The First Report and Phylogenetic Analysis of Canine Distemper Virus in Cerdocyon thous from Colombia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
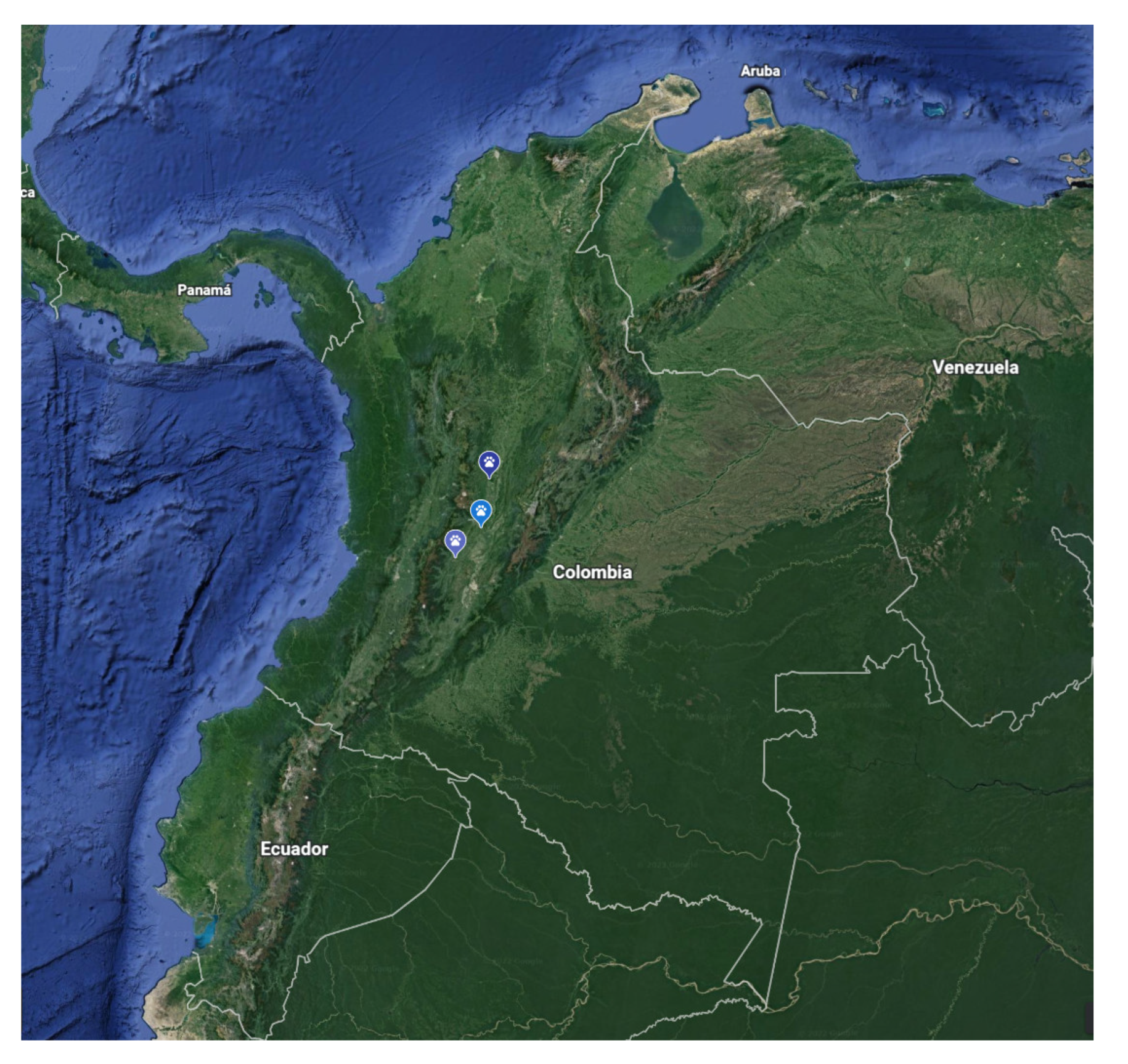
2.2. Sample Collection
2.3. Serological Rapid Test for CDV
2.4. RNA Extraction and cDNA Synthesis
2.5. PCR and Sequencing
2.6. Phylogenetic Analysis
3. Results
3.1. Animals
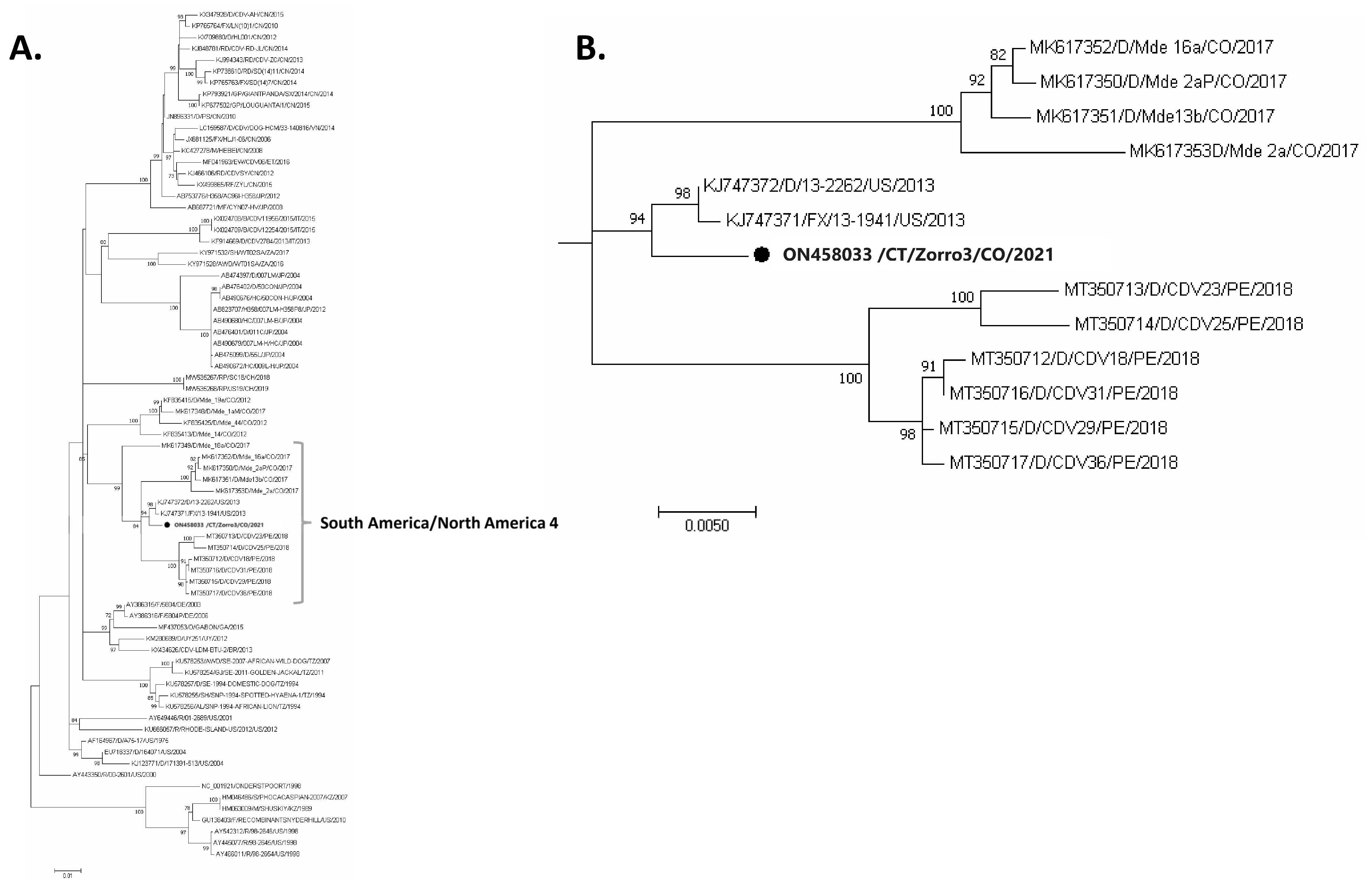
3.2. Molecular Confirmation and Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martinez-Gutierrez, M.; Ruiz-Saenz, J. Diversity of susceptible hosts in canine distemper virus infection: A systematic review and data synthesis. BMC Vet. Res. 2016, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Rendon-Marin, S.; da Fontoura Budaszewski, R.; Canal, C.W.; Ruiz-Saenz, J. Tropism and molecular pathogenesis of canine distemper virus. Virol. J. 2019, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Parks, G.D. Paramyxoviridae. In Fields Virology, 6th ed.; Wolters Kluwer Health: Philadelphia, PA, USA, 2013. [Google Scholar]
- MacLachlan, N.; Dubovi, E.; Fenner, F. Paramyxoviridae. In Fenner’s Veterinary Virology; Academic Press: Cambridge, MA, USA, 2011; pp. 299–325. [Google Scholar]
- Anis, E.; Newell, T.K.; Dyer, N.; Wilkes, R.P. Phylogenetic analysis of the wild-type strains of canine distemper virus circulating in the United States. Virol. J. 2018, 15, 118. [Google Scholar] [CrossRef]
- Bhatt, M.; Rajak, K.; Chakravarti, S.; Yadav, A.; Kumar, A.; Gupta, V.; Chander, V.; Mathesh, K.; Chandramohan, S.; Sharma, A. Phylogenetic analysis of haemagglutinin gene deciphering a new genetically distinct lineage of canine distemper virus circulating among domestic dogs in India. Transbound. Emerg. Dis. 2019, 66, 1252–1267. [Google Scholar] [CrossRef]
- Duque-Valencia, J.; Diaz, F.J.; Ruiz-Saenz, J. Phylogenomic Analysis of Two Co-Circulating Canine Distemper Virus Lineages in Colombia. Pathogens 2019, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Duque-Valencia, J.; Forero-Munoz, N.R.; Diaz, F.J.; Martins, E.; Barato, P.; Ruiz-Saenz, J. Phylogenetic evidence of the intercontinental circulation of a Canine distemper virus lineage in the Americas. Sci. Rep. 2019, 9, 15747. [Google Scholar] [CrossRef] [PubMed]
