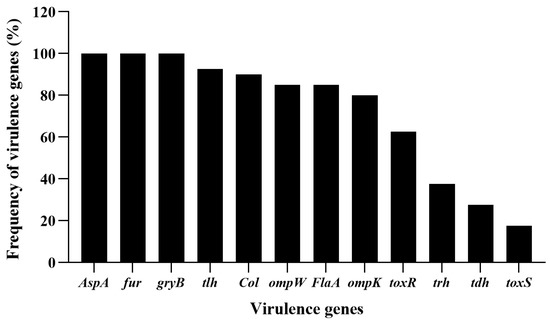
Figure 1.
Frequency of virulent genes detected in the isolates of V. alginolyticus. Twelve virulent genes, Col, AspA, tlh, tdh, trh, FlaA, ompK, ompW, fur, gyrB, toxR and toxS were selected for PCR detection for the V. alginolyticus isolates.
The virulent genes used in the work are not all co-existing in the genome of V. alginolyticus, and their expression levels will change due to gene mutations or changes in the surrounding environment [2,20,35]. Therefore, the absence of some virulent genes in the above result did not imply the absence of the related genes in the genome, but the result was helpful in finding genetically stable V. alginolyticus strains showing more virulent genes [20].
3.2. Isolation and Screening of Lytic Bacteriophages using V. alginolyticus VA15 and VA17 as Hosts
Using V. alginolyticus VA15 and VA17 as hosts, six bacteriophages were successfully isolated and purified from the collected water samples, which were named Vibrio phage PVA21, PVA22, PVA23, PVA24, PVA25 and PVA26, respectively. These phages could form clear plaques on the double-layer plates with their host strains after overnight incubation at 37 °C, indicating that they were likely to be lytic phages against the hosts.
The result of the host ranges of the six phages using the 40 V. alginolyticus isolates and the reference strain V. alginolyticus ATCC 17749 is shown in Figure 2 and Table 2. From the results, PVA23 had the broadest host range with the lysis rate of 63.41% (26/41), followed by PVA24 (19/41, 46.34%), PVA25 (17/41, 41.46%), PVA21 (12/41, 29.27), PVA22 (9/41, 21.95%) and PVA26 (6/41, 14.63%), respectively, and it also showed strong lytic ability to most of its hosts reflected by the maximum number of “+++” (Table 2). Therefore, phage PVA23 with V. alginolyticus VA15 as the host was selected for further experiments.
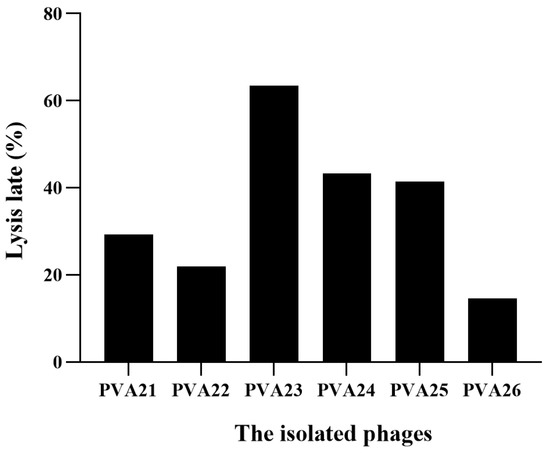
Figure 2.
Lysis rate of the six isolated phages using the 41 V. alginolyticus strains.

Table 2.
Host range of the isolated phages to the V. alginolyticus strains.
3.3. Host Ranges of Phage PVA23
To determine the inhibition activity of PVA23 more accurately, isolates of V. alginolyticus were further challenged by PVA23 to observe clear phage plaque using the EOP assay. According to the results of the spot inoculation test, about 63.41% (26/41) of the V. alginolyticus isolates could be inhibited by PVA23; however, the rate dropped to 56.1% (23/41) when EOP assay applied (Table 3). This difference could be because the EOP assay was more accurate in determining the host range of PVA23 as it excluded the abortive infections that may escape from the spot inoculation test. This result agreed with the recommendation on the use of progeny phages in the determination of host range for a given phage [24].

Table 3.
Host range of the isolated phages to the V. alginolyticus strains.
Furthermore, it was confirmed that isolate 15 was sensitive to PVA23 with relatively decent EOP of 1 (Table 3). Taking into account the 12 virulence genes carried, isolate 15 was used as the host strain of PVA23 hereafter.
In addition, the result of the host ranges of PVA23 using other species of Vibrio and non-Vibrio is shown in Table 4. Except V. alginolyticus, PVA23 could also lyse at least other two Vibrio species, and one of them was Vibrio parahaemolyticus. As expected, it did not exhibit any infectivity to bacterial species in genus other than Vibrio. This cross-species lytic ability and specificity for Vibrio species of phage PVA23 can be used to control different pathogenic Vibrio species and is a suitable choice for further study.

Table 4.
Host ranges of phage PVA23 to other Vibrio strains.
3.4. Morphology and Identification of Phage PVA23
Phage PVA23 formed clear plaques on the lawn of the V. alginolyticus VA15, with an average diameter of about 0.5 mm after 8 h incubation (Figure 3A). The transmission electron microscopy image revealed that PVA23 had an elongated head (prolate capsid) about 134.57 nm long and 89.08 nm wide and a contractile tail about 115.14 nm long (Figure 3B). Therefore, PVA23 was identified as one member of the Myoviridae family, according to the transmission-electron-microscopy based on classification standards used in the International Committee on Taxonomy of Viruses (ICTV).
