Pseudorabies Virus: From Pathogenesis to Prevention Strategies
Abstract
1. Introduction
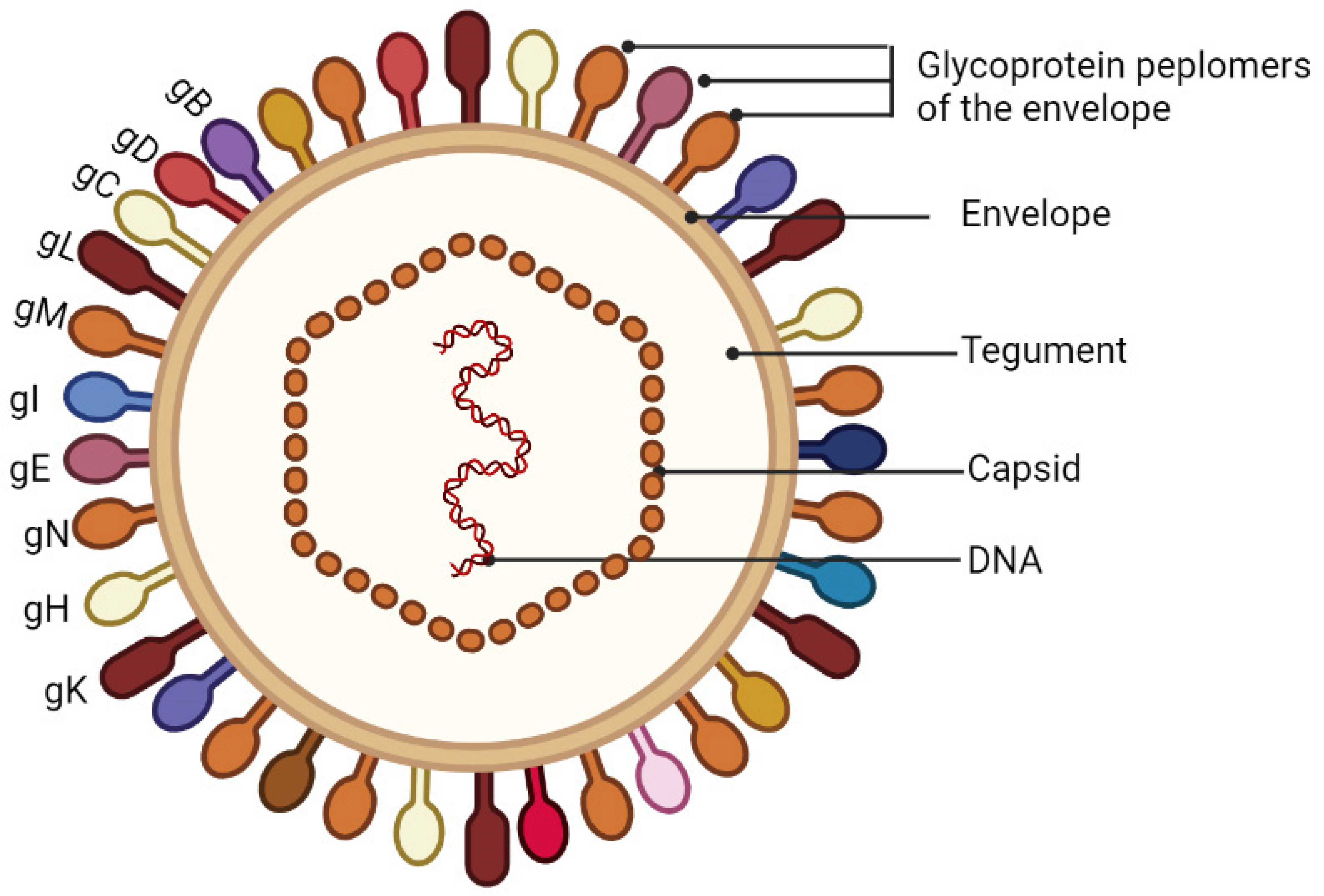
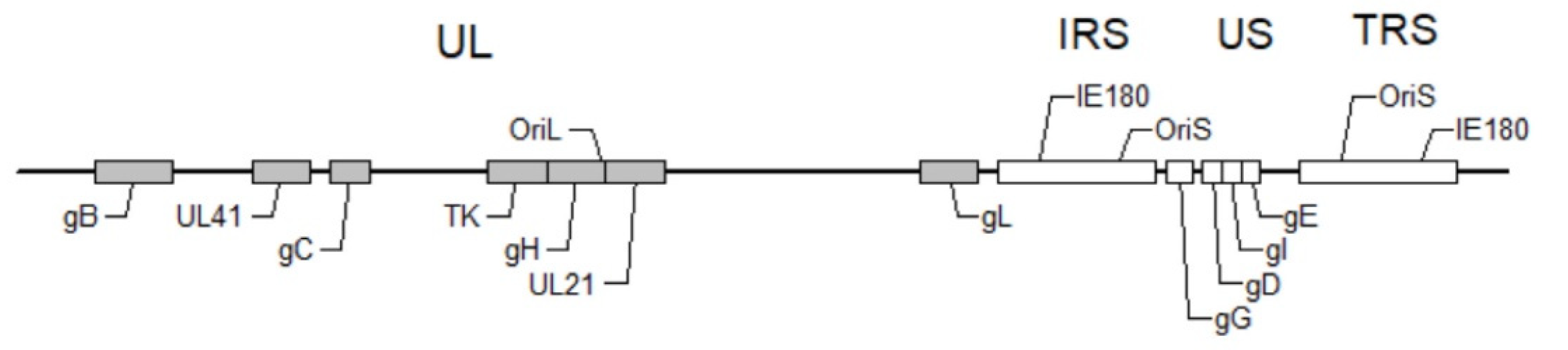
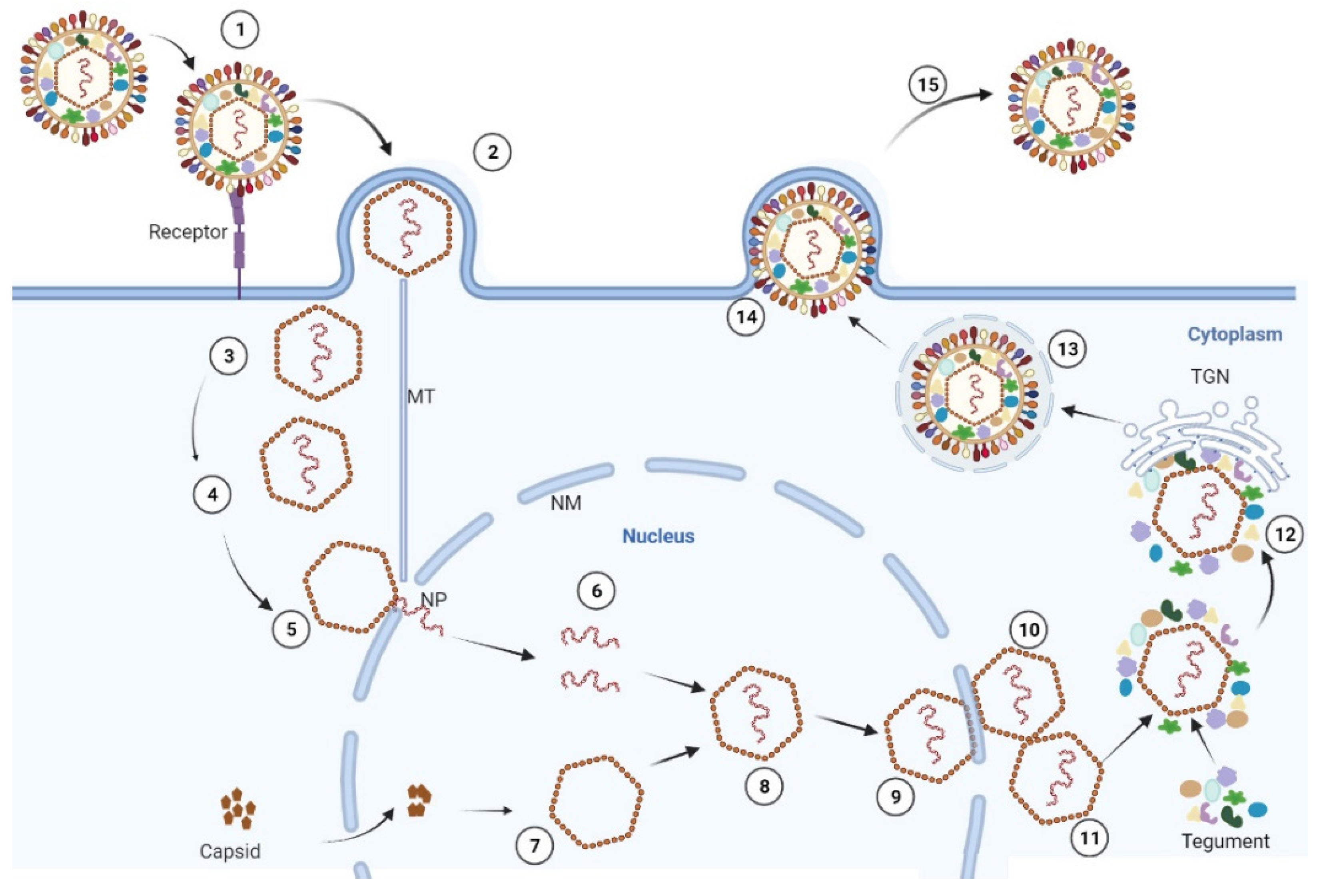
2. The Virion Structure, Genome Structure and the Life Cycle
2.1. The Virion Structure
2.2. Genome, Gene Content and Role in Viral Replication
2.3. The Life Cycle of PRV
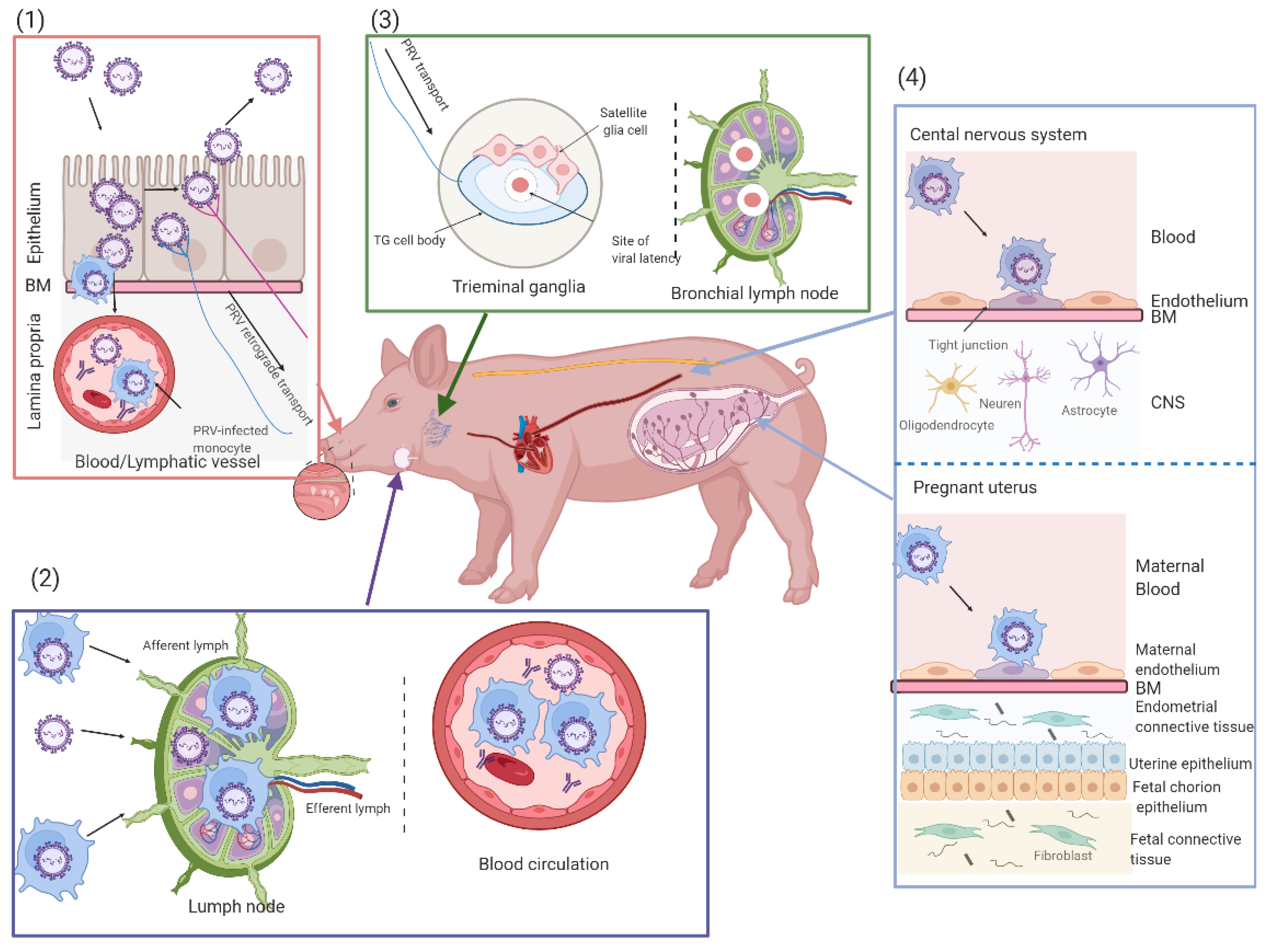
3. Occurrence and Development of PRV Infection
3.1. PRV Primary Replication in the Upper Respiratory Tract
3.2. PRV Replication in the Draining Lymph Nodes and Viremia
3.3. PRV Entry into the Peripheral Nervous System (PNS) Neurons and Spread to the Central Nervous System (CNS)
3.4. Secondary Replication in the Swine Pregnant Uterus
3.5. PRV Infection in Suckling and Weaned Piglets
3.6. PRV Infection in Humans
4. Genetic Evolution of PRV
5. Diagnostic Methods
5.1. Serological Approaches for the Detection of PRV Antibodies
5.2. Molecular Biology Approaches for the Detection of PRV Infection
5.3. Other Approaches for the Detection of PRV Infection
6. The Prevention of PR
6.1. Main Vaccines against PRV Infection
6.2. Chinese Herbal Medicines as Potential Anti-PRV Drugs
| Source | Mechanism | 50% Effective Concentration | 50% Cytotoxic Concentration | PRV Strain | In Vitro | In Vivo | References |
|---|---|---|---|---|---|---|---|
| Resveratrol (Res) | The inhibition of viral proliferation, IκB kinase activation | 17.17 ± 0.35 μM | Above 262.87 μM | Rong A strain | Yes | Yes | [142,209,211] |
| Kaempferol | The inhibition of viral proliferation | 25.57 μM of 50% inhibited concentration | No mention | Ra strain | Yes | Yes | [213] |
| Panax notoginseng polysaccharides | The inhibition of viral adsorption and replication | No mention | No mention | PRV XJ5 strain | Yes | No | [214] |
| Germacrone | The inhibition of viral proliferation | 54.51 μM for Vero cells and 88.78 μM for LLC-PK-1 cells | 233.5 μM for Vero cells and 184.1 μM for LLC-PK-1 cells | Variant PRV and PRV vaccine strain Barth K61 | Yes | No | [206] |
| Plantago | The inhibition of viral attachment and penetration; decreasing ROS (reactive oxygen species) production | No mention | No mention | PRV XJ5 | Yes | No | [215] |
