Susceptibility of Domestic Goat (Capra aegagrus hircus) to Experimental Infection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) B.1.351/Beta Variant
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Study
2.2. Cells and Virus
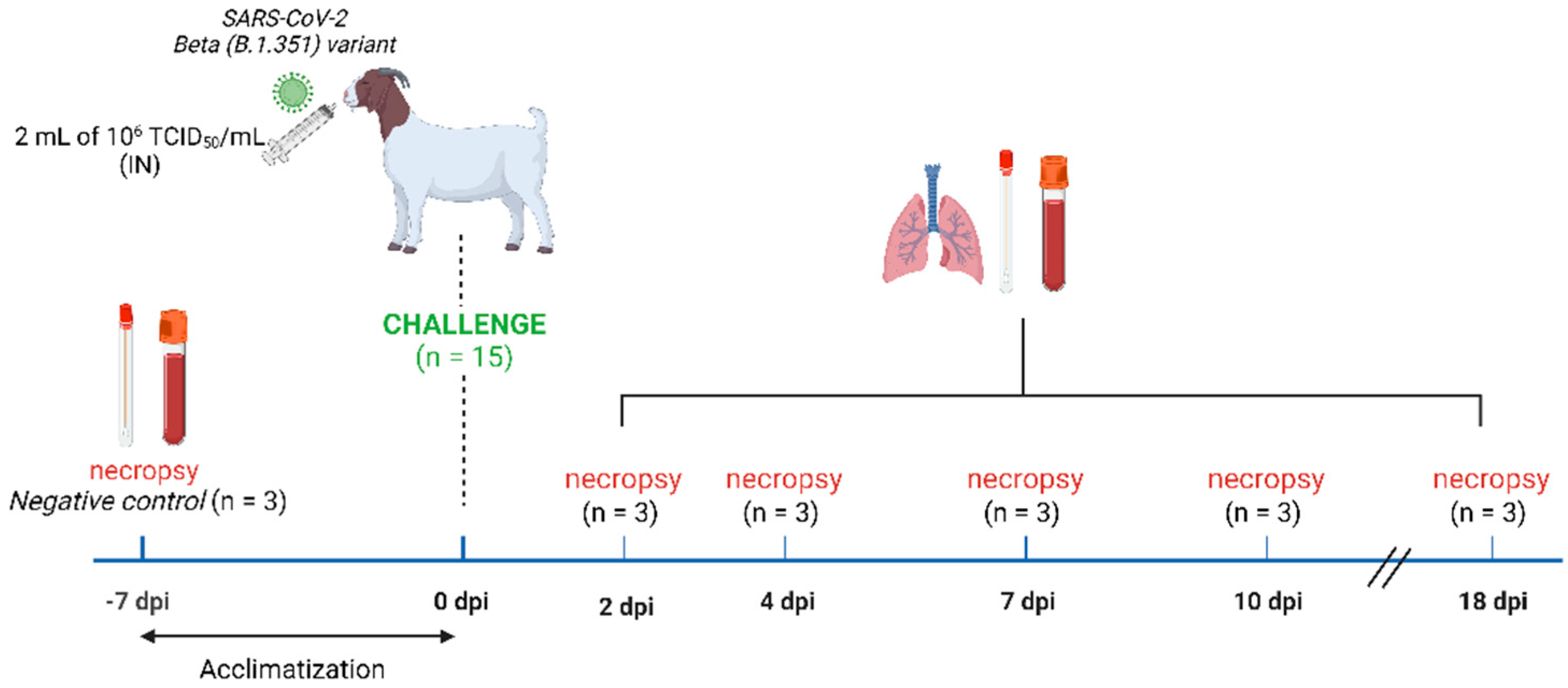
2.3. Experimental Study Design
2.4. Virus Detection and Isolation
2.5. Neutralising Antibodies
2.6. Histopathology
2.7. Statistical Analyses
3. Results
3.1. In Silico Predictions of Binding Affinity Changes in B.1.351/Beta, B.1.617.2/Delta, P.1/Gamma, and B.1.1.529/Omicron Variants
3.2. Clinical Signs and Pathological Findings
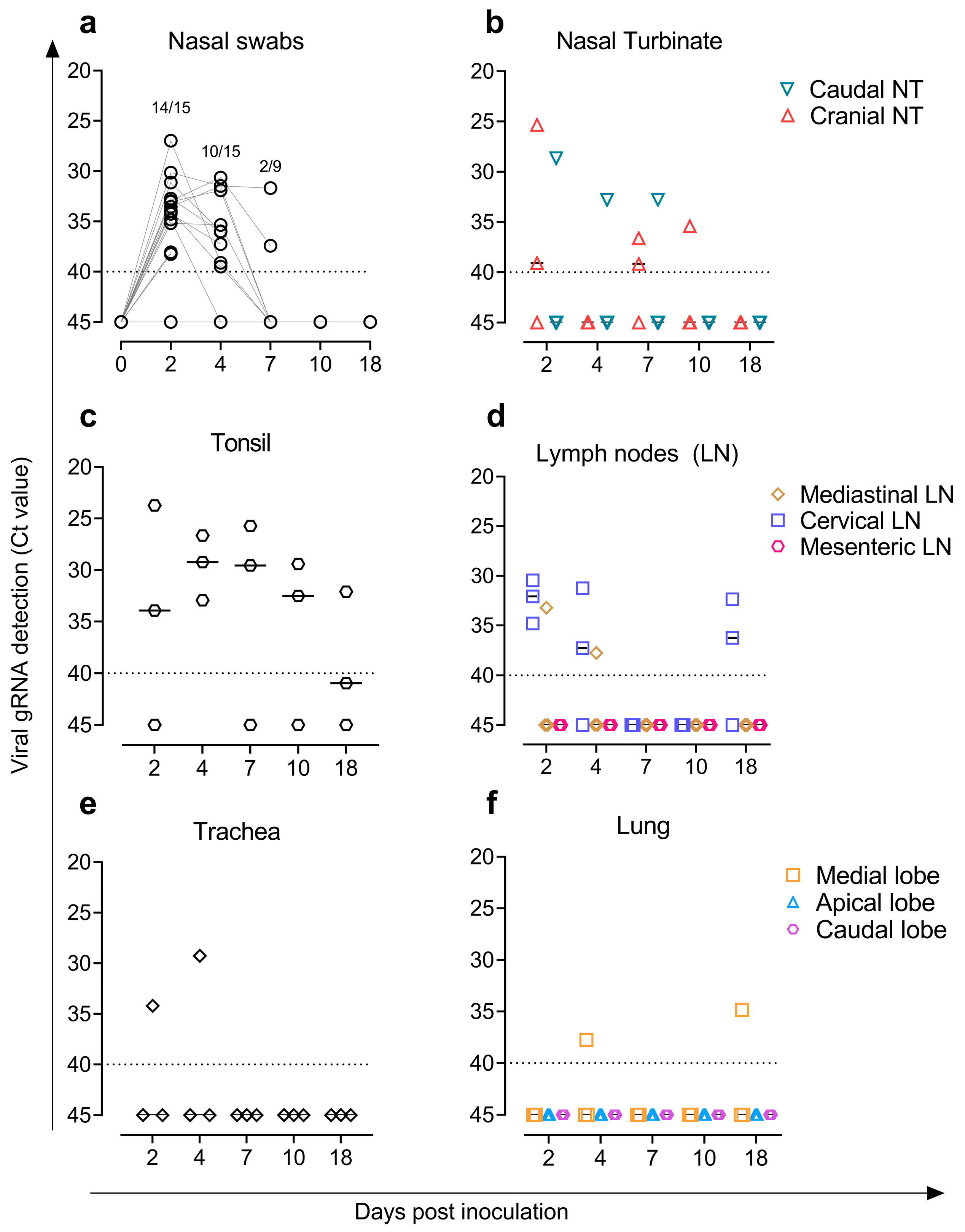
3.3. Virus Detection and Replicative Viral Isolation in Goat Samples
3.4. Detection of SARS-CoV-2 Nucleoprotein in Tissues by Immunohistochemistry
3.5. Inoculated Goats Developed Humoral Responses against SARS-CoV-2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jayaweera, M.; Perera, H.; Gunawardana, B.; Manatunge, J. Transmission of COVID-19 Virus by Droplets and Aerosols: A Critical Review on the Unresolved Dichotomy. Environ. Res. 2020, 188, 109819. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Vongphayloth, K.; Salazar, E.B.