Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Preparation for SAXS
2.2. Protein Purification for Activity Assay
2.3. DNA Synthesis
2.4. NMR Inhibition Assay
2.5. SAXS Data Acquisition and Analysis
3. Results
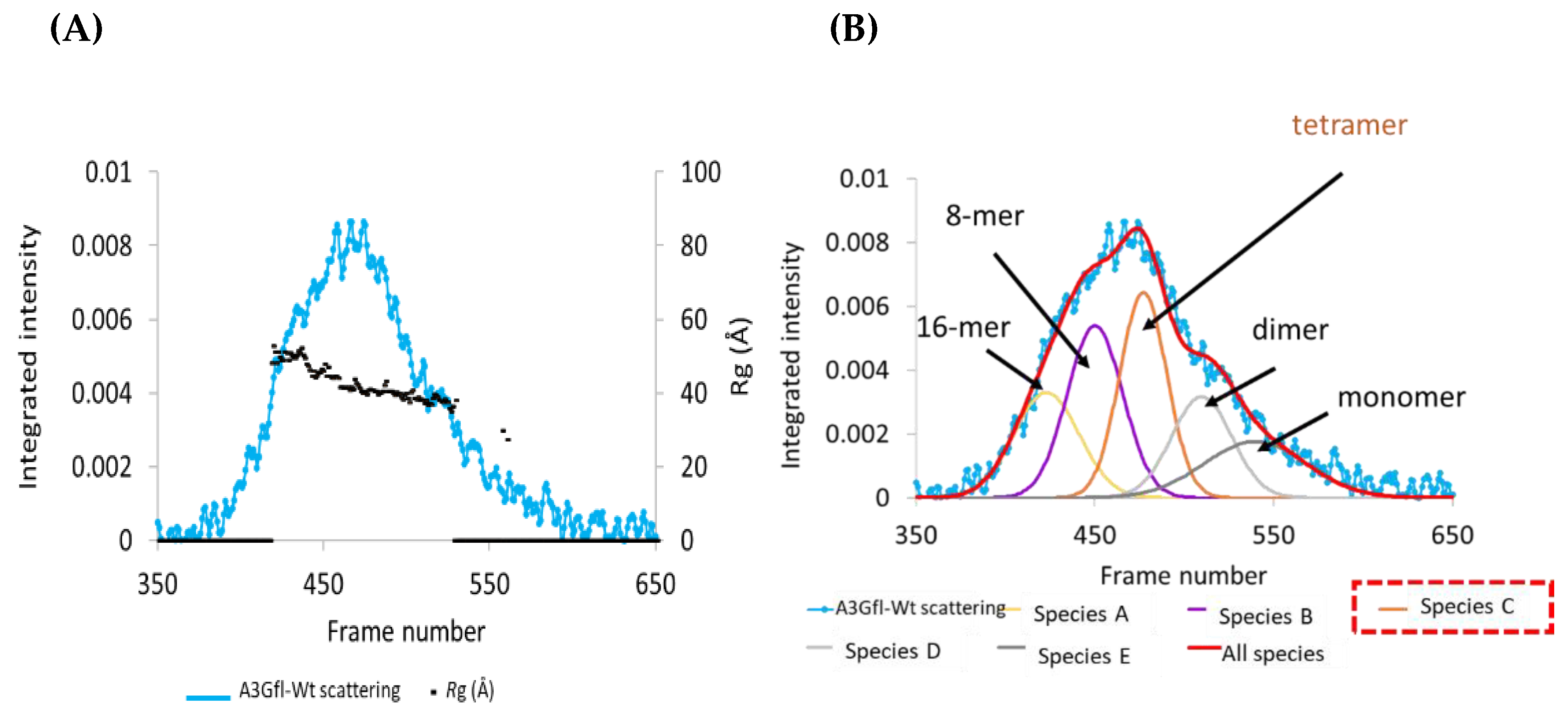
3.1. SEC-SAXS Analysis of Apo A3Gfl: Deconvolution of Scattering Data
3.2. SAXS Analysis of Apo A3Gfl: Analysis of the Scattering Curves
3.3. SAXS Model of A3Gfl in Complex with ssDNA
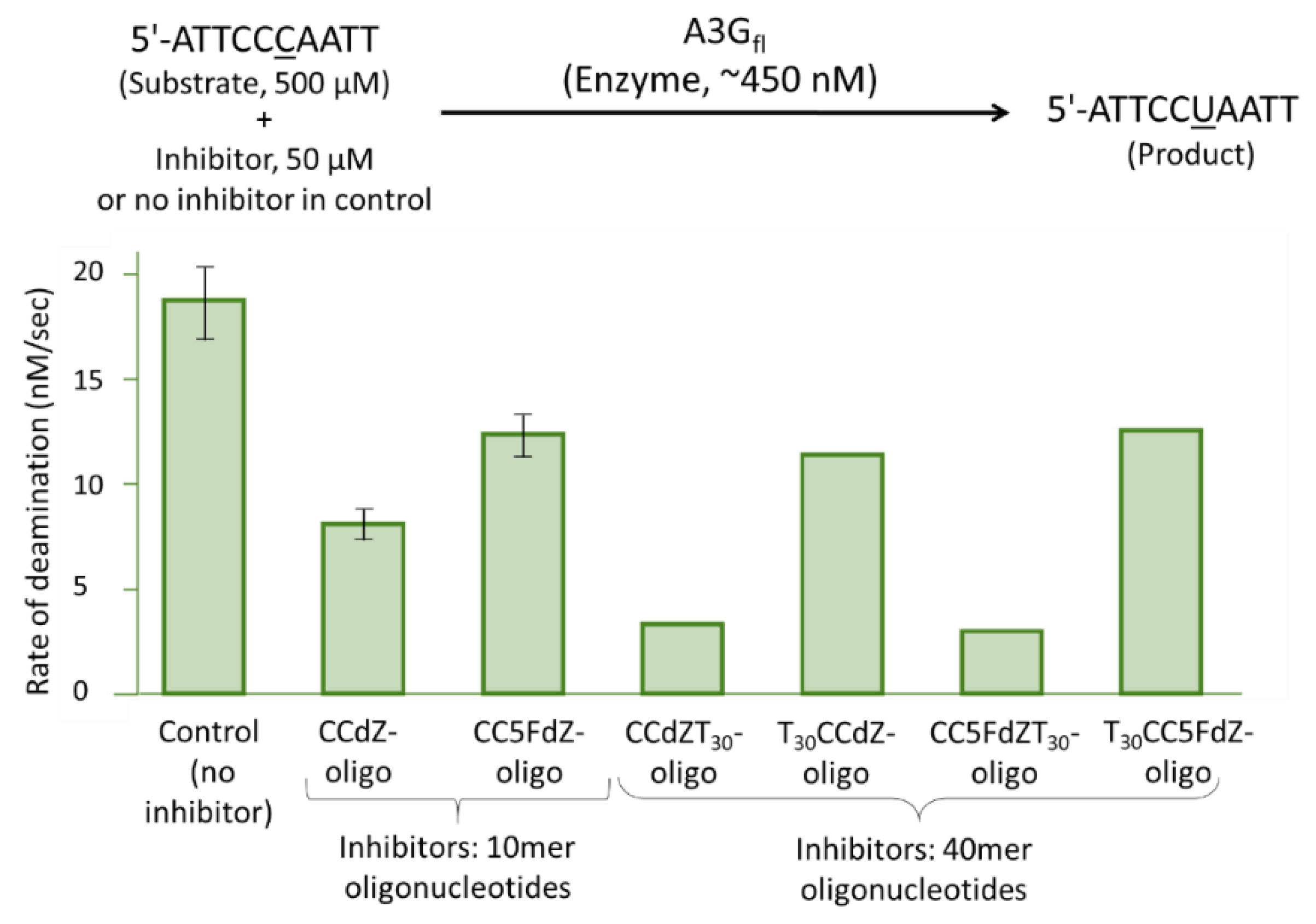
Selection of ssDNA for SAXS Studies
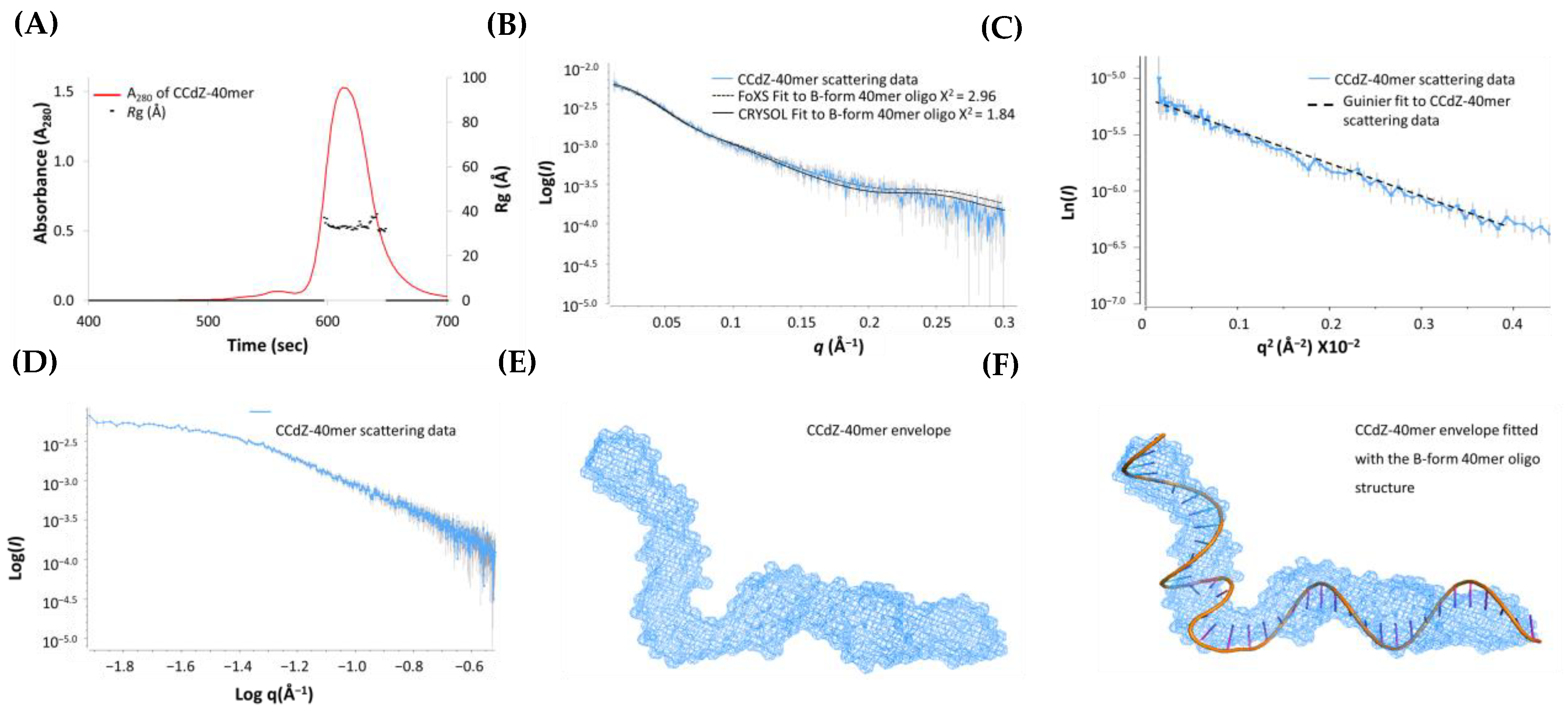
3.4. SAXS Studies of CCdZ-T30-Oligo
3.5. SAXS Model of A3Gfl in the Presence of dZ-Containing ssDNA
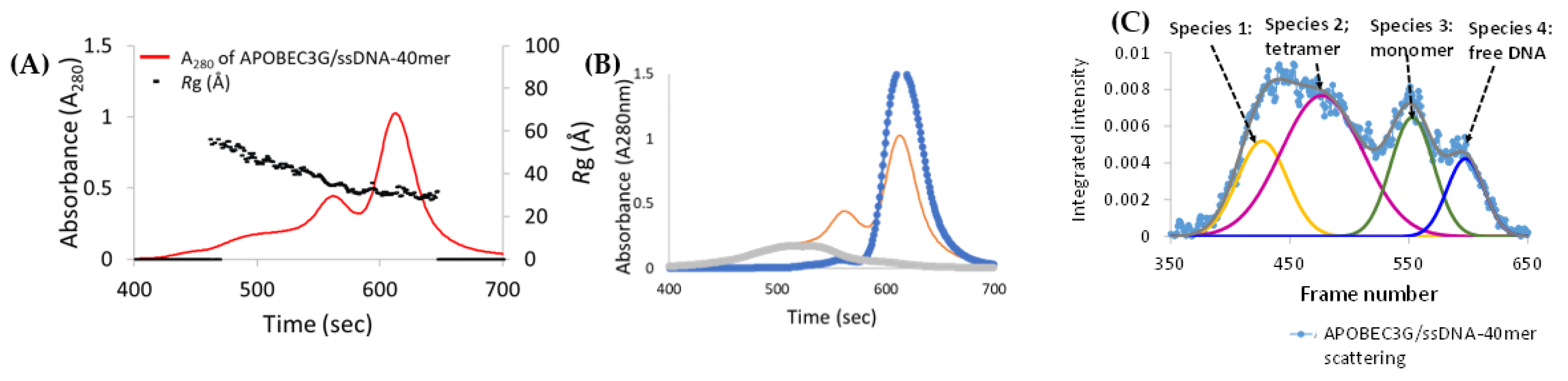
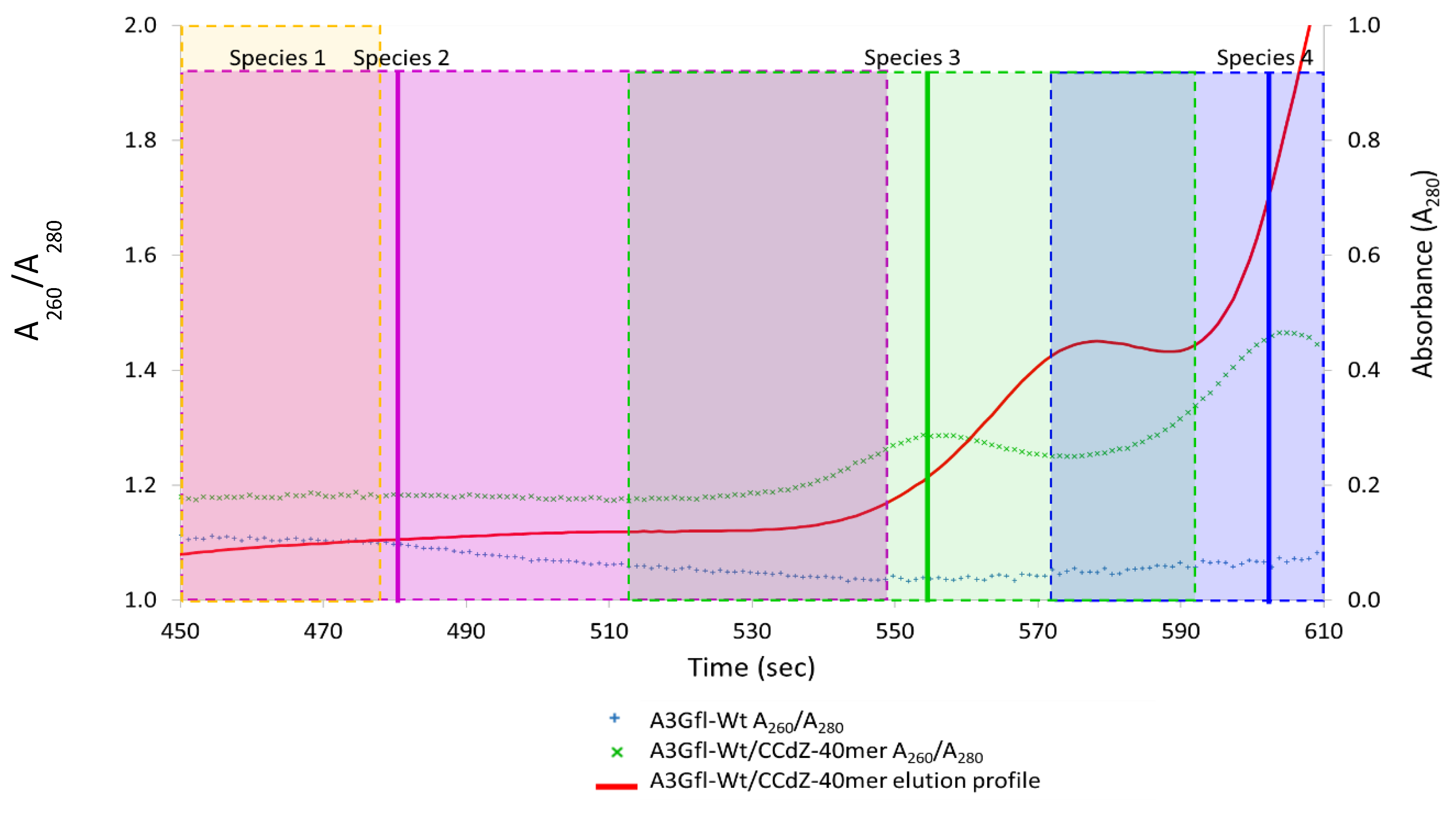
3.6. Analysis of Species-2 from A3Gfl/CCdZ-T30-Oligo SEC-SAXS
3.7. Model-Free Analysis of SEC-SAXS Data for Species-3 and -4 from A3Gfl/CCdZ-T30-Oligo
3.8. Modeling Species-3 of A3Gfl/CCdZ-T30-Oligo as a Monomer with DNA
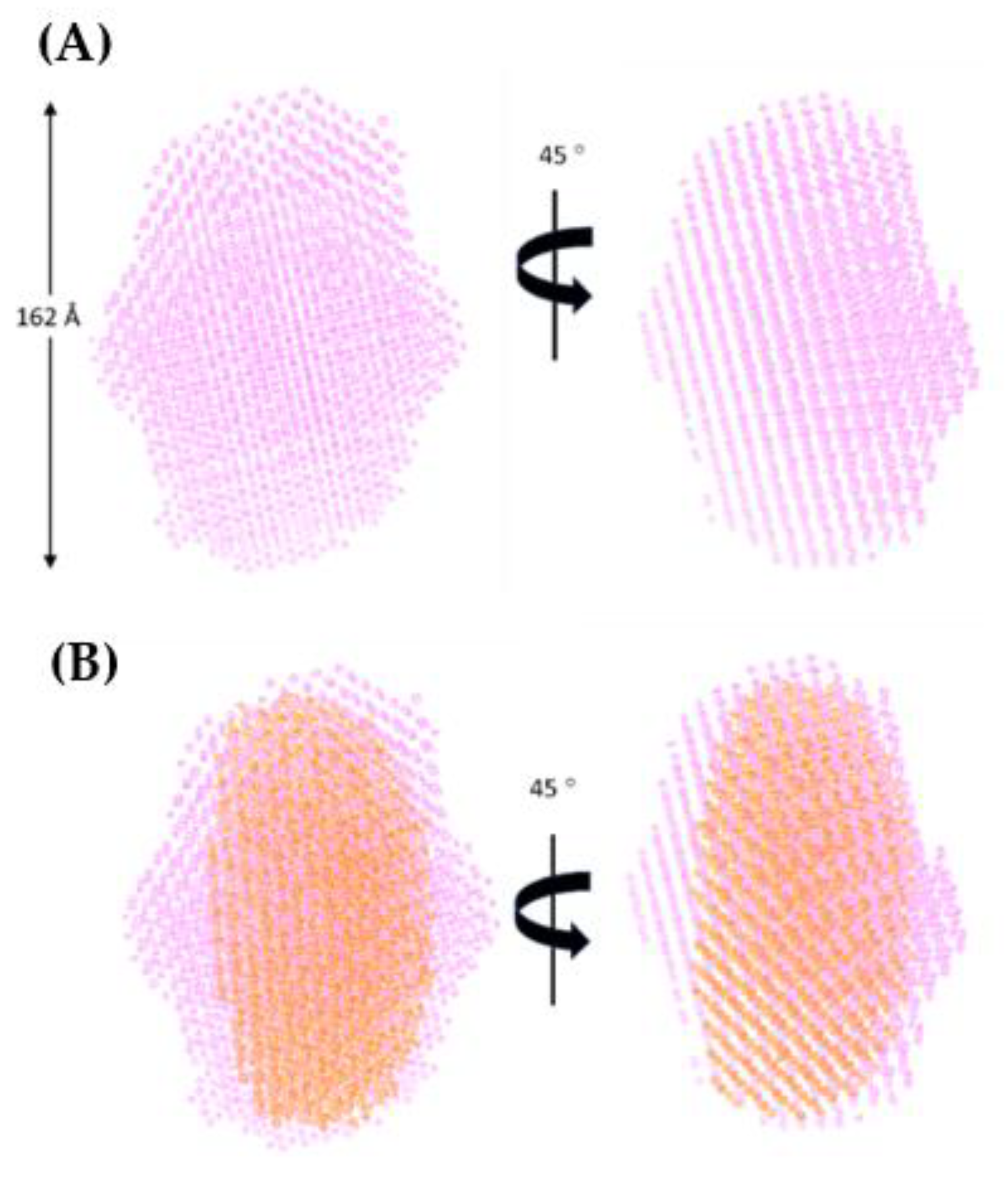
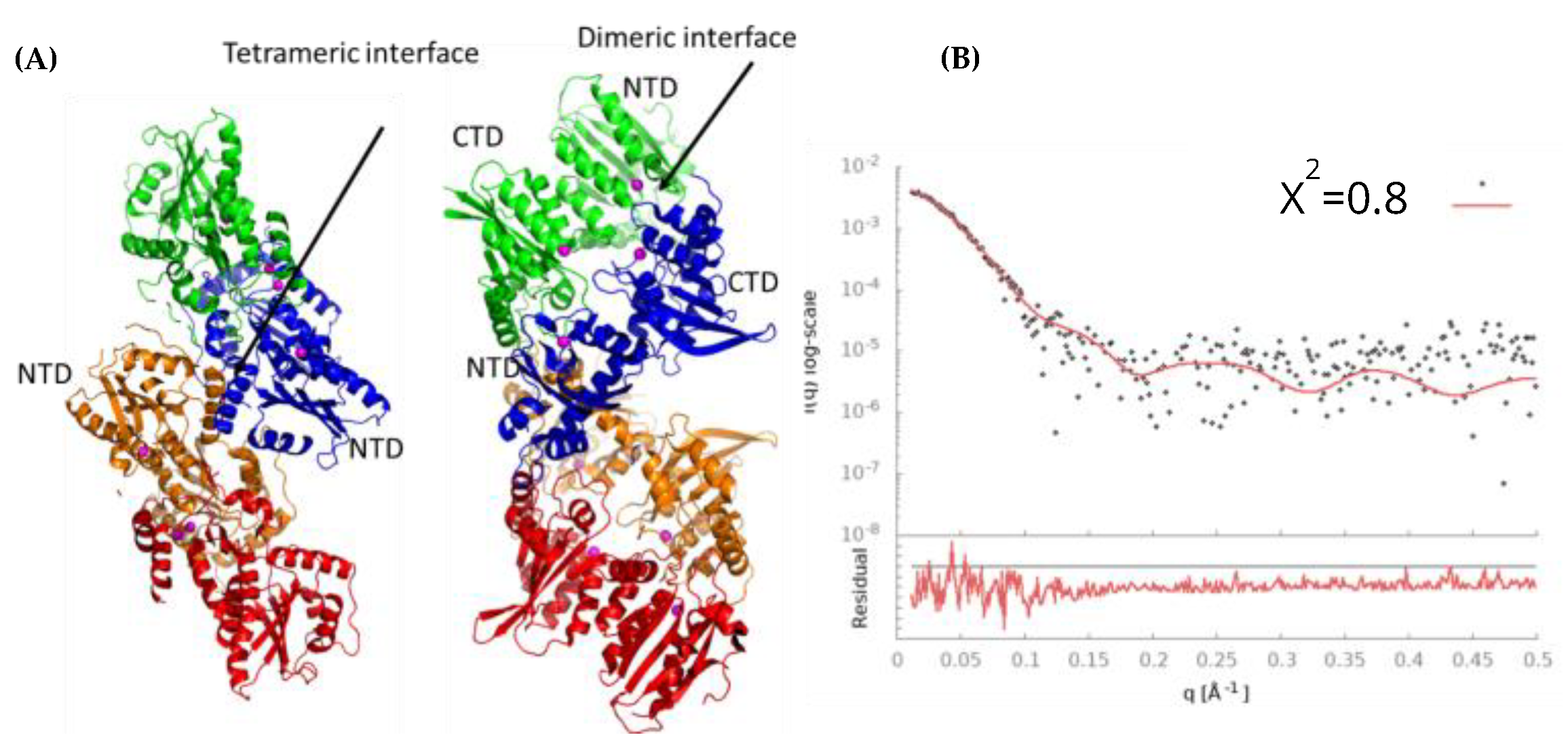
3.9. Model of Free A3Gfl Tetramer Based on Disappearance of Dimers and Preservation of Tetramers in the Presence of DNA
4. Discussion
4.1. Multimeric Associations of A3Gfl
4.2. Modeling the Interaction of CCdZ-T30-Oligo with Monomeric A3Gfl
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harris, R.S.; Bishop, K.N.; Sheehy, A.M.; Craig, H.M.; Petersen-Mahrt, S.K.; Watt, I.N.; Neuberger, M.S.; Malim, M.H. DNA Deamination Mediates Innate Immunity to Retroviral Infection. Cell 2003, 113, 803–809. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 6881. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Liddament, M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004, 4, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Izumi, T.; Shirakawa, K.; Takaori-Kondo, A. Cytidine deaminases as a weapon against retroviruses and a new target for antiviral therapy. Mini-Rev. Med. Chem. 2008, 8, 231–238. [Google Scholar]