- Espinal, M.A.; Diaz, F.J.; Ruiz-Saenz, J. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet. Microbiol. 2014, 172, 168–176. [Google Scholar] [CrossRef]
- Giacinti, J.A.; Pearl, D.L.; Ojkic, D.; Campbell, G.D.; Jardine, C.M. Genetic characterization of canine distemper virus from wild and domestic animal submissions to diagnostic facilities in Canada. Prev. Vet. Med. 2022, 198, 105535. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Zhai, J.; Zhang, P.; Irwin, D.M.; Shen, X.; Chen, W.; Shen, Y. A new canine distemper virus lineage identified from red pandas in China. Transbound. Emerg. Dis. 2022, 69, e944–e952. [Google Scholar] [CrossRef]
- Rendon-Marin, S.; Martinez-Gutierrez, M.; Suarez, J.A.; Ruiz-Saenz, J. Canine Distemper Virus (CDV) Transit Through the Americas: Need to Assess the Impact of CDV Infection on Species Conservation. Front. Microbiol. 2020, 11, 810. [Google Scholar] [CrossRef]
- Takeda, M.; Seki, F.; Yamamoto, Y.; Nao, N.; Tokiwa, H. Animal morbilliviruses and their cross-species transmission potential. Curr. Opin. Virol. 2020, 41, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Duque-Valencia, J.; Sarute, N.; Olarte-Castillo, X.A.; Ruiz-Saenz, J. Evolution and Interspecies Transmission of Canine Distemper Virus-An Outlook of the Diverse Evolutionary Landscapes of a Multi-Host Virus. Viruses 2019, 11, 582. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.; Cleaveland, S.; Matthiopoulos, J.; Halliday, J.; Packer, C.; Craft, M.E.; Hampson, K.; Czupryna, A.; Dobson, A.P.; Dubovi, E.J. Dynamics of a morbillivirus at the domestic–wildlife interface: Canine distemper virus in domestic dogs and lions. Proc. Natl. Acad. Sci. USA 2015, 112, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Visser, I.; Mamaev, L.; Goatley, L.; Van Bressem, M.-F.; Osterhaus, A. Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology 1993, 193, 1010–1012. [Google Scholar] [CrossRef]