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D.; et al. Bat Coronaviruses Related to SARS-CoV-2 and Infectious for Human Cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Worobey, M.; Levy, J.I.; Serrano, L.M.; Crits-Christoph, A.; Pekar, J.E.; Goldstein, S.A.; Rasmussen, A.L.; Kraemer, M.U.G.; Newman, C.; Koopmans, M.P.G.; et al. The Huanan Seafood Wholesale Market in Wuhan Was the Early Epicenter of the COVID-19 Pandemic. Science 2022, 377, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Dhama, K.; Pawde, A.M.; Gortázar, C.; Tiwari, R.; Katterine Bonilla-Aldana, D.; Rodriguez-Morales, A.J.; De La Fuente, J.; Michalak, I.; Attia, Y.A.; et al. SARS-CoV-2 in Animals: Potential for Unknown Reservoir Hosts and Public Health Implications. Vet. Q. 2021, 41, 181–201. [Google Scholar] [CrossRef]
- Dróżdż, M.; Krzyżek, P.; Dudek, B.; Makuch, S.; Janczura, A.; Paluch, E. Current State of Knowledge about Role of Pets in Zoonotic Transmission of SARS-CoV-2. Viruses 2021, 13, 1149. [Google Scholar] [CrossRef]
- World Health Organization Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 31 August 2022).