- Mangeat, B.; Turelli, P.; Caron, G.; Friedli, M.; Perrin, L.; Trono, D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 2003, 424, 99–103. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Green, A.M.; Weitzman, M.D. The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities. DNA Repair (Amst.) 2019, 83, 102700. [Google Scholar] [CrossRef]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479–480, 131–145. [Google Scholar] [CrossRef]
- Sheehy, A.; Gaddis, N.; Malim, M. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003, 9, 1404–1407. [Google Scholar] [CrossRef]
- Stopak, K.; de Noronha, C.; Yonemoto, W.; Greene, W. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 2003, 12, 591–601. [Google Scholar] [CrossRef]
- Akari, H.; Fujita, M.; Kao, S.; Khan, M.; Shehu-Xhilaga, M.; Adachi, A. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 2004, 279, 12355–12362. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Ruiz, A.; Moris, A.; Prado, J.G. Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 2018, 9, 2876. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, S.; Rosenthal, R.; Kanu, N.; McGranahan, N.; Bartek, J.; Quezada, S.A.; Hare, J.; Harris, R.S.; Swanton, C. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 2018, 29, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, N.; Love, R.P.; Gibson, R.; Arts, E.J.; Poon, A.F.Y.; Chelico, L. Role of co-expressed APOBEC3F and APOBEC3G in inducing HIV-1 drug resistance. Heliyon 2019, 5, e01498. [Google Scholar] [CrossRef]
- Bruner, K.M.; Murray, A.J.; Pollack, R.A.; Soliman, M.G.; Laskey, S.B.; Capoferri, A.A.; Lai, J.; Strain, M.C.; Lada, S.M.; Hoh, R.; et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016, 22, 1043–1049. [Google Scholar] [CrossRef]
- Hernandez, M.M.; Fahrny, A.; Jayaprakash, A.; Gers-Huber, G.; Dillon-White, M.; Audige, A.; Mulder, L.C.F.; Sachidanandam, R.; Speck, R.F.; Simon, V. Impact of Suboptimal APOBEC3G Neutralization on the Emergence of HIV Drug Resistance in Humanized Mice. J. Virol. 2020, 94, e01543. [Google Scholar] [CrossRef]
- Iwatani, Y.; Chan, D.S.; Wang, F.; Maynard, K.S.; Sugiura, W.; Gronenborn, A.M.; Rouzina, I.; Williams, M.C.; Musier-Forsyth, K.; Levin, J.G. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007, 35, 7096–7108. [Google Scholar] [CrossRef]