- Lan, N.T.; Yamaguchi, R.; Inomata, A.; Furuya, Y.; Uchida, K.; Sugano, S.; Tateyama, S. Comparative analyses of canine distemper viral isolates from clinical cases of canine distemper in vaccinated dogs. Vet. Microbiol. 2006, 115, 32–42. [Google Scholar] [CrossRef]
- Riley, M.C.; Wilkes, R.P. Sequencing of emerging canine distemper virus strain reveals new distinct genetic lineage in the United States associated with disease in wildlife and domestic canine populations. Virol. J. 2015, 12, 219. [Google Scholar] [CrossRef]
- Sarute, N.; Pérez, R.; Aldaz, J.; Alfieri, A.A.; Alfieri, A.F.; Name, D.; Llanes, J.; Hernández, M.; Francia, L.; Panzera, Y. Molecular typing of canine distemper virus strains reveals the presence of a new genetic variant in South America. Virus Genes 2014, 48, 474–478. [Google Scholar] [CrossRef]
- Loots, A.K.; Mitchell, E.; Dalton, D.L.; Kotze, A.; Venter, E.H. Advances in canine distemper virus pathogenesis research: A wildlife perspective. J. Gen. Virol. 2017, 98, 311–321. [Google Scholar] [CrossRef]
- Deem, S.L.; Spelman, L.H.; Yates, R.A.; Montali, R.J. Canine distemper in terrestrial carnivores: A review. J. Zoo Wildl. Med. 2000, 31, 441–451. [Google Scholar] [CrossRef]
- Nelson, R.W.; Couto, C.G. Small Animal Internal Medicine-E-Book; Elsevier Health Sciences: St. Louis, MO, USA, 2019. [Google Scholar]
- Evermann, J.F.; Kennedy, M.A. Chapter 16—Viral Infections. In Small Animal Pediatrics; Peterson, M.E., Kutzler, M.A., Eds.; W.B. Saunders: Saint Louis, MO, USA, 2011; pp. 119–129. [Google Scholar] [CrossRef]
- Beineke, A.; Puff, C.; Seehusen, F.; Baumgärtner, W. Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet. Immunol. Immunopathol. 2009, 127, 1–18. [Google Scholar] [CrossRef]
- Fiorello, C.V.; Noss, A.J.; Deem, S.L.; Maffei, L.; Dubovi, E.J. Serosurvey of small carnivores in the Bolivian Chaco. J. Wildl. Dis. 2007, 43, 551–557. [Google Scholar] [CrossRef]