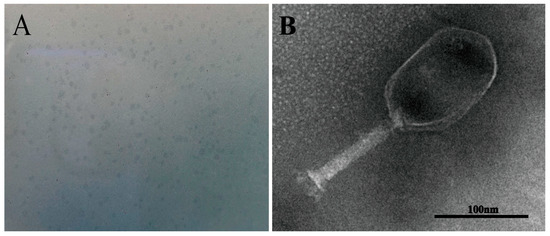
Figure 3.
Phage plaques and transmission electron microscopy of phage PVA23. (A) Phage plagues of PVA23 formed on double-layer agar plate; (B) Transmission electron microscopy of PVA23, scale bar = 100 nm.
3.5. Biological Characteristics of Phage PVA23
The highest titer (4.26 × 1012 PFU/mL) of PVA23 was achieved at MOI of 0.001 (Figure 4A). Lowest MOI of PVA23 observed was as low as 0.000001, indicating that the phage had strong proliferative capacity.
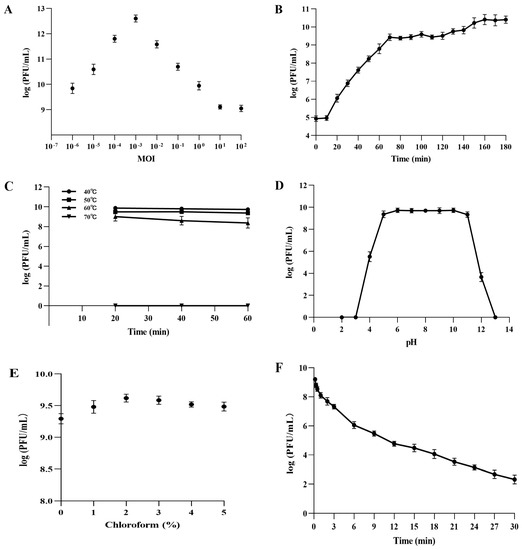
Figure 4.
Biological characteristics of PVA23. (A) The MOI test of PVA23; (B) The one-step growth curve of PVA23; (C) The thermal stability test of PVA23; (D) The pH stability test of PVA23; (E) Chloroform sensitivity test of PVA23, and (F) Ultraviolet (UV) sensitivity test of PVA23.
PVA23 was confirmed to have relatively strong lysis ability by the one-step growth curve determination, in which the short latent time (10 min) and the large burst size (203 PFUs/infected cell) were demonstrated, respectively (Figure 4B).
The thermal stability assay showed that PVA23 was fairly stable at temperatures below 60 °C, but completely inactivated in temperatures at 70 °C (Figure 4C).
The pH stability test showed that PVA23 was stable over a broad pH range, from 5.0 to 11.0, for at least 2 h, but it was completely inactivated under extreme pH conditions (below pH 3.0 or above pH 13.0) (Figure 4D).
As shown in Figure 4E, PVA23 was not sensitive to the chloroform treatment, which indicated the phage capsid protein contained very limited or no lipids.
The survival curve of PVA23 under UV irradiation was depicted in Figure 4F. The results demonstrated that PVA23 was sensitive to the UV irradiation, by following a time based PFU declination trend as observed.
3.6. Lysis Activity in vitro of Phage PVA23
To evaluate the lysis potential of PVA23, in vitro tests were conducted using strain VA15 as the host with four MOIs at 0.01, 0.1, 1 and 10 applied, respectively (Figure 5). It was observed that the OD600 kept increasing and amounted to 1.399 within 300 min in the control, whereas the OD600 of the treatments reduced significantly after the initial growth in the first few hours, indicating the lysis of the cells was caused by PVA23. It was also noted that the lysis time was negatively correlated to the MOIs, with a rough S-shape OD600 trend observed for each MOI, respectively. The OD600 rebounding might reflect the establishment of host resistance to the phage infection via some undefined escaping ways, such as mutation or alteration. It was of interest that the higher MOI lead to faster rebounding in OD600 of the host.
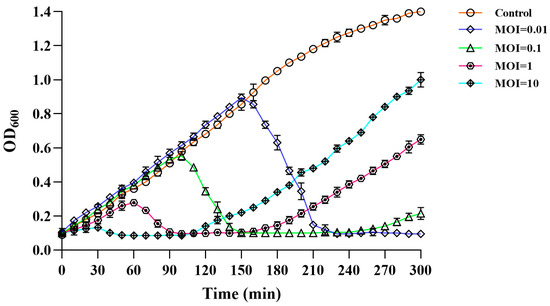
Figure 5.
Lysis activity in vitro of phage PVA23 against the host organism VA15 at four different MOIs (10, 1, 0.1 and 0.01).
3.7. Bacterial Phage-Insensitive Mutation Frequency (BIMF) of V. alginolyticus VA15 to Phage PVA23
To investigate the rebounding of the host bacteria, the BIMF test was conducted. As shown in Table 5, the BIMF of V. alginolyticus VA15 to PVA23 was about (8.48 ± 0.63) × 10−7, similar to those seen in other references [27]. Above results reasoned why the rebounding of the host bacteria in the phage application experiment were as observed, which implied that a certain mutation might occur when the target Vibrio strain was challenged with the phage. Thus, for the control of V. alginolyticus VA15, phage cocktails were suggested in practice to reduce the possible resistance derived due to mutations in the targeted Vibrio.

Table 5.
The result of the BIMF of V. alginolyticus VA15 to phage PVA23.
3.8. Genome Sequencing, Characterization and Analysis of Phage PVA23
The complete genome of PVA23 was 246, 962 bp long, and 99.49% of the reads was matched to the complete genome (8,357,362 out of 8,399,976).