- Morse, M.; Huo, R.; Feng, Y.; Rouzina, I.; Chelico, L.; Williams, M.C. Dimerization regulates both deaminase-dependent and deaminase-independent HIV-1 restriction by APOBEC3G. Nat. Commun. 2017, 8, 597. [Google Scholar] [CrossRef]
- Morse, M.; Naufer, M.N.; Feng, Y.; Chelico, L.; Rouzina, I.; Williams, M.C. HIV restriction factor APOBEC3G binds in multiple steps and conformations to search and deaminate single-stranded DNA. eLife 2019, 8, e52649. [Google Scholar] [CrossRef]
- Chen, X.S. Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions. Viruses 2021, 13, 497. [Google Scholar] [CrossRef]
- Pollpeter, D.; Parsons, M.; Sobala, A.E.; Coxhead, S.; Lang, R.D.; Bruns, A.M.; Papaioannou, S.; McDonnell, J.M.; Apolonia, L.; Chowdhury, J.A.; et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat. Microbiol. 2018, 3, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Chelico, L.; Pham, P.; Calabrese, P.; Goodman, M.F. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006, 13, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.; Bollman, B.; Chen, H.; Konig, R.; Yu, Q.; Chiles, K.; Landau, N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology 2005, 333, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chelico, L. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J. Biol. Chem. 2011, 286, 11415–11426. [Google Scholar] [CrossRef]
- Schröfelbauer, B.; Chen, D.; Landau, N.R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 2004, 101, 3927–3932. [Google Scholar] [CrossRef]
- Nowarski, R.; Britan-Rosich, E.; Shiloach, T.; Kotler, M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 2008, 15, 1059–1066. [Google Scholar] [CrossRef]
- Chelico, L.; Prochnow, C.; Erie, D.A.; Chen, X.S.; Goodman, M.F. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010, 285, 16195–16205. [Google Scholar] [CrossRef]
- Senavirathne, G.; Jaszczur, M.; Auerbach, P.A.; Upton, T.G.; Chelico, L.; Goodman, M.F.; Rueda, D. Single-stranded DNA Scanning and Deamination by APOBEC3G Cytidine Deaminase at Single Molecule Resolution. J. Biol. Chem. 2012, 287, 15826–15835. [Google Scholar] [CrossRef]