- World Organisation for Animal Health (WOAH) Events in Animals. Available online: https://www.woah.org/en/what-we-offer/emergency-and-resilience/covid-19/#ui-id-3 (accessed on 31 August 2022).
- Fernández-Bellon, H.; Rodon, J.; Fernández-Bastit, L.; Almagro, V.; Padilla-Solé, P.; Lorca-Oró, C.; Valle, R.; Roca, N.; Grazioli, S.; Trogu, T.; et al. Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses. Viruses 2021, 13, 1683. [Google Scholar] [CrossRef]
- Goraichuk, I.V.; Arefiev, V.; Stegniy, B.T.; Gerilovych, A.P. Zoonotic and Reverse Zoonotic Transmissibility of SARS-CoV-2. Virus Res. 2021, 302, 198473. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural Sars-Cov-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220-20. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; Van Der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on Mink Farms between Humans and Mink and Back to Humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; Van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory Disease in Rhesus Macaques Inoculated with SARS-CoV-2. Nature 2020, 585, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of Ferrets, Cats, Dogs, and Other Domesticated Animals to SARS-Coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.V.; Martins, M.; Falkenberg, S.; Buckley, A.; Caserta, L.C.; Mitchell, P.K.; Cassmann, E.D.; Rollins, A.; Zylich, N.C.; Renshaw, R.W.; et al. Susceptibility of White-Tailed Deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 2021, 95, e00083-21. [Google Scholar] [CrossRef]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in Fruit Bats, Ferrets, Pigs, and Chickens: An Experimental Transmission Study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Root, J.J.; Porter, S.M.; Walker, A.E.; Guilbert, L.; Hawvermale, D.; Pepper, A.; Maison, R.M.; Hartwig, A.E.; Gordy, P.; et al. Peridomestic Mammal Susceptibility to Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Emerg. Infect. Dis. 2021, 27, 2073–2080. [Google Scholar] [CrossRef] [PubMed]
- Freuling, C.M.; Breithaupt, A.; Müller, T.; Sehl, J.; Bakema-Buschmann, A.; Rissmann, M.; Klein, A.; Wylezich, C.; Höper, D.; Wernike, K.; et al. Susceptibility of Raccoon Dogs for SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2982. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Lamers, M.M.; Okba, N.M.A.; Breugem, T.I.; Schipper, D.; van den Doel, P.B.; van Run, P.; van Amerongen, G.; de Waal, L.; Koopmans, M.P.G.; et al. Susceptibility of Rabbits to SARS-CoV-2. Emerg. Microbes Infect. 2021, 10, 1–7. [Google Scholar] [CrossRef]
- Tarrés-Freixas, F.; Trinité, B.; Pons-Grífols, A.; Romero-Durana, M.; Riveira-Muñoz, E.; Ávila-Nieto, C.; Pérez, M.; Garcia-Vidal, E.; Perez-Zsolt, D.; Muñoz-Basagoiti, J.; et al. Heterogeneous Infectivity and Pathogenesis of SARS-CoV-2 Variants Beta, Delta and Omicron in Transgenic K18-HACE2 and Wildtype Mice. Front. Microbiol. 2022, 1, 1382. [Google Scholar] [CrossRef]