The genome circle map of PVA23 was shown in Figure 6. Sequencing analysis showed that the genome was a linear, double-stranded DNA molecule of 246, 962 bp in length with a GC content of 41.25%. The BLASTn comparison result showed that the whole genome sequences of PVA23 had a maximum nucleotide homology with Vibrio phage VH7D (98% identity, 97% coverage), and following the higher homology with four other Vibrio phages, which were vB_ValM_R10Z, vB_ValM_R11Z, Va3 and phi-ST2 (97% identity, 96% coverage). The ANI heatmap showed that the ANIm percentage identity of PVA23 with VH7D, vB_ValM_R10Z, vB_ValM_R11Z, Va3 and phi-ST2 were 98.02%, 97.97%, 97.90%, 97.87% and 97.62%, respectively (Figure 7). A total of 379 ORFs encoded by the genome were predicted, and 25.33% (96/379) were annotated to have corresponding functions by Blastp analysis against the NCBI nr database. The ORFs that were assigned to the basic phage-related functions were identified, including DNA replication/modification, structure protein, packing protein, metabolism/regulation and host lysis.
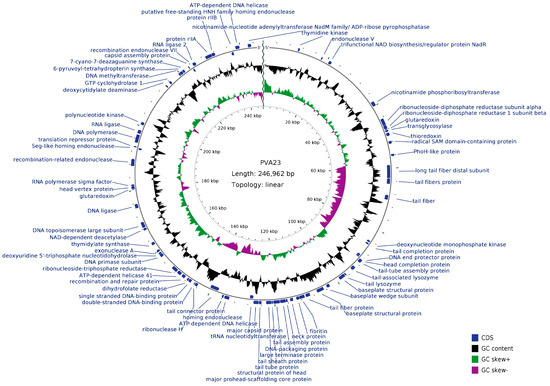
Figure 6.
Genome circle map of PVA23. The outermost circle represents ORFs encoded in the genome. The two innermost circles represent GC-skew [(G − C)/(G + C)] and G + C content. The black circle represents the G + C content. The outward direction indicates that the G + C content of this region is larger than the average G + C content of the whole genome, and the inward direction indicates that the G + C content of that region is lower than the average G + C content. The gray circle represents the GC-skew [(G − C)/(G + C)]. The outward direction indicates that the GC-skew [(G − C)/(G + C)] is larger than zero, and the inward direction indicates that the GC-skew [(G − C)/(G + C)] is lower than zero.
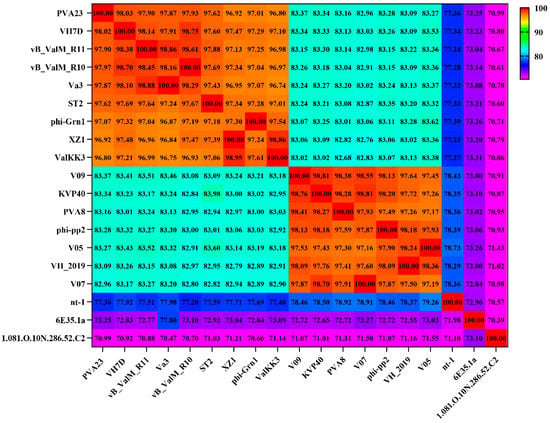
Figure 7.
Average nucleotide identity heatmap. The percentage identity values range from 70% (purple) to 100% (red).
In the DNA replication/modification module, several typical enzymes and proteins were found, which were similar to those of other Vibrio phages. ORF212 (100% identity to phi-ST2) was a main ORF, encoded single stranded DNA-binding protein. DNA polymerase and DNA helicase were encoded by ORF29 (99.53% identity to phi-ST2) and ORF363 (99.76% identity to VH7D), respectively. ORF251 (100% identity to VH7D) and ORF108 (100% identity to ValKK3) were predicted to encode DNA topoisomerase large subunit and medium subunit, respectively. DNA primase was encoded by ORF227 (100% identity to ValKK3) and ORF264 (100% identity to VH7D) encoded DNA ligase. In addition, phage PVA23 also encoded proteins involved in nucleotide metabolism, including the exonuclease encoded by ORF230 (100% identity to VH7D) and the endonuclease encoded by ORF43 (96.27% identity to ValKK3). These enzymes were responsible for generating deoxyribonucleotides for phage DNA synthesis by hydrolyzing host genomic DNA.
3.9. Phylogenetic Analysis and Comparative Genomic Analysis of Phage PVA23
Structure proteins of phages are commonly used for phages taxonomy; among them, major capsid protein is a highly conserved structural protein [36]. Terminase enzyme can specifically catalyze the packaging reaction of phages, and the larger subunit of terminase holoenzyme is involved in translocation of the cleaved DNA into the empty prohead [36]. Therefore, in order to decipher the evolutionary relationships of PVA23 with the other closely related homologous phages, sequences of the major capsid protein and the large terminase subunit of PVA23 were aligned with that of the other closely related phages downloaded from NCBI database. The phylogenetic trees of the major capsid protein (Figure 8A) and the large terminase subunit (Figure 8B) both confirm that phage PVA23 was more closely related to Vibrio phages of phi-ST2 and VH7D, forming a clear clade and showing high homologous coverage (≥91%), while Vibrio phages of vB_ValM_R11Z, Va3 and vB_ValM_R10Z were in other clades.