- Siriwardena, S.U.; Guruge, T.A.; Bhagwat, A.S. Characterization of the Catalytic Domain of Human APOBEC3B and the Critical Structural Role for a Conserved Methionine. J. Mol. Biol. 2015, 427, 3042–3055. [Google Scholar] [CrossRef]
- Fu, Y.; Ito, F.; Zhang, G.; Fernandez, B.; Yang, H.; Chen, X.S. DNA Cytosine and Methylcytosine Deamination by APOBEC3B: Enhancing Methylcytosine Deamination by Engineering APOBEC3B. Biochem. J. 2015, 471, 25–35. [Google Scholar] [CrossRef]
- Hakata, Y.; Landau, N.R. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J. Biol. Chem. 2006, 281, 36624–36631. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A.; Myint, W.; Delviks-Frankenberry, K.A.; Hou, S.; Kanai, T.; Balachandran, V.; Sierra Rodriguez, C.; Tripathi, R.; Kurt Yilmaz, N.; Pathak, V.K.; et al. Crystal Structure of a Soluble APOBEC3G Variant Suggests ssDNA to Bind in a Channel that Extends between the Two Domains. J. Mol. Biol. 2020, 432, 6042–6060. [Google Scholar] [CrossRef]
- Yang, H.; Ito, F.; Wolfe, A.D.; Li, S.; Mohammadzadeh, N.; Love, R.P.; Yan, M.; Zirkle, B.; Gaba, A.; Chelico, L.; et al. Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G. Nat. Commun. 2020, 11, 632. [Google Scholar] [CrossRef]
- Maiti, A.; Myint, W.; Kanai, T.; Delviks-Frankenberry, K.; Sierra Rodriguez, C.; Pathak, V.K.; Schiffer, C.A.; Matsuo, H. Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA. Nat. Commun. 2018, 9, 2460. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.M.; Harjes, E.; Gross, P.J.; Fahmy, A.; Lu, Y.; Shindo, K.; Harris, R.S.; Matsuo, H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 2008, 452, 116–119. [Google Scholar] [CrossRef]
- Harjes, E.; Gross, P.J.; Chen, K.M.; Lu, Y.; Shindo, K.; Nowarski, R.; Gross, J.D.; Kotler, M.; Harris, R.S.; Matsuo, H. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 2009, 389, 819–832. [Google Scholar] [CrossRef]
