Detection of Cetacean Poxvirus in Peruvian Common Bottlenose Dolphins (Tursiops truncatus) Using a Pan-Poxvirus PCR
Abstract
1. Introduction
2. Materials and Methods
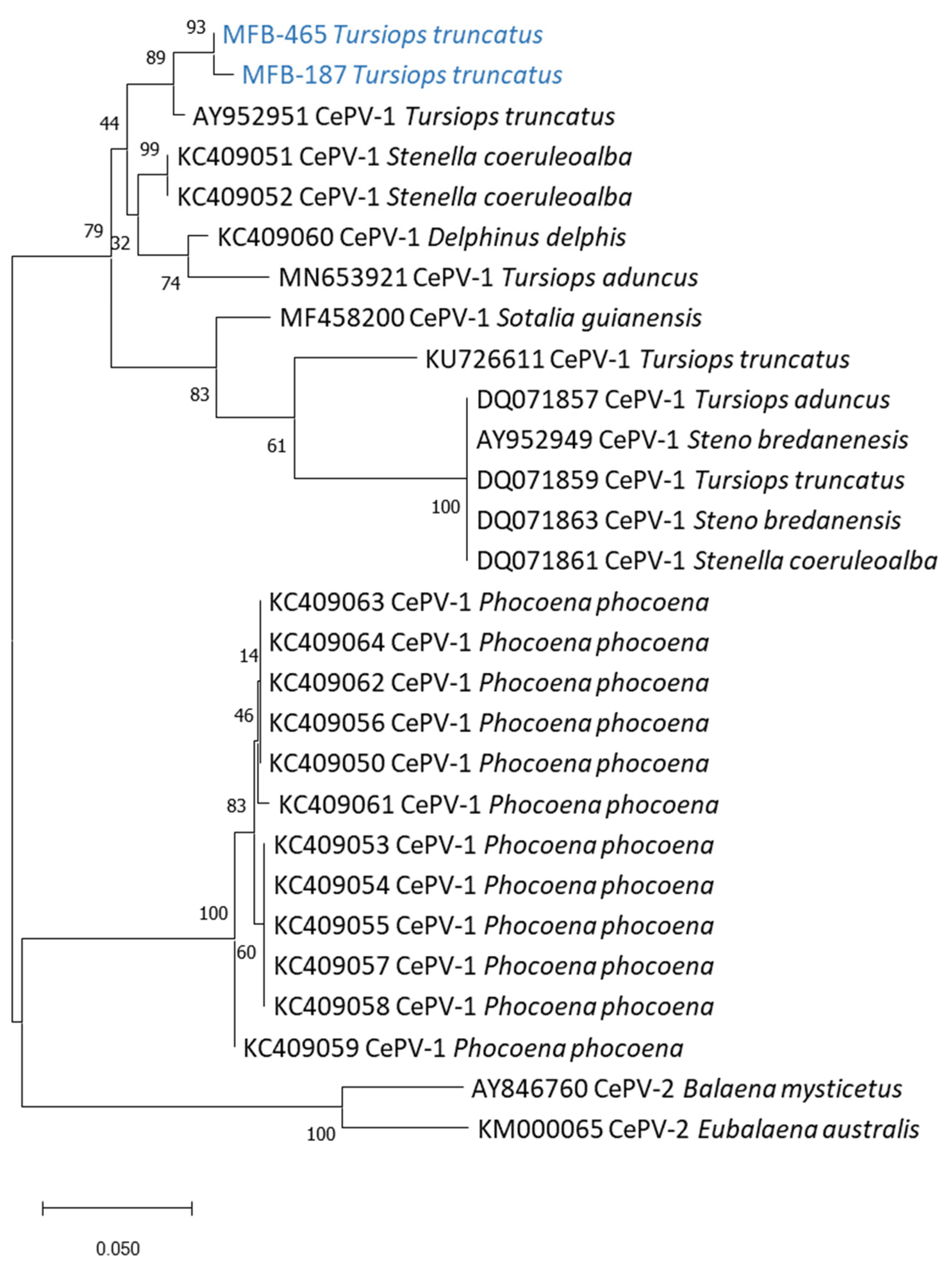
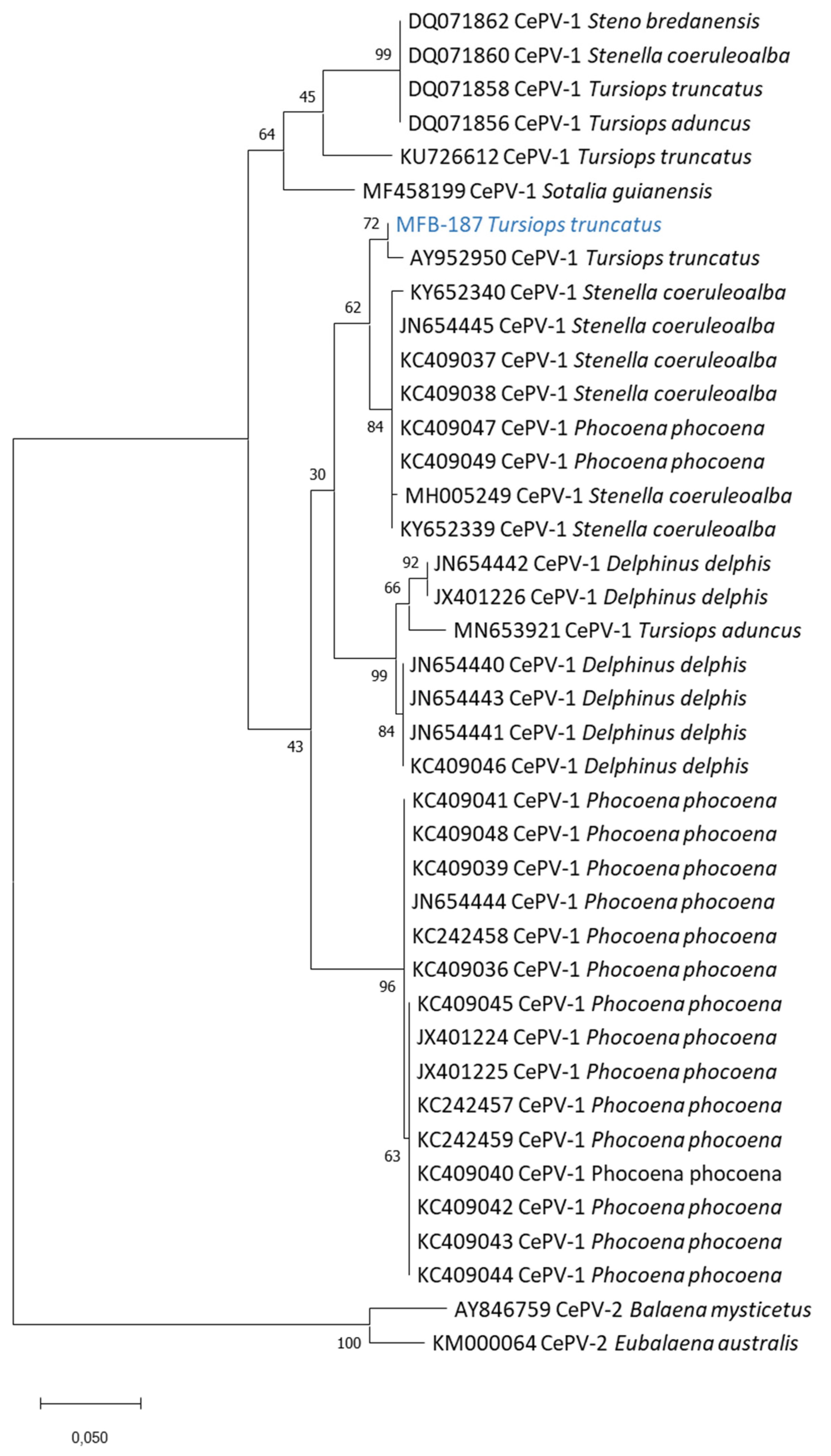
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.L.; Peng, C.; McFadden, G.; Rothenburg, S. Poxviruses and the evolution of host range and virulence. Infect. Genet. Evol. 2013, 21, 15–40. [Google Scholar] [CrossRef] [PubMed]
- Duignan, P.J.; Van Bressem, M.F.; Cortés-Hinojosa, G.; Kennedy-Stoskopf, S. Viruses of Marine Mammals. In CRC Handbook of Marine Mammal Medicine: Health, Disease, and Rehabilitation, 3rd ed.; Dierauf, L.A., Gulland, F.M.D., Eds.; Taylor and Francis Publishers: Boca Raton, FL, USA, 2018. [Google Scholar]
- Osterhaus, A.D.M.E.; Broeders, H.W.J.; Visser, I.K.G.; Teppema, J.S.; Vedder, E.J. Isolation of an orthopoxvirus from pox-like lesions of a grey seal (Halichoerus grypus). Vet. Rec. 1990, 127, 91–92. [Google Scholar] [PubMed]
- Burek, K.A.; Beckmen, K.; Gelatt, T.; Fraser, W.; Bracht, A.J.; Smolarek, K.A.; Romero, C.H. Poxvirus Infection of Steller Sea Lions (Eumetopias jubatus) in Alaska. J. Wildl. Dis. 2005, 41, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.M.; Subramaniam, K.; Tu, S.-L.; Nielsen, O.; Tuomi, P.A.; Upton, C.; Waltzek, T.B. Complete genome sequence of a novel sea otterpox virus. Virus Genes 2018, 54, 756–767. [Google Scholar] [CrossRef]
- Bracht, A.J.; Brudek, R.L.; Ewing, R.Y.; Manire, C.A.; Burek, K.A.; Rosa, C.; Beckmen, K.B.; Maruniak, J.E.; Romero, C.H. Genetic identification of novel poxviruses of cetaceans and pinnipeds. Arch. Virol. 2006, 151, 423–438. [Google Scholar] [CrossRef]
- Blacklaws, B.A.; Gajda, A.M.; Tippelt, S.; Jepson, P.D.; Deaville, R.; Van Bressem, M.-F.; Pearce, G.P. Molecular Characterization of Poxviruses Associated with Tattoo Skin Lesions in UK Cetaceans. PLoS ONE 2013, 8, e71734. [Google Scholar] [CrossRef]
- Rodrigues, T.C.; Subramaniam, K.; Varsani, A.; McFadden, G.; Schaefer, A.M.; Bossart, G.D.; Romero, C.H.; Waltzek, T.B. Genome characterization of cetaceanpox virus from a managed Indo-Pacific bottlenose dolphin (Tursiops aduncus). Virus Res. 2020, 278, 197861. [Google Scholar] [CrossRef]
- Geraci, J.R.; Hicks, B.D.; St Aubin, D.J. Dolphin pox: A skin disease of cetaceans. Can. J. Comp. Med. 1979, 43, 399–404. [Google Scholar]