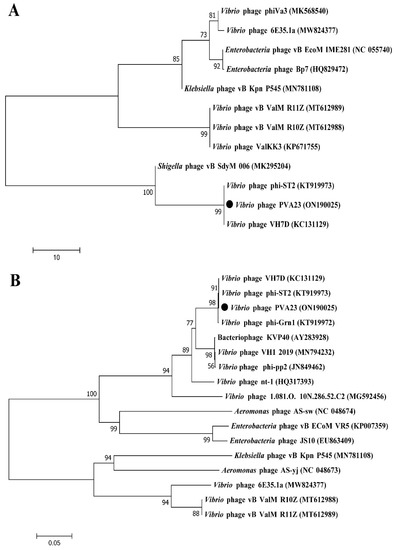
Figure 8.
Phylogenetic analysis of phage PVA23. The phylogenetic tree was constructed using the Maximum Likelihood method with 1000 bootstrap replicates. (A) Phylogenetic tree was constructed based on the major capsid protein; (B) Phylogenetic tree was constructed based on the terminase large subunit.
Furthermore, the comparison of whole genomic sequence alignment of PVA23, phi-ST2 and VH7D was conducted, which could enable the depiction of the relationships of the phages at the genomic levels. As aforementioned, the ORFs assigned to basic phage-related functions can be identified and categorized. Therefore, we categorize the genome sequence of PVA23 into five groups: DNA replication/modification, structure and packing proteins, metabolism/regulation, host lysis and hypothetical protein. Similarly, the genome sequences of phi-ST2 and VH7D were downloaded from NCBI and grouped as PVA23. As shown in Figure 9, the genome of PVA23 highly resembled that of phi-ST2 and VH7D, and the nucleotide identity was not less than 66%.
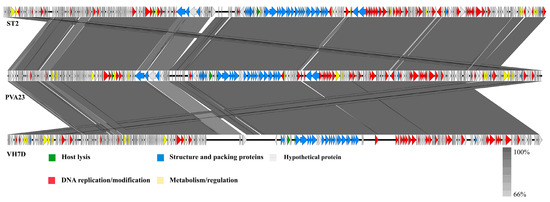
Figure 9.
Comparative genome map of a multiple-genome comparison (amino acid level) performed using Easyfig software. Open reading frames (ORFs) are shown as arrows to indicate the direction of transcription and are colored in accordance with their predicted functions: DNA replication/modification (red arrows), structure protein and packing proteins (blue arrows), metabolism/regulation (yellow arrows) and host lysis (green arrows). Gray arrows represent ORFs of unknown functions (hypothetical protein). Gray shading indicates nucleotide identity between sequences (66–100%).
3.10. Application Evaluation of Phage PVA23 for Vibrio Control in Laboratory Shrimp Culture Trials
Results of phage PVA23 application for Vibrio control in laboratory shrimp culture trials were shown in Figure 10, and no shrimps died through all the trials. It was observed that in both the phage treatment groups (G-3, G-4 and G-5 in Figure 10) and the iodine disinfectant treatment group (G-2 in Figure 10), the level of V. alginolyticu VA15 was reduced by at least three logs at 2 h after supplementation, which is significantly lower than that of the control (G-1 in Figure 10, ~2.14 × 106 CFU/mL). Following this and until 12 h, the level of V. alginolyticu VA15 in the iodine treatment group had a tendency to go up, and it eventually bounced up by almost two logs. However, the levels of V. alginolyticu VA15 in the three phage treatment groups did not go up, with negligible rebounding observed, which clearly revealed that phage treatment outperformed the iodine treatment in the control of V. alginolyticu VA15. The bouncing of VA15 in iodine treatment group may be due to the chemical not being stable with time, e.g., being oxidized when exposed to the continuous airing facilitated by the pump during farming practice. Apparently, the airing process had no effects on the phage, thus no V. alginolyticu bouncing was observed with all three treatments receiving phage PVA23. It was concluded that the use of chemical iodine agent could inhibit VA15 in a limited period of time, but its effectiveness was inferior to that of the phage treatments, which could constantly maintain the V. alginolyticus VA 15 in a low level during the trials. The above results indicate that PVA23 can rapidly and effectively inhibit the growth of the V. alginolyticus VA 15 in shrimp culture practice on laboratory scale, which means that it may have promising application potential in commercial farming practice for the control of the V. alginolyticus strains that may cause disease to aquatic animals.
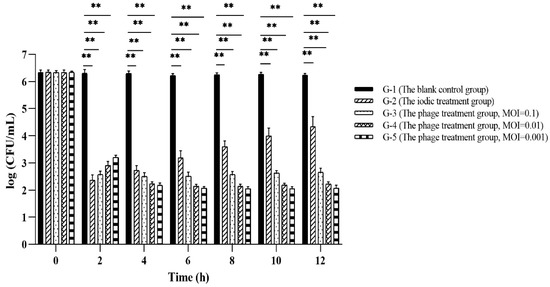
Figure 10.
The result of phage PVA23 application in laboratory shrimp culture trials. **, p < 0.01.
3.11. Application Evaluation of Phage PVA23 for Vibrio Control in Shrimp Farming Plants
To evaluate the effectiveness of PVA23 in the commercial shrimp farming practice, application trials were designed similarly to the lab scale trials, and results were shown in Figure 11. It was found that Vibrio content in each blank control (G-1 in Figure 11) were kept constant or increased with time, which reflected the challenge presented by Vibrio if no intervention applied. The application of chemical disinfectant iodin (G-2 in Figure 11) showed little effect and the Vibrio kept bouncing and elevated to the same level as the control at 24 h. In contrast, Vibrio (V. alginolyticus for specifical) in the ponds with phages (G-3 and G-4 in Figure 11) maintained a tendency of decreasing during the whole trial, with negligible rebounding observed.