- Shandilya, S.M.; Nalam, M.N.; Nalivaika, E.A.; Gross, P.J.; Valesano, J.C.; Shindo, K.; Li, M.; Munson, M.; Royer, W.E.; Harjes, E.; et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 2010, 18, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, A.; Nagata, T.; Matsugami, A.; Habu, Y.; Sugiyama, R.; Hayashi, F.; Kobayashi, N.; Yokoyama, S.; Takaku, H.; Katahira, M. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009, 28, 440–451. [Google Scholar] [CrossRef]
- Kouno, T.; Luengas, E.M.; Shigematsu, M.; Shandilya, S.M.; Zhang, J.; Chen, L.; Hara, M.; Schiffer, C.A.; Harris, R.S.; Matsuo, H. Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat. Struct. Mol. Biol. 2015, 22, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Holden, L.G.; Prochnow, C.; Chang, Y.P.; Bransteitter, R.; Chelico, L.; Sen, U.; Stevens, R.C.; Goodman, M.F.; Chen, X. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 2008, 456, 121–124. [Google Scholar] [CrossRef]
- Chelico, L.; Sacho, E.J.; Erie, D.A.; Goodman, M.F. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. J. Biol. Chem. 2008, 283, 13780–13791. [Google Scholar] [CrossRef]
- McDougall, W.M.; Okany, C.; Smith, H.C. Deaminase Activity on Single-stranded DNA (ssDNA) Occurs in Vitro when APOBEC3G Cytidine Deaminase Forms Homotetramers and Higher-order Complexes. J. Biol. Chem. 2011, 286, 30655–30661. [Google Scholar] [CrossRef] [PubMed]
- Salter, J.D.; Krucinska, J.; Raina, J.; Smith, H.C.; Wedekind, J.E. A hydrodynamic analysis of APOBEC3G reveals a monomer-dimer-tetramer self-association that has implications for anti-HIV function. Biochemistry 2009, 48, 10685–10687. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirby, N.; Cowieson, N.; Hawley, A.M.; Mudie, S.T.; McGillivray, D.J.; Kusel, M.; Samardzic-Boban, V.; Ryan, T.M. Improved radiation dose efficiency in solution SAXS using a sheath flow sample environment. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72 Pt 12, 1254–1266. [Google Scholar] [CrossRef]
- Barzak, F.M.; Ryan, T.M.; Kvach, M.V.; Kurup, H.M.; Aihara, H.; Harris, R.S.; Filichev, V.V.; Harjes, E.; Jameson, G.B. Small-Angle X-ray Scattering Models of APOBEC3B Catalytic Domain in a Complex with a Single-Stranded DNA Inhibitor. Viruses 2021, 13, 290. [Google Scholar] [CrossRef] [PubMed]