- Van Bressem, M.F.; Van Waerebeek, K. Epidemiology of Poxvirus in Small Cetaceans from the Eastern South Pacific. Mar. Mammal. Sci. 1996, 12, 371–382. [Google Scholar] [CrossRef]
- Van Bressem, M.F.; Raga, J.A.; Di Guardo, G.; Jepson, P.D.; Duignan, P.J.; Siebert, U.; Van Waerebeek, K. Emerging infectious diseases in cetaceans worldwide and the possible role of environmental stressors. Dis. Aquat. Org. 2009, 86, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.; Dastjerdi, A.; Davison, N.; Deaville, R.; Everest, D.; Peake, J.; Finnegan, C.; Jepson, P.; Steinbach, F. Identification of Novel Cetacean Poxviruses in Cetaceans Stranded in South West England. PLoS ONE 2015, 10, e0124315. [Google Scholar] [CrossRef] [PubMed]
- Fiorito, C.; Palacios, C.; Golemba, M.; Bratanich, A.; Argüelles, M.; Fazio, A.; Bertellotti, M.; Lombardo, D. Identification, molecular and phylogenetic analysis of poxvirus in skin lesions of southern right whale. Dis. Aquat. Org. 2015, 116, 157–163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sacristán, C.; Esperón, F.; Marigo, J.; Ewbank, A.C.; de Carvalho, R.R.; Groch, K.R.; de Castilho, P.V.; Sánchez-Sarmiento, A.M.; Costa-Silva, S.; Ferreira-Machado, E.; et al. Molecular identification and microscopic characterization of poxvirus in a Guiana dolphin and a common bottlenose dolphin, Brazil. Dis. Aquat. Org. 2018, 130, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Sanino, G.P.; Van Waerebeek, K.; Van Bressem, M.F.; Pastene, L.A. A preliminary note on population structure in eastern South Pacific common bottlenose dolphins, Tursiops truncatus. J. Cetacean. Res. Manag. 2005, 7, 65. [Google Scholar]
- Van Bressem, M.F.; Van Waerebeek, K.; Reyes, J.C.; Dekegel, D.; Pastoret, P.P. Evidence of Poxvirus in Dusky Dolphin (Lagenorhynchus obscurus) and Burmeister’s Porpoise (Phocoena spinipinnis) from Coastal Peru. J. Wildl. Dis. 1993, 29, 109–113. [Google Scholar] [CrossRef]
- Van Bressem, M.F.; Van Waerebeek, K.; Bennett, M. Orthopoxvirus neutralising antibodies in small cetaceans from the Southeast Pacific. Lat. Am. J. Aquat. Mamm. 2006, 5, 49–54. [Google Scholar] [CrossRef]