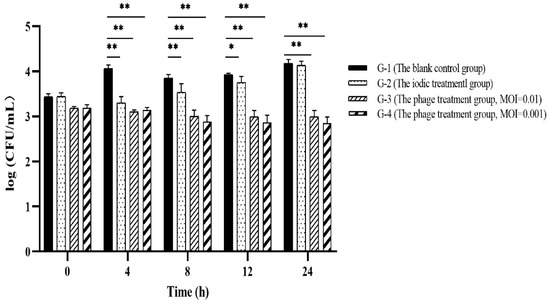
Figure 11.
Application evaluation of PVA23 against Vibrio in commercial shrimp farming plants. In experiment, it was hard to manage to get identical Vibrio counts in different ponds, the ponds with Vibrio ≥ 1000 CFU/mL were selected for the trials and each trial repeated. Vibrio counting in the chosen ponds were real, though they might be varied among ponds to some extent. *, p < 0.05, **, p < 0.01.
4. Discussion
Since 2006, the FDA approved the direct use of bacteriophages for food applications; thereafter, many commercial products using bacteriophages have been released to control food-borne pathogens [37]. However, the application of phage therapy in aquaculture as well as in clinics is still greatly limited. This may be due to (i) the lack of standardized workflow to prepare phage agents for controlling pathogen infections in practical applications [13], (ii) the lack of documentations supporting the safety, effectiveness and so on 47, (iii) the unpredictable influences on the development of multidrug-resistant bacteria pathogens facilitated by transduction of genetic material through horizontal gene transfer [12,38].
In this work, we demonstrated a systematic workflow for a V. alginolyticus lytic bacteriophage, starting from its isolation, identification, then propagation, characterization & sequencing, evaluation, all the way to practical applications. Rather than using V. alginolyticus model strains, VA15 and VA17, two V. alginolyticus strains isolated from diseased animals in ponds were applied as the hosts for phage isolation, which largely eases the screening of the bacteriophage that kills Vibrio isolates and may cause disease in shrimp farming.
With V. alginolyticus VA15 as the host strain, a potent V. alginolyticus lytic bacteriophage PVA23 was screened and developed in this work. It showed high lytic capability and lytic rate against a panel of V. alginolyticus isolates. It also exhibited promising application potentials such as cross-species lysis ability, low MOI, short latent time, large burst size and decent tolerances to adverse conditions.
Generally, for the phage life cycle, DNA packaging is an important process that requires multiple proteins to interact with the phage genome. Terminase enzymes that can specifically catalyze the packaging reaction of phage are composed of large and small subunits [39]. The large subunit is a multifunctional enzyme, which plays a major role in packaging, and the small subunit blinds specifically to the genome and directs the packaging process. In gene annotating of phage PVA23, the large terminase subunit and the small terminase subunit were found encoded by ORF179 and ORF178, respectively. The annotation results also identified other packaging proteins, such as capsid assembly proteins encoded by ORF337. The phage tail is an important component, and the tail adsorption devices specialize in identifying and binding receptors on the surface of bacteria [36]. In the genome of phage PVA23, the tail-associated ORFs that assigned to tail structure protein were identified and categorized, including tail fiber proteins (ORF125, ORF127, ORF172 and ORF207), tail competition protein (ORF150), tail sheath protein (ORF180), tail tube protein (ORF181), tail-tube assembly protein (ORF157), tail connector protein (ORF206) and baseplate structural proteins (ORF158, ORF165, ORF168 and ORF169), which were involved in tail assembly or in facilitating the penetration of the outer membrane of the host cell upon infection [40]. Except these tail structure proteins, the annotation results also identified other phage structure protein, including the major capsid protein (ORF187), head structural protein (ORF182), neck proteins (ORF174 and ORF175) and head vertex protein (ORF270). There are some ORFs in the genome of PVA23 that were assigned to metabolic/regulated functions. For example, ORF25 and ORF238 were predicted to encode thymidylate kinase and thymidylate synthase, respectively, which were involved in the metabolism of nucleic acids, while several other enzymes predicted were involved in the metabolism of tricarboxylic acid or related compounds. In addition, protein rllA, encoded by ORF349, is one of the major players affecting or avoiding the lysis of another phage, which enables the cell infected by the first invader to avoid or exclude the invasion of another phage virion [41]. For double-stranded DNA bacteriophages, the bacterial lysis is achieved by phage-encoded muralytic enzyme called endolysin that hydrolyze the peptidoglycan (PG) layer present in the bacterial cell wall during the final stage of the phage reproduction events [42]. Based on the specificity, endolysins have at least four different hydrolase activities that degrade the cell wall, which are glycoside transferase, lysozyme, amidase and endopeptidase, respectively [43]. The lytic system of phage PVA23 were composed of tail lysozyme and transglycosylase. ORF160 and ORF164 were predicted to encode tail-associated lysozyme and tail lysozyme, respectively, and transglycosylase was encoded by ORF105. In addition, it was noteworthy that up to thirty-four tRNAs were found in the genome of PVA23 by the tRNAscan-SE, which was on the high end of tRNA numbers found in other similar Vibrio phages. The high number of tRNA benefits the synthesis of phage capsid and tail proteins, thus may enhance the phage infection process [44]. In general, virulent phages contain more tRNAs than lysogenic phages. The reason might be that there is a significant association between tRNA distribution and codon usage which leads to an enhanced translational efficiency [45]. The high number of tRNA might also benefit the phage in synthesis of its own proteins [46].