- Anis, E.; Holford, A.L.; Galyon, G.D.; Wilkes, R.P. Antigenic analysis of genetic variants of Canine distemper virus. Vet. Microbiol. 2018, 219, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Oviedo Centeno, Y.N. Diferenciación de Cepas de Campo y Vacunales del Virus del Distemper Canino en Perros Infectados Naturalmente; Universidad Nacional Mayor de San Marcos Lima: Lima, Peru, 2021. [Google Scholar]
- Fuques, E.; Tomás, G.; Grecco, S.; Condon, E.; Techera, C.; Marandino, A.; Sarute, N.; Aldaz, J.; Enciso, J.; Benech, A.; et al. Origin and spreading of canine morbillivirus in South America. Virus Res. 2022, 319, 198858. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.J.; Shaw, M.-A.; Goodman, S.J. Pathogen evolution and disease emergence in carnivores. Proc. R. Soc. B Biol. Sci. 2007, 274, 3165–3174. [Google Scholar] [CrossRef] [PubMed]
- Nikolin, V.M.; Wibbelt, G.; Michler, F.-U.F.; Wolf, P.; East, M.L. Susceptibility of carnivore hosts to strains of canine distemper virus from distinct genetic lineages. Vet. Microbiol. 2012, 156, 45–53. [Google Scholar] [CrossRef]
- Messling, V.v.; Springfeld, C.; Devaux, P.; Cattaneo, R. A Ferret Model of Canine Distemper Virus Virulence and Immunosuppression. J. Virol. 2003, 77, 12579–12591. [Google Scholar] [CrossRef]
- Nikolin, V.M.; Osterrieder, K.; von Messling, V.; Hofer, H.; Anderson, D.; Dubovi, E.; Brunner, E.; East, M.L. Antagonistic Pleiotropy and Fitness Trade-Offs Reveal Specialist and Generalist Traits in Strains of Canine Distemper Virus. PLoS ONE 2012, 7, e50955. [Google Scholar] [CrossRef]
- Weckworth, J.K.; Davis, B.W.; Roelke-Parker, M.E.; Wilkes, R.P.; Packer, C.; Eblate, E.; Schwartz, M.K.; Mills, L.S. Identifying Candidate Genetic Markers of CDV Cross-Species Pathogenicity in African Lions. Pathogens 2020, 9, 872. [Google Scholar] [CrossRef]
- Sillero-Zubiri, C.; Hoffmann, M.; Macdonald, D.W. Canids: Foxes, Wolves, Jackals, and Dogs: Status Survey and Conservation Action Plan; IUCN: Gland, Switzerland, 2004; Volume 95.