| Quercetin | The inhibition of viral adsorption | 2.618 ± 0.673 μM of 50% inhibited concentration | Above 599 μM | HNX strain | Yes | Yes | [216] |
| Isatis indigotica | The inhibition of viral proliferation | 11 μg/mL | 299 μg/mL | TNL strain | Yes | No | [217] |
| Radix isatidis | The inhibition of viral proliferation; killing virus directly | The inhibition rate of viral replication by 14.674–30.84% | No mention | Min A strain | Yes | No | [207] |
| Marine Bacillus S-12–86 lysozyme | The inhibition of viral proliferation; killing virus directly | 0.46 mg/L | 100 mg/L | Min A strain | Yes | No | [218] |
| Diammonium glycyrrhizin | Killing virus directly | No mention | Above 1250 μg/mL | Bartha K-61 | Yes | No | [219] |
| Vanadium-substituted Heteropolytungstate | Killing virus directly | 3.5–5.0 mg/L | 400–420 mg/L | Bartha strain | Yes | No | [220] |
| Graphene Oxide | Killing virus directly | No mention | No mention | HNX strain | Yes | No | [223] |
| Ivermectin | The inhibition of viral DNA polymerase UL42 in entering the nucleus | No mention | No mention | No mention | Yes | Yes | [208] |
| Phosphonoformate sodium | Inhibition of viral DNA polymerase | Nearly 60 μg/mL of 50% inhibited concentration | No mention | Kaplan | Yes | No | [221] |
6.3. Novel Small RNAs
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, H.H.; Jin, Y.; Hou, C.Y.; Li, X.S.; Zhao, L.; Wang, Z.Y.; Chen, H.Y. Seroprevalence investigation and genetic analysis of pseudorabies virus within pig populations in Henan province of China during 2018–2019. Infect. Genet. Evol. 2021, 92, 104835. [Google Scholar] [CrossRef]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. MMBR 2005, 69, 462–500. [Google Scholar] [CrossRef] [PubMed]
- Laval, K.; Vernejoul, J.; Van Cleemput, J.; Koyuncu, O.; Enquist, L. Virulent Pseudorabies Virus Infection Induces a Specific and Lethal Systemic Inflammatory Response in Mice. J. Virol. 2018, 92, e01614-18. [Google Scholar] [CrossRef]