- Luciani, L.; Inchauste, L.; Ferraris, O.; Charrel, R.; Nougairède, A.; Piorkowski, G.; Peyrefitte, C.; Bertagnoli, S.; de Lamballerie, X.; Priet, S. A novel and sensitive real-time PCR system for universal detection of poxviruses. Sci. Rep. 2021, 11, 1798, Erratum in 2022, 12, 5961. [Google Scholar] [CrossRef]
- Van Waerebeek, K.; Reyes, J. Post-Ban Small Cetacean Takes off Peru: A Review. In Report of the International Whaling Commission; International Whaling Commission: Cambridge, UK, 1994; pp. 503–519. [Google Scholar]
- Upton, C.; Slack, S.; Hunter, A.L.; Ehlers, A.; Roper, R.L. Poxvirus Orthologous Clusters: Toward Defining the Minimum Essential Poxvirus Genome. J. Virol. 2003, 77, 7590–7600. [Google Scholar] [CrossRef]
- Kulesh, D.A.; Baker, R.O.; Loveless, B.M.; Norwood, D.; Zwiers, S.H.; Mucker, E.; Hartmann, C.; Herrera, R.; Miller, D.; Christensen, D.; et al. Smallpox and pan -Orthopox Virus Detection by Real-Time 3′-Minor Groove Binder TaqMan Assays on the Roche LightCycler and the Cepheid Smart Cycler Platforms. J. Clin. Microbiol. 2004, 42, 601–609. [Google Scholar] [CrossRef]
- Nitsche, A.; Büttner, M.; Wilhelm, S.; Pauli, G.; Meyer, H. Real-Time PCR Detection of Parapoxvirus DNA. Clin. Chem. 2006, 52, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Cocumelli, C.; Fichi, G.; Marsili, L.; Senese, M.; Cardeti, G.; Cersini, A.; Ricci, E.; Garibaldi, F.; Scholl, F.; Di Guardo, G.; et al. Cetacean Poxvirus in Two Striped Dolphins (Stenella coeruleoalba) Stranded on the Tyrrhenian Coast of Italy: Histopathological, Ultrastructural, Biomolecular, and Ecotoxicological Findings. Front. Veter. Sci. 2018, 5, 219. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Van Waerebeek, K.; Reyes, J.C.; Read, A.J.; McKinnon, J.S. Preliminary Observations of Bottlenose Dolphins from the Pacific Coast of South America. In The Bottlenose Dolphin; Leatherwood, S., Reeves, R.R., Eds.; Elsevier: Amsterdam, The Netherlands, 1990; pp. 143–154. [Google Scholar] [CrossRef]
- Koshiba, M.; Ogawa, K.; Hamazaki, S.; Sugiyama, T.; Ogawa, O.; Kitajima, T. The Effect of Formalin Fixation on DNA and the Extraction of High-molecular-weight DNA from Fixed and Embedded Tissues. Pathol. Res. Pract. 1993, 189, 66–72. [Google Scholar] [CrossRef]
- Babkin, I.V.; Babkina, I.N. Molecular Dating in the Evolution of Vertebrate Poxviruses. Intervirology 2011, 54, 253–260. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Wang, C.; Upton, C. Poxviruses: Past, present and future. Virus Res. 2006, 117, 105–118. [Google Scholar] [CrossRef]