- Guerrero, Y.M.; Cadena, A. Caracterización, evaluación y uso de hábitats del zorro perruno (Cerdocyon thous) en los llanos orientales de Colombia. Rev. Acad. Colomb. Cienc. Exactas Físicas Nat. 2000, 24, 383–392. [Google Scholar]
- Muñoz Mazo, S.S. Agentes Infecciosos en el Zorro Cangrejero (Cerdocyon Thous) en las Áreas Protegidas Urbanas del Valle de Aburrá, Colombia; Universidad Nacional de Costa Rica: Heredia, Costa Rica, 2021. [Google Scholar]
- Costanzi, L.; Brambilla, A.; Di Blasio, A.; Dondo, A.; Goria, M.; Masoero, L.; Gennero, M.S.; Bassano, B. Beware of dogs! Domestic animals as a threat for wildlife conservation in Alpine protected areas. Eur. J. Wildl. Res. 2021, 67, 70. [Google Scholar] [CrossRef]
- Kapil, S.; Yeary, T.J. Canine Distemper Spillover in Domestic Dogs from Urban Wildlife. Vet. Clin. N. Am. Small Anim. Pract. 2011, 41, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- Pope, J.P.; Miller, D.L.; Riley, M.C.; Anis, E.; Wilkes, R.P. Characterization of a novel Canine distemper virus causing disease in wildlife. J. Vet. Diagn. Investig. 2016, 28, 506–513. [Google Scholar] [CrossRef]
- Bohm, J.; Blixenkrone-Møller, M.; Lund, E. A serious outbreak of canine distemper among sled-dogs in northern Greenland. Arct. Med. Res. 1989, 48, 195–203. [Google Scholar]
- Frölich, K.; Czupalla, O.; Haas, L.; Hentschke, J.; Dedek, J.; Fickel, J. Epizootiological investigations of canine distemper virus in free-ranging carnivores from Germany. Vet. Microbiol. 2000, 74, 283–292. [Google Scholar] [CrossRef]
- Ricci, I.; Cersini, A.; Manna, G.; Marcario, G.A.; Conti, R.; Brocherel, G.; Grifoni, G.; Eleni, C.; Scicluna, M.T. A Canine Distemper Virus Retrospective Study Conducted from 2011 to 2019 in Central Italy (Latium and Tuscany Regions). Viruses 2021, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Zecchin, B.; Fusaro, A.; Schivo, A.; Ormelli, S.; Bregoli, M.; Citterio, C.V.; Obber, F.; Dellamaria, D.; Trevisiol, K.; et al. Two waves of canine distemper virus showing different spatio-temporal dynamics in Alpine wildlife (2006–2018). Infect. Genet. Evol. 2020, 84, 104359. [Google Scholar] [CrossRef]
- Furtado, M.M.; Hayashi, E.M.K.; Allendorf, S.D.; Coelho, C.J.; de Almeida Jácomo, A.T.; Megid, J.; Ramos Filho, J.D.; Silveira, L.; Tôrres, N.M.; Ferreira Neto, J.S. Exposure of Free-Ranging Wild Carnivores and Domestic Dogs to Canine Distemper Virus and Parvovirus in the Cerrado of Central Brazil. EcoHealth 2016, 13, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Hübner, S.d.O.; Pappen, F.G.; Ruas, J.L.; Vargas, G.D.Á.; Fischer, G.; Vidor, T. Exposure of pampas fox (Pseudalopex gymnocercus) and crab-eating fox (Cerdocyon thous) from the Southern region of Brazil to Canine distemper virus (CDV), Canine parvovirus (CPV) and Canine coronavirus (CCoV). Braz. Arch. Biol. Technol. 2010, 53, 593–597. [Google Scholar] [CrossRef]