- Yang, H.; Han, H.; Wang, H.; Cui, Y.; Liu, H.; Ding, S. A Case of Human Viral Encephalitis Caused by Pseudorabies Virus Infection in China. Front. Neurol. 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Weng, S.; Cheng, Q.; Cui, P.; Li, Y.; Wu, H.; Zhu, Y.; Xu, B.; Zhang, W. Human Endophthalmitis Caused By Pseudorabies Virus Infection, China, 2017. Emerg. Infect. Dis. 2018, 24, 1087–1090. [Google Scholar] [CrossRef]
- Li, X.; Fu, S.; Chen, L.; Li, F.; Deng, J.; Lu, X.; Wang, H.; Tian, K. Detection of Pseudorabies Virus Antibodies in Human Encephalitis Cases. Biomed. Environ. Sci. BES 2020, 33, 444–447. [Google Scholar] [CrossRef]
- Wang, Y.; Nian, H.; Li, Z.; Wang, W.; Wang, X.; Cui, Y. Human encephalitis complicated with bilateral acute retinal necrosis associated with pseudorabies virus infection: A case report. Int. J. Infect. Dis. IJID 2019, 89, 51–54. [Google Scholar] [CrossRef]
- Ying, M.; Hu, X.; Wang, M.; Cheng, X.; Zhao, B.; Tao, Y. Vitritis and retinal vasculitis caused by pseudorabies virus. J. Int. Med. Res. 2021, 49, 3000605211058990. [Google Scholar] [CrossRef]
- Freuling, C.M.; Muller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Yuan, J.; Qin, H.Y.; Luo, Y.; Cong, X.; Li, Y.; Chen, J.; Li, S.; Sun, Y.; Qiu, H.J. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine 2014, 32, 3379–3385. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, S.; Wang, X.; Zou, M.; Gao, S. Bartha-k61 vaccine protects growing pigs against challenge with an emerging variant pseudorabies virus. Vaccine 2017, 35, 1161–1166. [Google Scholar] [CrossRef]
- Mettenleiter, T. Aujeszky’s disease (pseudorabies) virus: The virus and molecular pathogenesis--state of the art, June 1999. Vet. Res. 2000, 31, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Masić, M.; Ercegan, M.; Petrović, M. The significance of the tonsils in the pathogenesis and diagnosis of Aujeszyk’s disease in pigs. Zent. Fur Veterinarmedizin. Reihe B J. Vet. Medicine. Ser. B 1965, 12, 398–405. [Google Scholar]
- Tirabassi, R.; Townley, R.; Eldridge, M.; Enquist, L. Molecular mechanisms of neurotropic herpesvirus invasion and spread in the CNS. Neurosci. Biobehav. Rev. 1998, 22, 709–720. [Google Scholar] [CrossRef]
- Gutekunst, D.; Pirtle, E.; Miller, L.; Stewart, W. Isolation of pseudorabies virus from trigeminal ganglia of a latently infected sow. Am. J. Vet. Res. 1980, 41, 1315–1316. [Google Scholar]