- Hughes, A.L.; Irausquin, S.; Friedman, R. The evolutionary biology of poxviruses. Infect. Genet. Evol. 2010, 10, 50–59. [Google Scholar] [CrossRef]

| Sample | Species | Year | Locality | Conservation | EM | Pan-Poxvirus | |
|---|---|---|---|---|---|---|---|
| AGG-574 | Long-beaked common dolphin | Delphinus cf. capensis | 1991 | Ancon | GA | ND | Neg |
| MFB-195 | Long-beaked common dolphin | Delphinus cf. capensis | 1993 | Cerro Azul | Formalin | ND | Neg |
| MFB-297 | Long-beaked common dolphin | Delphinus cf. capensis | 1993 | Cerro Azul | GA | ND | Neg |
| MFB-364 | Short-finned pilot whale | Globicephala macrorhynchus | 1993 | Cerro Azul | DMSO | ND | Neg |
| MFB-001 | Dusky dolphin | Lagenorhynchus obscurus | 1989 | Pucusana | Formalin | Poxvirus | Neg |
| KVW-1899 | Dusky dolphin | Lagenorhynchus obscurus | 1989 | Pucusana | Formalin | ND | Neg |
| AGG-580 | Dusky dolphin | Lagenorhynchus obscurus | 1991 | Ancon | GA | ND | Neg |
| AGG-663 | Dusky dolphin | Lagenorhynchus obscurus | 1992 | Ancon | GA | ND | Neg |
| MFB-146 | Dusky dolphin | Lagenorhynchus obscurus | 1993 | Cerro Azul | GA | ND | Neg |
| KVW-1864 | Burmeister’s porpoise | Phocoena spinipinnis | 1989 | Pucusana | Formalin | ND | Neg |
| KVW-1863 | Burmeister’s porpoise | Phocoena spinipinnis | 1989 | Pucusana | Formalin | ND | Neg |
| KVW-2283 | Burmeister’s porpoise | Phocoena spinipinnis | 1990 | Pucusana | Formalin | Poxvirus | Neg |
| KOS-83 | Burmeister’s porpoise | Phocoena spinipinnis | 1993 | Cerro Azul | Formalin | ND | Neg |
| KVW-2427 | Burmeister’s porpoise | Phocoena spinipinnis | 1995 | San Jose | Formalin | ND | Neg |
| MFB-465 | Common bottlenose dolphin | Tursiops truncatus | 1993 | Cerro Azul | DMSO | ND | 21CT |
| MFB-187 | Common bottlenose dolphin | Tursiops truncatus | 1993 | Cerro Azul | GA | Poxvirus | 16CT |
| Species | Scientific Name | Location | Sample type | Dna Polymerase Genbank # | Dna Topoisomerase Genbank # | References | Clade |
|---|---|---|---|---|---|---|---|
| bowhead whale | B. mysticetus | USA (Alaska) | Killed | AY846759 | AY846760 | [7] | CePV-2 |
| southern right whale | E. australis | Argentina | stranded | KM000064 | KM000065 | [14] | CePV-2 |
| common dolphin | D. delphis | UK (Cornwall) | stranded | KC409046 | KC409060 | [8] | CePV-1 |