In the genome of phage PVA23, the genes for virulence, antibiotic resistance and lysogeny were not detected, indicating that it could be safely used to control V. alginolyticus. The biological and genomic features guarantee that PVA23 is a suitable candidate for phage therapy in the aquaculture farming.
In the evaluation trials, phage PVA23 could rapidly yet effectively reduce the level of V. alginolyticus both in the lab and plant shrimp framing applications. The results of phage PVA23 against Vibrio (V. alginolyticus for specifical) in farming plant trials was consistent with the positive results observed in laboratory tests, which confirmed that PVA23 could serve as an effective biological agent for the control of V. alginolyticus in farming plant practice. Not only that, unlike the challenge of shrimp supplemented only with V. alginolyticus in the laboratory trial, the bacteria were not added in the field experiments, so any reductions observed were related to Vibrio (in general) and both V. alginolyticus (specifically). This finding is actually more interesting as it shows that phage PVA23 can also be used to against total Vibrio in farming practice, which broadens its application in aquaculture farming. It was noteworthy that phage application evidently outperformed the chemical disinfectant iodine treatment in the prevention of V. alginolyticus propagation in pond. Moreover, unlike the immediate rebounds of V. alginolyticus in iodine treatment, the levels of V. alginolyticus were kept controlled at lower levels than the initial levels, with no rebounding observed. This finding is the first case reporting that phage treatment performs better than the chemical disinfectant in Vibrio control in farming practice.
The main challenge of phage therapy observed recently is the resistance issue, which is spontaneous mutation associated and developed by the bacterial defense system [40]. Phage-resistant bacterial variants often emerge when mutations occur spontaneously after treatment with single phage [47,48]. Although phage resistance develops about ten times slower than antibiotic resistance, it does limit the therapeutic applications of phage therapy [12].
Phage cocktails, or mixtures of multiple phages, may help overcome the problem by delaying the appearance of resistant variants during phage treatment. It resolves the problems of low efficacy, narrow lysis spectrum and fast emergence of phage-resistant bacterial mutants when using a single phage, making treatment more effective against a specific strain or when targeting multiple strains [13]. However, from the industrial points of view, the phage cocktails might be too costly; as it asks for the preparation of an array of phages, each may need separate production line, not to mention the processing challenges like sterile mixing and the input in formulations.
In this work, fresh cells of V. alginolyticus VA15 culture were challenged with PVA23 to check the host resistance development. It was of interest that the addition of phage at higher MOI lead to faster rebounding of the Vibrio in pond, though it demonstrated fast killing. This could be that the lytic phage itself functions as the selective pressure, and the higher level of the phage simply leads to the faster presence of stronger resistant bacteria. Therefore, in practice phage application, the optimal MOI of phage should be carefully chosen to ensure achieving faster killing yet slow rebounding of the target Vibrio. Low-end MOI was chosen in the evaluation trials in this work, and negligible rebounding in Vibrio was noticed. It appeared to be that the choice of phage application at low MOI might be an economic yet practical way to retard the Vibrio resistance in aquatic farming.
In addition, the combination of phages with other antimicrobials derived from probiotics has also been attempted to control the Vibrio in the farming practice by our group. Since the antimicrobials killing mechanism are different with that of phage, the application of the combination of phages with other antimicrobials has efficiently reduced the Vibrio rebounding and demonstrated decent Vibrio control effects (data unpublished).
In conclusion, PVA23, a potent V. alginolyticus lytic bacteriophage, was isolated and identified. The phage belongs to the Myoviridae family, and it demonstrated a panel of promising application potentials, including high lytic capability, high lytic rate, cross-species lysis ability, low MOI, short latent time, large burst size and decent tolerances to adverse conditions. Genome analysis revealed that there was neither virulence nor antibiotic resistance genes in the genome, thus the phage is environmentally friendly and safe for application. In the pond application trails, PVA23 could rapidly yet effectively reduce the level of V. alginolyticus and significantly outperform the chemical disinfectant iodine in Vibrio controlling. Furthermore, it was confirmed that the approach for application of phage at low MOI could be a good choice to effectively control the immediate Vibrio rebounding caused by resistant Vibrio variants or mutants. In this study, a systematic workflow on the discovery and evaluation of a potent phage was exemplified, with PVA23 being successfully evaluated as a promising Vibrio control agent in V. alginolyticus controlling. Our work confirmed that phage PVA23 can be used as an alternative to antibiotics or chemical disinfectants in shrimp farming practice.