- Verpoest, S.; Cay, B.; Favoreel, H.; De Regge, N. Age-Dependent Differences in Pseudorabies Virus Neuropathogenesis and Associated Cytokine Expression. J. Virol. 2017, 91, e02058-16. [Google Scholar] [CrossRef] [PubMed]
- Nawynck, H.; Pensaert, M. Cell-free and cell-associated viremia in pigs after oronasal infection with Aujeszky’s disease virus. Vet. Microbiol. 1995, 43, 307–314. [Google Scholar] [CrossRef]
- Nauwynck, H.; Pensaert, M. Abortion induced by cell-associated pseudorabies virus in vaccinated sows. Am. J. Vet. Res. 1992, 53, 489–493. [Google Scholar] [PubMed]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: An update. Virus Res. 2009, 143, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Daniel, G.R.; Pegg, C.E.; Smith, G.A.; Sandri-Goldin, R.M. Dissecting the Herpesvirus Architecture by Targeted Proteolysis. J. Virol. 2018, 92, e00738-18. [Google Scholar] [CrossRef] [PubMed]
- Kramer, T.; Greco, T.M.; Enquist, L.W.; Cristea, I.M. Proteomic characterization of pseudorabies virus extracellular virions. J. Virol. 2011, 85, 6427–6441. [Google Scholar] [CrossRef] [PubMed]
- Romero, C.H.; Meade, P.; Santagata, J.; Gillis, K.; Lollis, G.; Hahn, E.C.; Gibbs, E.P. Genital infection and transmission of pseudorabies virus in feral swine in Florida, USA. Vet. Microbiol. 1997, 55, 131–139. [Google Scholar] [CrossRef]
- Fuchs, W.; Ehrlich, C.; Klupp, B.; Mettenleiter, T. Characterization of the replication origin (Ori(S)) and adjoining parts of the inverted repeat sequences of the pseudorabies virus genome. J. Gen. Virol. 2000, 81, 1539–1543. [Google Scholar] [CrossRef]
- Wu, C.; Harper, L.; Ben-Porat, T. Molecular basis for interference of defective interfering particles of pseudorabies virus with replication of standard virus. J. Virol. 1986, 59, 308–317. [Google Scholar] [CrossRef]
- Wesley, R.D.; Cheung, A.K. A Pseudorabies Virus Mutant with Deletions in the Latency and Early Protein O Genes: Replication, Virulence, and Immunity in Neonatal Piglets. J. Vet. Diagn. Investig. 1996, 8, 21–24. [Google Scholar] [CrossRef]