| common dolphin | D. delphis | UK (Cornwall) | stranded | JN654440 | NA | [13] | CePV-1 |
| common dolphin | D. delphis | UK (Cornwall) | stranded | JN654441/42 | NA | [13] | CePV-1 |
| common dolphin | D. delphis | UK (Cornwall) | stranded | JN654443 | NA | [13] | CePV-1 |
| common dolphin | D. delphis | UK (Cornwall) | stranded | JX401226 | NA | [13] | CePV-1 |
| rough-toothed dolphin | S. bredanensis | USA (FL) | stranded | AY463004 | AY952949 | [7] | CePV-1 |
| rough-toothed dolphin | S. bredanensis | USA (FL) | stranded | DQ071862 | DQ071863 | [7] | CePV-1 |
| striped dolphin | S. coeruleoalba | UK (Dorset) | stranded | KC409037 | KC409051 | [8] | CePV-1 |
| striped dolphin | S. coeruleoalba | USA (FL) | stranded | DQ071860 | DQ071861 | [7] | CePV-1 |
| striped dolphin | S. coeruleoalba | UK (Cornwall) | stranded | JN654445 | NA | [13] | CePV-1 |
| striped dolphin | S. coeruleoalba | UK (Dorset) | stranded | KC409038 | KC409052 | [8] | CePV-1 |
| striped dolphin | S. coeruleoalba | Italy (Toscana) | stranded | KY652339 | NA | [26] | CePV-1 |
| striped dolphin | S. coeruleoalba | Italy (Lazio) | stranded | KY652340 | NA | [26] | CePV-1 |
| striped dolphin | S. coeruleoalba | Spain (Mediterranean) | stranded | MH005249 | NA | [15] | CePV-1 |
| guiana dolphin | S. guianensis | Brazil (Rio de Janeiro) | stranded | MF458199 | MF458200 | [15] | CePV-1 |
| indo-pacific bottlenose dolphin | T. aduncus | Hong Kong | captive | MN653921 | MN653921 | [9] | CePV-TA |
| indo-pacific bottlenose dolphin | T. aduncus | Hong Kong | captive | DQ071856 | DQ071857 | [7] | CePV-1 |
| common bottlenose dolphin | T. truncatus | Brazil (Santa Catarina) | stranded | KU726612 | KU726611 | [15] | CePV-1 |
| common bottlenose dolphin | T. truncatus | USA (FL) | stranded | AY952950 | AY952951 | [7] | CePV-1 |
| common bottlenose dolphin | T. truncatus | USA (FL) | NA | DQ071858 | DQ071859 | [7] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | KC409047 | KC409064 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Dorset) | stranded | KC409045 | KC409059 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (West Sussex) | stranded | KC409043 | KC409057 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Devon) | stranded | KC409044 | KC409058 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | KC409042 | KC409056 