Author Contributions
J.F.: conceptualization, methodology, formal analysis, validation, investigation, software, writing-original draft, writing-review and editing, data curation; Y.L.: methodology, formal analysis, software; L.Z.: methodology, software; C.W.: methodology, software; Z.H.: conceptualization, writing-review and editing, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Special Fund for TaiShan Industrial Experts Program, grant number tscy202006017. This research was also supported by Science and Technology Projects in Key Areas of Guangdong, grant number 2020B0202010001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated for this study can be found in the phage was preserved in the China General Microbiological Culture Collection Center (Beijing, China) with CGMCC number of No.24119. The complete genome sequences of the phage PVA23 have been submitted to the NCBI GenBank database with accession number of ON190025.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baffone, W.; Citterio, B.; Vittoria, E.; Casaroli, A.; Pianetti, A.; Campana, R.; Bruscolini, F. Determination of several potential virulence factors in Vibrio spp. isolated from sea water. Food Microbiol. 2001, 1, 479–488. [Google Scholar] [CrossRef]
- Kahla-Nakbi, A.B.; Chaieb, K.; Besbes, A.; Zmantar, T.; Bakhrouf, A. Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet. Microbiol. 2006, 117, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.W.; Kim, H.J.; Yun, S.K.; Chai, J.Y.; Park, S.C. Eating oysters without risk of vibriosis: Application of a bacteriophage against Vibrio parahaemolyticus in oysters. Int. J. Food Microbiol. 2014, 188, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Letchumanan, V.; Yin, W.F.; Lee, L.H.; Chan, K.G. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 2015, 6, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.A.; El Bayomi, R.M.; Hussein, M.A.; Khedr, M.H.E.; Abo Remela, E.M.; El-Ashram, A.M.M. Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int. J. Food Microbiol. 2018, 274, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Boonyawiwat, V.; Patanasatienkul, T.; Kasornchandra, J.; Poolkhet, C.; Yaemkasem, S.; Hammell, L.; Davidson, J. Impact of farm management on expression of early mortality syndrome/acute hepatopancreatic necrosis disease (EMS/AHPND) on penaeid shrimp farms in Thailand. J. Fish Dis. 2017, 40, 649–659. [Google Scholar] [CrossRef]
- Lu, H.; Han, W.; Lei, L. Research progress of therapeutic bacteriophage preparations. Chin. J. Vet. Drug 2002, 36, 39–41. [Google Scholar]
- Oliveira, J.; Castilho, F.; Cunha, A.; Pereira, M.J. Bacteriophage therapy as a bacterial control strategy in aquaculture. Aquac. Int. 2012, 20, 879–910. [Google Scholar] [CrossRef]
- Letchumanan, V.; Chan, K.G.; Pusparajah, P.; Saokaew, S.; Duangjai, A.; Goh, B.H.; Ab Mutalib, N.S.; Lee, L.H. Insights into Bacteriophage Application in Controlling Vibrio Species. Front. Microbiol. 2016, 7, 1114. [Google Scholar] [CrossRef]
- Wu, L.; Tian, Y.; Pang, M.; Yang, Z.; Bao, H.; Zhou, Y.; Sun, L.; Wang, R.; Zhang, H. A novel vibriophage vB_VhaS_PcB-1G capable of inhibiting virulent Vibrio harveyi pathogen. Aquaculture 2021, 542, 736854. [Google Scholar] [CrossRef]
- Kalatzis, P.G.; Castillo, D.; Katharios, P.; Middelboe, M. Bacteriophage Interactions with Marine Pathogenic Vibrios: Implications for Phage Therapy. Antibiotics 2018, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Doss, J.; Culbertson, K.; Hahn, D.; Camacho, J.; Barekzi, N. A Review of Phage Therapy against Bacterial Pathogens of Aquatic and Terrestrial Organisms. Viruses 2017, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fan, J.; Yan, T.; Liu, Q.; Yuan, S.; Zhang, H.; Yang, J.; Deng, D.; Huang, S.; Ma, Y. Isolation and Characterization of Specific Phages to Prepare a Cocktail Preventing Vibrio sp. Va-F3 Infections in Shrimp (Litopenaeus vannamei). Front. Microbiol. 2019, 10, 2337–2347. [Google Scholar] [CrossRef] [PubMed]
- Lomeli-Ortega, C.O.; Martinez-Sandez, A.J.; Barajas-Sandoval, D.R.; Reyes, A.G.; Magallon-Barajas, F.; Veyrand-Quiros, B.; Gannon, L.; Harrison, C.; Michniewski, S.; Millard, A.; et al. Isolation and characterization of vibriophage vB_Vc_SrVc9: An effective agent in preventing Vibrio campbellii infections in brine shrimp nauplii (Artemia franciscana). J. Appl. Microbiol. 2021, 131, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Stalin, N.; Srinivasan, P. Efficacy of potential phage cocktails against Vibrio harveyi and closely related Vibrio species isolated from shrimp aquaculture environment in the south east coast of India. Vet. Microbiol. 2017, 207, 83–96. [Google Scholar] [CrossRef]
- Kalatzis, P.G.; Rorbo, N.I.; Castillo, D.; Mauritzen, J.J.; Jorgensen, J.; Kokkari, C.; Zhang, F.; Katharios, P.; Middelboe, M. Stumbling across the Same Phage: Comparative Genomics of Widespread Temperate Phages Infecting the Fish Pathogen Vibrio anguillarum. Viruses 2017, 9, 122. [Google Scholar] [CrossRef]
- Li, Z.; Ren, H.; Li, Q.; Murtaza, B.; Li, X.; Zhang, J.; Xu, Y. Exploring the effects of phage cocktails in preventing Vibrio infections in juvenile sea cucumber (Apostichopus japonicus) farming. Aquaculture 2020, 515, 734599. [Google Scholar] [CrossRef]