- Kit, S.; Kit, M.; Pirtle, E. Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. Am. J. Vet. Res. 1985, 46, 1359–1367. [Google Scholar] [PubMed]
- Xu, L.; Wei, J.; Zhao, J.; Xu, S.; Lee, F.; Nie, M.; Xu, Z.; Zhou, Y.; Zhu, L. The Immunity Protection of Central Nervous System Induced by Pseudorabies Virus DelgI/gE/TK in Mice. Front. Microbiol. 2022, 13, 862907. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.; Lei, J.L.; Xia, S.L.; Wang, Y.M.; Li, Y.; Li, S.; Luo, Y.; Sun, Y.; Qiu, H.J. Pathogenicity and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant in susceptible animals. Vet. Microbiol. 2016, 182, 170–177. [Google Scholar] [CrossRef]
- Rauh, I.; Mettenleiter, T.C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 1991, 65, 5348–5356. [Google Scholar] [CrossRef] [PubMed]
- Peeters, B.; de Wind, N.; Hooisma, M.; Wagenaar, F.; Gielkens, A.; Moormann, R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 1992, 66, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Hayashi, S.; Tanioka, Y.; Matsumoto, Y.; Otsuka, H. Pseudorabies virus (PRV) is protected from complement attack by cellular factors and glycoprotein C (gC). Virus Res. 2002, 84, 79–87. [Google Scholar] [CrossRef]
- Rue, C.A.; Ryan, P. Pseudorabies virus glycoprotein C attachment-proficient revertants isolated through a simple, targeted mutagenesis scheme. J. Virol. Methods 2008, 151, 101–106. [Google Scholar] [CrossRef]
- Sapir, A.; Avinoam, O.; Podbilewicz, B.; Chernomordik, L.V. Viral and developmental cell fusion mechanisms: Conservation and divergence. Dev. Cell 2008, 14, 11–21. [Google Scholar] [CrossRef]
- Vallbracht, M.; Brun, D.; Tassinari, M.; Vaney, M.C.; Pehau-Arnaudet, G.; Guardado-Calvo, P.; Haouz, A.; Klupp, B.G.; Mettenleiter, T.C.; Rey, F.A.; et al. Structure-Function Dissection of Pseudorabies Virus Glycoprotein B Fusion Loops. J. Virol. 2018, 92, e01203-17. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Lukàcs, N.; Rziha, H.J. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J. Virol. 1985, 56, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Lu, G.; Qi, J.; Wu, L.; Tian, K.; Luo, T.; Shi, Y.; Yan, J.; Gao, G.F. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. PLoS Pathog. 2017, 13, e1006314. [Google Scholar] [CrossRef]