- Kvach, M.V.; Barzak, F.M.; Harjes, S.; Schares, H.A.M.; Kurup, H.M.; Jones, K.F.; Sutton, L.; Donahue, J.; D’Aquila, R.T.; Jameson, G.B.; et al. Differential Inhibition of APOBEC3 DNA-Mutator Isozymes by Fluoro- and Non-Fluoro-Substituted 2′-Deoxyzebularine Embedded in Single-Stranded DNA. Chembiochem 2020, 21, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Durocher, Y.; Perret, S.; Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002, 30, E9. [Google Scholar] [CrossRef]
- Kvach, M.V.; Barzak, F.M.; Harjes, S.; Schares, H.A.M.; Jameson, G.B.; Ayoub, A.M.; Moorthy, R.; Aihara, H.; Harris, R.S.; Filichev, V.V.; et al. Inhibiting APOBEC3 Activity with Single-Stranded DNA Containing 2′-Deoxyzebularine Analogues. Biochemistry 2019, 58, 391–400. [Google Scholar] [CrossRef]
- Hopkins, J.B.; Gillilan, R.E.; Skou, S. BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 2017, 50 Pt 5, 1545–1553. [Google Scholar] [CrossRef]
- Brookes, E.; Pérez, J.; Cardinali, B.; Profumo, A.; Vachette, P.; Rocco, M. Fibrinogen species as resolved by HPLC-SAXS data processing within the UltraScan Solution Modeler (US-SOMO) enhanced SAS module. J. Appl. Crystallogr. 2013, 46, 1823–1833. [Google Scholar] [CrossRef]
- Brookes, E.; Vachette, P.; Rocco, M.; Pérez, J. US-SOMO HPLC-SAXS module: Dealing with capillary fouling and extraction of pure component patterns from poorly resolved SEC-SAXS data. J. Appl. Crystallogr. 2016, 49, 1827–1841. [Google Scholar] [PubMed]
- Brookes, E.; Rocco, M. Recent advances in the UltraScan SOlution MOdeller (US-SOMO) hydrodynamic and small-angle scattering data analysis and simulation suite. Eur. Biophys. J. 2018, 47, 855–864. [Google Scholar] [PubMed]
- Svergun, D.I.; Koch, M.H.J. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003, 66, 1735–1782. [Google Scholar]
- Burger, V.M.; Arenas, D.J.; Stultz, C.M. A Structure-free Method for Quantifying Conformational Flexibility in proteins. Sci. Rep. 2016, 6, 29040. [Google Scholar]