- Pan, T.; Chen, R.; He, X.; Yuan, Y.; Deng, X.; Li, R.; Yan, H.; Yan, S.; Liu, J.; Zhang, Y.; et al. Infection of Wild-Type Mice by SARS-CoV-2 B.1.351 Variant Indicates a Possible Novel Cross-Species Transmission Route. Signal. Transduct. Target. Ther. 2021, 6, 1–12. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Zhang, Z.R.; Zhang, H.Q.; Li, N.; Zhang, Q.Y.; Li, X.D.; Deng, C.L.; Deng, F.; Shen, S.; Zhu, B.; et al. Different Pathogenesis of SARS-CoV-2 Omicron Variant in Wild-Type Laboratory Mice and Hamsters. Signal. Transduct. Target. Ther. 2022, 7, 1–3. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.W.; Yuen, T.T.T.; Yoon, C.; Hu, J.C.; Wen, L.; Hu, B.; Yang, D.; Wang, Y.; Hou, Y.; et al. Emerging SARS-CoV-2 Variants Expand Species Tropism to Murines. EBioMedicine 2021, 73, 103643. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Zhai, X.; Sun, J.; Yan, Z.; Zhang, J.; Zhao, J.; Zhao, Z.; Gao, Q.; He, W.-T.; Veit, M.; Su, S. Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts. J. Virol. 2020, 94, e00831-20. [Google Scholar] [CrossRef]
- Jori, F.; Hernandez-Jover, M.; Magouras, I.; Dürr, S.; Brookes, V.J. Wildlife-Livestock Interactions in Animal Production Systems: What Are the Biosecurity and Health Implications? Anim. Front. 2021, 11, 8–19. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; Mcbride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 Infection in Free-Ranging White-Tailed Deer. Nature 2022, 602, 481. [Google Scholar] [CrossRef]
- Cool, K.; Gaudreault, N.N.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; McDowell, C.; Carossino, M.; Bold, D.; Kwon, T.; Balaraman, V.; et al. Infection and Transmission of Ancestral SARS-CoV-2 and Its Alpha Variant in Pregnant White-Tailed Deer. bioRxiv 2022, 11, 95–112. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Walker, A.; Guilbert, L.; Porter, S.; Hartwig, A.; Mcvicker, E.; Bielefeldt-Ohmann, H.; Bowen, R.A. Susceptibility of Livestock to SARS-CoV-2 Infection. Emerg. Mcirobes Infect. 2021, 10, 2199–2201. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Cool, K.; Trujillo, J.D.; Morozov, I.; Meekins, D.A.; McDowell, C.; Bold, D.; Carossino, M.; Balaraman, V.; Mitzel, D.; et al. Susceptibility of Sheep to Experimental Co-Infection with the Ancestral Lineage of SARS-CoV-2 and Its Alpha Variant. Emerg. Microbes Infect. 2022, 11, 662–675. [Google Scholar] [CrossRef]
- Gilbert, M.; Nicolas, G.; Cinardi, G.; Van Boeckel, T.P.; Vanwambeke, S.O.; Wint, G.R.W.; Robinson, T.P. Global Distribution Data for Cattle, Buffaloes, Horses, Sheep, Goats, Pigs, Chickens and Ducks in 2010. Nat. Sci. Data 2018, 5, 180227. [Google Scholar] [CrossRef] [PubMed]
- Montagutelli, X.; Prot, M.; Levillayer, L.; Salazar, E.B.; Jouvion, G.; Conquet, L.; Donati, F.; Albert, M.; Gambaro, F.; Behillil, S.; et al. The B1.351 and P.1 Variants Extend SARS-CoV-2 Host Range to Mice. bioRxiv 2021, 10, 18-436013. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Brustolin, M.; Rodon, J.; Rodríguez de la Concepción, M.L.; Ávila-Nieto, C.; Cantero, G.; Pérez, M.; Te, N.; Noguera-Julián, M.; Guallar, V.; Valencia, A.; et al. Protection against Reinfection with D614- or G614-SARS-CoV-2 Isolates in Golden Syrian Hamster. Emerg. Microbes Infect. 2021, 10, 797–809. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581, 465. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty per Cent Endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Munnink, B.B.O.; De Meulder, D.; Van Amerongen, G.; Van Den Brand, J.; Okba, N.M.A.; et al. Comparative Pathogenesis of COVID-19, MERS, and SARS in a Nonhuman Primate Model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.