- Thomsen, D.R.; Marchioli, C.C.; Yancey, R.J., Jr.; Post, L.E. Replication and virulence of pseudorabies virus mutants lacking glycoprotein gX. J. Virol. 1987, 61, 229–232. [Google Scholar] [CrossRef]
- Lerma, L.; Muñoz, A.L.; García Utrilla, R.; Sainz, B., Jr.; Lim, F.; Tabarés, E.; Gómez-Sebastián, S. Partial complementation between the immediate early proteins ICP4 of herpes simplex virus type 1 and IE180 of pseudorabies virus. Virus Res. 2020, 279, 197896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; El Omari, K.; Duman, R.; Liu, S.; Haider, S.; Wagner, A.; Parkinson, G.N.; Wei, D. Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths. Nucleic Acids Res. 2020, 48, 9886–9898. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wang, P.; Wang, Y.; Chen, T.; Xu, Z.; Zou, X.; Ou, X.; Li, Y.; Chen, D.; Peng, T.; et al. Identification of the molecular determinants for nuclear import of PRV EP0. Biol. Chem. 2019, 400, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Liu, J.; Guan, X.; Yin, Y.X.; Peng, H.; Chen, H.C.; Liu, Z.F. The Carboxyl Terminus of Tegument Protein pUL21 Contributes to Pseudorabies Virus Neuroinvasion. J. Virol. 2019, 93, e02052-18. [Google Scholar] [CrossRef]
- Liu, Y.F.; Tsai, P.Y.; Chulakasian, S.; Lin, F.Y.; Hsu, W.L. The pseudorabies virus vhs protein cleaves RNA containing an IRES sequence. FEBS J. 2016, 283, 899–911. [Google Scholar] [CrossRef]
- Liu, Y.F.; Tsai, P.Y.; Lin, F.Y.; Lin, K.H.; Chang, T.J.; Lin, H.W.; Chulakasian, S.; Hsu, W.L. Roles of nucleic acid substrates and cofactors in the vhs protein activity of pseudorabies virus. Vet. Res. 2015, 46, 141. [Google Scholar] [CrossRef]
- Granzow, H.; Klupp, B.G.; Mettenleiter, T.C. Entry of pseudorabies virus: An immunogold-labeling study. J. Virol. 2005, 79, 3200–3205. [Google Scholar] [CrossRef] [PubMed]
- Luxton, G.W.; Haverlock, S.; Coller, K.E.; Antinone, S.E.; Pincetic, A.; Smith, G.A. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 5832–5837. [Google Scholar] [CrossRef]