- Wedekind, J.E.; Gillilan, R.; Janda, A.; Krucinska, J.; Salter, J.D.; Bennett, R.P.; Raina, J.; Smith, H.C. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J. Biol. Chem. 2006, 281, 38122–38126. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, E.; Svergun, D.I. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J. Appl. Crystallogr. 2007, 40, s245–s249. [Google Scholar] [CrossRef]
- Harjes, S.; Solomon, W.C.; Li, M.; Chen, K.M.; Harjes, E.; Harris, R.S.; Matsuo, H. Impact of H216 on the DNA binding and catalytic activities of the HIV restriction factor APOBEC3G. J. Virol. 2013, 87, 7008–7014. [Google Scholar] [CrossRef]
- Barzak, F.M.; Harjes, S.; Kvach, M.V.; Kurup, H.M.; Jameson, G.B.; Filichev, V.V.; Harjes, E. Selective inhibition of APOBEC3 enzymes by single-stranded DNAs containing 2′-deoxyzebularine. Org. Biomol. Chem. 2019, 17, 9435–9441. [Google Scholar] [CrossRef]
- Barzak, F.M.Y.; Filichev, V.V.; Harjes, E.; Harjes, S.; Jameson, G.B.; Kurup, H.M.; Kvach, M.V.; Su, Y. Single Stranded DNA Enzyme Inhibitors. WO2022162536, 4 August 2022. [Google Scholar]
- Maiti, A.; Hedger, A.K.; Myint, W.; Balachandran, V.; Watts, J.K.; Schiffer, C.A.; Matsuo, H. Structure of the catalytically active APOBEC3G bound to a DNA oligonucleotide inhibitor reveals tetrahedral geometry of the transition state. bioRxiv 2022. [Google Scholar] [CrossRef]
- Roth, E.; Glick Azaria, A.; Girshevitz, O.; Bitler, A.; Garini, Y. Measuring the Conformation and Persistence Length of Single-Stranded DNA Using a DNA Origami Structure. Nano Lett. 2018, 18, 6703–6709. [Google Scholar] [CrossRef]
- Rayment, I. Diffraction and Scattering by X-Rays and Neutrons. In Molecular Biophysics for the Life Sciences; Allewell, N., Narhi, L.O., Rayment, I., Eds.; Springer: New York, NY, USA, 2013; pp. 91–112. [Google Scholar]
- Kozin, M.B.; Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001, 34, 33–41. [Google Scholar] [CrossRef]
- Bohn, M.F.; Shandilya, S.M.; Albin, J.S.; Kouno, T.; Anderson, B.D.; McDougle, R.M.; Carpenter, M.A.; Rathore, A.; Evans, L.; Davis, A.N.; et al. Crystal Structure of the DNA Cytosine Deaminase APOBEC3F: The Catalytically Active and HIV-1 Vif-Binding Domain. Structure 2013, 21, 1042–1050. [Google Scholar]