-P.; Pfenning, A.R.; Zhao, H.; et al. Broad Host Range of SARS-CoV-2 Predicted by Comparative and Structural Analysis of ACE2 in Vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Kapczynski, D.R.; Sweeney, R.; Spackman, E.; Pantin-Jackwood, M.; Suarez, D.L. Development of an in Vitro Model for Animal Species Susceptibility to SARS-CoV-2 Replication Based on Expression of ACE2 and TMPRSS2 in Avian Cells. Virology 2022, 569, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Li, Y.M.; Sun, J.; Zhang, Y.Y.; Wang, T.Y.; Sun, M.X.; Wang, M.H.; Yang, Y.L.; Hu, X.L.; Tang, Y.D.; et al. Evaluating Angiotensin-Converting Enzyme 2-Mediated SARS-CoV-2 Entry across Species. J. Biol. Chem. 2021, 296, 100435. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, L.; Wernike, K.; Hoffmann, D.; Mettenleiter, T.C.; Beer, M. Experimental Infection of Cattle with SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2979–2981. [Google Scholar] [CrossRef] [PubMed]
- Niles, M.A.; Gogesch, P.; Kronhart, S.; Ortega Iannazzo, S.; Kochs, G.; Waibler, Z.; Anzaghe, M. Macrophages and Dendritic Cells Are Not the Major Source of Pro-Inflammatory Cytokines Upon SARS-CoV-2 Infection. Front. Immunol. 2021, 12, 647824. [Google Scholar] [CrossRef] [PubMed]
- Perez-Zsolt, D.; Muñoz-Basagoiti, J.; Rodon, J.; Elosua-Bayes, M.; Raïch-Regué, D.; Risco, C.; Sachse, M.; Pino, M.; Gumber, S.; Paiardini, M.; et al. SARS-CoV-2 Interaction with Siglec-1 Mediates Trans-Infection by Dendritic Cells. Joaquim Segalés 2021, 18, 2676–2678. [Google Scholar] [CrossRef]
- Li, L.; Han, P.; Huang, B.; Xie, Y.; Li, W.; Zhang, D.; Han, P.; Xu, Z.; Bai, B.; Zhou, J.; et al. Cell Discovery Broader-Species Receptor Binding and Structural Bases of Omicron SARS-CoV-2 to Both Mouse and Palm-Civet ACE2s. Cell Discov. 2022, 8, 65. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Bastit, L.; Roca, N.; Romero-Durana, M.; Rodon, J.; Cantero, G.; García, Ó.; López, C.; Pérez, M.; López, R.; Carrillo, J.; et al. Susceptibility of Domestic Goat (Capra aegagrus hircus) to Experimental Infection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) B.1.351/Beta Variant. Viruses 2022, 14, 2002. https://doi.org/10.3390/v14092002
Fernández-Bastit L, Roca N, Romero-Durana M, Rodon J, Cantero G, García Ó, López C, Pérez M, López R, Carrillo J, et al. Susceptibility of Domestic Goat (Capra aegagrus hircus) to Experimental Infection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) B.1.351/Beta Variant. Viruses. 2022; 14(9):2002. https://doi.org/10.3390/v14092002
Chicago/Turabian StyleFernández-Bastit, Leira, Núria Roca, Miguel Romero-Durana, Jordi Rodon, Guillermo Cantero, Óscar García, Carlos López, Mònica Pérez, Rosa López, Jorge Carrillo, and et al. 2022. "Susceptibility of Domestic Goat (Capra aegagrus hircus) to Experimental Infection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) B.1.351/Beta Variant" Viruses 14, no. 9: 2002. https://doi.org/10.3390/v14092002
APA StyleFernández-Bastit, L., Roca, N., Romero-Durana, M., Rodon, J., Cantero, G., García, Ó., López, C., Pérez, M., López, R., Carrillo, J., Izquierdo-Useros, N., Blanco, J., Clotet, B., Pujols, J., Vergara-Alert, J., Segalés, J., & Lorca-Oró, C. (2022). Susceptibility of Domestic Goat (Capra aegagrus hircus) to Experimental Infection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) B.1.351/Beta Variant. Viruses, 14(9), 2002. https://doi.org/10.3390/v14092002