- Megid, J.; de Souza, V.A.F.; Teixeira, C.R.; Cortez, A.; Amorin, R.L.; Heinemman, M.B.; Cagnini, D.Q.; Richtzenhain, L.J. Canine Distemper Virus in a Crab-eating Fox (Cerdocyon thous) in Brazil: Case Report and Phylogenetic Analyses. J. Wildl. Dis. 2009, 45, 527–530. [Google Scholar] [CrossRef]
- Ferreyra, H.; Calderón, M.G.; Marticorena, D.n.; Marull, C.; Leonardo, B.C. Canine Distemper Infection in Crab-eating Fox (Cerdocyon thous) from Argentina. J. Wildl. Dis. 2009, 45, 1158–1162. [Google Scholar] [CrossRef]
- Weber, M.N.; Mosena, A.C.S.; da Silva, M.S.; Canova, R.; de Lorenzo, C.; Olegário, J.C.; Budaszewski, R.F.; Baumbach, L.F.; Soares, J.F.; Sonne, L.; et al. Virome of crab-eating (Cerdocyon thous) and pampas foxes (Lycalopex gymnocercus) from southern Brazil and Uruguay. Infect. Genet. Evol. 2020, 85, 104421. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.; Miquelle, D.G.; Goodrich, J.M.; Reeve, R.; Cleaveland, S.; Matthews, L.; Joly, D.O. Estimating the Potential Impact of Canine Distemper Virus on the Amur Tiger Population (Panthera tigris altaica) in Russia. PLoS ONE 2014, 9, e110811. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.; Sulikhan, N.; Uphyrkina, O.; Goncharuk, M.; Kerley, L.; Castro, E.H.; Reeve, R.; Seimon, T.; McAloose, D.; Seryodkin, I.V.; et al. Distemper, extinction, and vaccination of the Amur tiger. Proc. Natl. Acad. Sci. USA 2020, 117, 31954–31962. [Google Scholar] [CrossRef] [PubMed]
- Mulia, B.H.; Mariya, S.; Bodgener, J.; Iskandriati, D.; Liwa, S.R.; Sumampau, T.; Manansang, J.; Darusman, H.S.; Osofsky, S.A.; Techakriengkrai, N.; et al. Exposure of Wild Sumatran Tiger (Panthera tigris sumatrae) to Canine Distemper Virus. J. Wildl. Dis. 2021, 57, 464–466. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Echeverry-Bonilla, D.F.; Buriticá-Gaviria, E.F.; Orjuela-Acosta, D.; Chinchilla-Cardenas, D.J.; Ruiz-Saenz, J. The First Report and Phylogenetic Analysis of Canine Distemper Virus in Cerdocyon thous from Colombia. Viruses 2022, 14, 1947. https://doi.org/10.3390/v14091947
Echeverry-Bonilla DF, Buriticá-Gaviria EF, Orjuela-Acosta D, Chinchilla-Cardenas DJ, Ruiz-Saenz J. The First Report and Phylogenetic Analysis of Canine Distemper Virus in Cerdocyon thous from Colombia. Viruses. 2022; 14(9):1947. https://doi.org/10.3390/v14091947
Chicago/Turabian StyleEcheverry-Bonilla, Diego Fernando, Edwin Fernando Buriticá-Gaviria, Delio Orjuela-Acosta, Danny Jaír Chinchilla-Cardenas, and Julian Ruiz-Saenz. 2022. "The First Report and Phylogenetic Analysis of Canine Distemper Virus in Cerdocyon thous from Colombia" Viruses 14, no. 9: 1947. https://doi.org/10.3390/v14091947
APA StyleEcheverry-Bonilla, D. F., Buriticá-Gaviria, E. F., Orjuela-Acosta, D., Chinchilla-Cardenas, D. J., & Ruiz-Saenz, J. (2022). The First Report and Phylogenetic Analysis of Canine Distemper Virus in Cerdocyon thous from Colombia. Viruses, 14(9), 1947. https://doi.org/10.3390/v14091947