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | KC409041 | KC409055 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | KC409040 | KC409054 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Kent) | stranded | KC409039 | KC409053 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Northumberland) | stranded | KC409036 | KC409050 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | Not used | KC409061 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | Not used | KC409062 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Humberside) | stranded | KC409049 | KC409063 | [8] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Cornwall) | stranded | KC242457 | NA | [13] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Cornwall) | stranded | KC242458 | NA | [13] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Cornwall) | stranded | KC242459 | NA | [13] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Cornwall) | stranded | JN654444 | NA | [13] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Cornwall) | stranded | JX401224 | NA | [13] | CePV-1 |
| harbor porpoise | P. phocoena | UK (Cornwall) | stranded | JX401225 | NA | [13] | CePV-1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luciani, L.; Piorkowski, G.; De Lamballerie, X.; Van Waerebeek, K.; Van Bressem, M.-F. Detection of Cetacean Poxvirus in Peruvian Common Bottlenose Dolphins (Tursiops truncatus) Using a Pan-Poxvirus PCR. Viruses 2022, 14, 1850. https://doi.org/10.3390/v14091850
Luciani L, Piorkowski G, De Lamballerie X, Van Waerebeek K, Van Bressem M-F. Detection of Cetacean Poxvirus in Peruvian Common Bottlenose Dolphins (Tursiops truncatus) Using a Pan-Poxvirus PCR. Viruses. 2022; 14(9):1850. https://doi.org/10.3390/v14091850
Chicago/Turabian StyleLuciani, Léa, Géraldine Piorkowski, Xavier De Lamballerie, Koen Van Waerebeek, and Marie-Françoise Van Bressem. 2022. "Detection of Cetacean Poxvirus in Peruvian Common Bottlenose Dolphins (Tursiops truncatus) Using a Pan-Poxvirus PCR" Viruses 14, no. 9: 1850. https://doi.org/10.3390/v14091850
APA StyleLuciani, L., Piorkowski, G., De Lamballerie, X., Van Waerebeek, K., & Van Bressem, M.-F. (2022). Detection of Cetacean Poxvirus in Peruvian Common Bottlenose Dolphins (Tursiops truncatus) Using a Pan-Poxvirus PCR. Viruses, 14(9), 1850. https://doi.org/10.3390/v14091850