- Granzow, H.; Weiland, F.; Jöns, A.; Klupp, B.G.; Karger, A.; Mettenleiter, T.C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: A reassessment. J. Virol. 1997, 71, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Granzow, H.; Klupp, B.G.; Kopp, M.; Mettenleiter, T.C. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 2002, 76, 6729–6742. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porat, T.; Veach, R.A.; Ihara, S. Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology 1983, 127, 194–204. [Google Scholar] [CrossRef]
- Lehman, I.; Boehmer, P. Replication of herpes simplex virus DNA. J. Biol. Chem. 1999, 274, 28059–28062. [Google Scholar] [CrossRef]
- De Wind, N.; Peeters, B.P.; Zuderveld, A.; Gielkens, A.L.; Berns, A.J.; Kimman, T.G. Mutagenesis and characterization of a 41-kilobase-pair region of the pseudorabies virus genome: Transcription map, search for virulence genes, and comparison with homologs of herpes simplex virus type 1. Virology 1994, 200, 784–790. [Google Scholar] [CrossRef]
- Jöns, A.; Mettenleiter, T.C. Identification and characterization of pseudorabies virus dUTPase. J. Virol. 1996, 70, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Klupp, B.G.; Granzow, H.; Rziha, H.J.; Mettenleiter, T.C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 1996, 70, 3517–3527. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Klupp, B.G.; Granzow, H.; Osterrieder, N.; Mettenleiter, T.C. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 2002, 76, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.E.; Wang, Y.F.; Meredith, D.M. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 1998, 72, 7108–7114. [Google Scholar] [CrossRef]
- Hahn, E.C.; Page, G.R.; Hahn, P.S.; Gillis, K.D.; Romero, C.; Annelli, J.A.; Gibbs, E.P. Mechanisms of transmission of Aujeszky’s disease virus originating from feral swine in the USA. Vet. Microbiol. 1997, 55, 123–130. [Google Scholar] [CrossRef]
- Romero, C.H.; Meade, P.N.; Shultz, J.E.; Chung, H.Y.; Gibbs, E.P.; Hahn, E.C.; Lollis, G. Venereal transmission of pseudorabies viruses indigenous to feral swine. J. Wildl. Dis. 2001, 37, 289–296. [Google Scholar] [CrossRef]
- Nauwynck, H.; Glorieux, S.; Favoreel, H.; Pensaert, M. Cell biological and molecular characteristics of pseudorabies virus infections in cell cultures and in pigs with emphasis on the respiratory tract. Vet. Res. 2007, 38, 229–241. [Google Scholar] [CrossRef]