- Yang, M.; Chen, H.; Guo, S.; Tan, S.; Xie, Z.; Zhang, J.; Wu, Q.; Tan, Z. Characterization and genome analysis of a novel Vibrio parahaemolyticus phage vB_VpP_DE17. Virus Res. 2022, 307, 198580. [Google Scholar] [CrossRef]
- George, M.R.; John, K.R.; Iyappan, T.; Jeyaseelan, M.J. Genetic heterogeneity among Vibrio alginolyticus isolated from shrimp farms by PCR fingerprinting. Lett. Appl. Microbiol. 2005, 40, 369–372. [Google Scholar] [CrossRef]
- Fethi, B.A.; Ali, E.; Rihab, L.; Heacute la, K.; Amina, B. Virulence gene expression, proteins secreted and morphological alterations of Vibrio parahaemolyticus and Vibrio alginolyticus in response to long-term starvation in seawater. Afr. J. Microbiol. Res. 2011, 5, 792–801. [Google Scholar] [CrossRef]
- Xiong, P.; Peng, X.; Wei, S.; Chen, Y.; Zhao, H.; Tang, S.; Wu, X. Virulence-related genes of Vibrio alginolyticus and its virulence in mice. Acta Microbiol. Sin. 2014, 54, 80–88. [Google Scholar]
- Feng, T.; Leptihn, S.; Dong, K.; Loh, B.; Zhang, Y.; Stefan, M.I.; Li, M.; Guo, X.; Cui, Z. JD419, a Staphylococcus aureus Phage with a Unique Morphology and Broad Host Range. Front. Microbiol. 2021, 12, 602902. [Google Scholar] [CrossRef] [PubMed]
- Moodley, A.; Kot, W.; Nalgard, S.; Jakociune, D.; Neve, H.; Hansen, L.H.; Guardabassi, L.; Vogensen, F.K. Isolation and characterization of bacteriophages active against methicillin-resistant Staphylococcus pseudintermedius. Res. Vet. Sci. 2019, 122, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ku, H.J.; Lee, D.H.; Kim, Y.T.; Shin, H.; Ryu, S.; Lee, J.H. Characterization and Genomic Study of the Novel Bacteriophage HY01 Infecting Both Escherichia coli O157:H7 and Shigella flexneri: Potential as a Biocontrol Agent in Food. PLoS ONE 2016, 11, e0168985. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F.; Fleming, H.P.; Altermann, E.; Klaenhammer, T.R. Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003, 84, 225–235. [Google Scholar] [CrossRef]
- Gutierrez, D.; Vandenheuvel, D.; Martinez, B.; Rodriguez, A.; Lavigne, R.; Garcia, P. Two Phages, phiIPLA-RODI and phiIPLA-C1C, Lyse Mono- and Dual-Species Staphylococcal Biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, 451–454. [Google Scholar] [CrossRef]
- Song, D.; Yang, Y.; Yu, B.; Zheng, B.; Deng, Z.; Lu, B.L.; Chen, X.; Jiang, T. Computational prediction of novel non-coding RNAs in Arabidopsis thaliana. BMC Bioinform. 2009, 10, S36. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Underwood, A.P.; Mulder, A.; Gharbia, S.; Green, J. Virulence Searcher: A tool for searching raw genome sequences from bacterial genomes for putative virulence factors. Clin. Microbiol. Infect. 2005, 11, 770–772. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.; Amal, M.N.A.; Saad, M.Z.; Yasin, I.S.M.; Zulkiply, N.A.; Mustafa, M.; Nasruddin, N.S. Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet. Res. 2019, 15, 176. [Google Scholar] [CrossRef]
- Ding, T.; Sun, H.; Pan, Q.; Zhao, F.; Zhang, Z.; Ren, H. Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res. 2020, 286, 198080. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, Y.T.; Kim, H.B.; Choi, S.H.; Lee, J.H. Characterization of bacteriophage VVP001 and its application for the inhibition of Vibrio vulnificus causing seafood-borne diseases. Food Microbiol. 2021, 94, 103630. [Google Scholar] [CrossRef]
- Ninawe, A.S.; Sivasankari, S.; Ramasamy, P.; Kiran, G.S.; Selvin, J. Bacteriophages for aquaculture disease control. Aquac. Int. 2020, 28, 1925–1938. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, F.; Sun, H.; Wang, Q.; Zhang, C.; Liu, W.; Zou, L.; Pan, Q.; Ren, H. Isolation and characterization of the Staphylococcus aureus bacteriophage vB_SauS_SA2. AIMS Microbiol. 2019, 5, 285–307. [Google Scholar] [CrossRef]
- Huang, L.; Xiang, Y. Structures of the tailed bacteriophages that infect Gram-positive bacteria. Curr. Opin. Virol. 2020, 45, 65–74. [Google Scholar] [CrossRef]
- Moussa, S.H.; Lawler, J.L.; Young, R. Genetic dissection of T4 lysis. J. Bacteriol. 2014, 196, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- O'Flaherty, S.; Ross, R.P.; Coffey, A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 2009, 33, 801–819. [Google Scholar] [CrossRef] [PubMed]
- Matamp, N.; Bhat, S.G. Phage Endolysins as Potential Antimicrobials against Multidrug Resistant Vibrio alginolyticus and Vibrio parahaemolyticus: Current Status of Research and Challenges Ahead. Microorganisms 2019, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Zhu, H. Multi-label multi-instance transfer learning for simultaneous reconstruction and cross-talk modeling of multiple human signaling pathways. BMC Bioinform. 2015, 16, 417. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, P.; Chen, B.; Song, Z.; Song, Y.; Yang, Y.; Ma, P.; Wang, H.; Ying, J.; Ren, P.; Yang, L.; et al. Bioinformatic analysis of the Acinetobacter baumannii phage AB1 genome. Gene 2012, 507, 125–134. [Google Scholar] [CrossRef]
- Lu, H.; Yan, P.; Xiong, W.; Wang, J.; Liu, X. Genomic characterization of a novel virulent phage infecting Shigella fiexneri and isolated from sewage. Virus Res. 2020, 283, 197983. [Google Scholar] [CrossRef]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Duarte, J.; Pereira, C.; Costa, P.; Almeida, A. Bacteriophages with Potential to Inactivate Aeromonas hydrophila in Cockles: In Vitro and In Vivo Preliminary Studies. Antibiotics 2021, 10, 710. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).