- Li, J.; Chen, Y.; Li, M.; Carpenter, M.A.; McDougle, R.M.; Luengas, E.M.; Macdonald, P.J.; Harris, R.S.; Mueller, J.D. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J. Mol. Biol. 2014, 426, 1296–1307. [Google Scholar]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar]
- Ara, A.; Love, R.P.; Chelico, L. Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms. PLoS Pathog. 2014, 10, e1004024. [Google Scholar]
- Polevoda, B.; McDougall, W.M.; Tun, B.N.; Cheung, M.; Salter, J.D.; Friedman, A.E.; Smith, H.C. RNA binding to APOBEC3G induces the disassembly of functional deaminase complexes by displacing single-stranded DNA substrates. Nucleic Acids Res. 2015, 43, 9434–9445. [Google Scholar]
- Salter, J.D.; Polevoda, B.; Bennett, R.P.; Smith, H.C. Regulation of Antiviral Innate Immunity Through APOBEC Ribonucleoprotein Complexes. Subcell. Biochem. 2019, 93, 193–219. [Google Scholar]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50 Pt 4, 1212–1225. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barzak, F.M.; Ryan, T.M.; Mohammadzadeh, N.; Harjes, S.; Kvach, M.V.; Kurup, H.M.; Krause, K.L.; Chelico, L.; Filichev, V.V.; Harjes, E.; et al. Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity. Viruses 2022, 14, 1974. https://doi.org/10.3390/v14091974
Barzak FM, Ryan TM, Mohammadzadeh N, Harjes S, Kvach MV, Kurup HM, Krause KL, Chelico L, Filichev VV, Harjes E, et al. Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity. Viruses. 2022; 14(9):1974. https://doi.org/10.3390/v14091974
Chicago/Turabian StyleBarzak, Fareeda M., Timothy M. Ryan, Nazanin Mohammadzadeh, Stefan Harjes, Maksim V. Kvach, Harikrishnan M. Kurup, Kurt L. Krause, Linda Chelico, Vyacheslav V. Filichev, Elena Harjes, and et al. 2022. "Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity" Viruses 14, no. 9: 1974. https://doi.org/10.3390/v14091974
APA StyleBarzak, F. M., Ryan, T. M., Mohammadzadeh, N., Harjes, S., Kvach, M. V., Kurup, H. M., Krause, K. L., Chelico, L., Filichev, V. V., Harjes, E., & Jameson, G. B. (2022). Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity. Viruses, 14(9), 1974. https://doi.org/10.3390/v14091974