- Crandell, R.A. Pseudorabies (Aujeszky’s disease). Vet. Clin. N. Am. Large Anim. Pract. 2014, 4, 321. [Google Scholar] [CrossRef]
- Glorieux, S.; Van den Broeck, W.; van der Meulen, K.M.; Van Reeth, K.; Favoreel, H.W.; Nauwynck, H.J. In vitro culture of porcine respiratory nasal mucosa explants for studying the interaction of porcine viruses with the respiratory tract. J. Virol. Methods 2007, 142, 105–112. [Google Scholar] [CrossRef]
- Glorieux, S.; Favoreel, H.W.; Meesen, G.; de Vos, W.; Van den Broeck, W.; Nauwynck, H.J. Different replication characteristics of historical pseudorabies virus strains in porcine respiratory nasal mucosa explants. Vet. Microbiol. 2009, 136, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Kawashima, K.; Matsuura, S.; Uchimura, A.; Miura, Y. Pneumonia in pigs infected with pseudorabies virus and Haemophilus parasuis serovar 4. J. Comp. Pathol. 1994, 110, 329–339. [Google Scholar] [CrossRef]
- Maes, R.K.; Kanitz, C.L.; Gustafson, D.P. Shedding patterns in swine of virulent and attenuated pseudorabies virus. Am. J. Vet. Res. 1983, 44, 2083–2086. [Google Scholar] [PubMed]
- Lamote, J.A.S.; Glorieux, S.; Nauwynck, H.J.; Favoreel, H.W. The US3 Protein of Pseudorabies Virus Drives Viral Passage across the Basement Membrane in Porcine Respiratory Mucosa Explants. J. Virol. 2016, 90, 10945–10950. [Google Scholar] [CrossRef]
- Glorieux, S.; Favoreel, H.W.; Steukers, L.; Vandekerckhove, A.P.; Nauwynck, H.J. A trypsin-like serine protease is involved in pseudorabies virus invasion through the basement membrane barrier of porcine nasal respiratory mucosa. Vet. Res. 2011, 42, 58. [Google Scholar] [CrossRef]
- Jamrichová, O.; Skoda, R. Multiplication of pseudorabies virus in the inguinal lymph nodes of pigs. Acta Virol. 1968, 12, 555. [Google Scholar]
- Wittmann, G.; Ohlinger, V.; Rziha, H.J. Occurrence and reactivation of latent Aujeszky’s disease virus following challenge in previously vaccinated pigs. Arch. Virol. 1983, 75, 29–41. [Google Scholar] [CrossRef]
- Mulder, W.A.; Jacobs, L.; Priem, J.; Kok, G.L.; Wagenaar, F.; Kimman, T.G.; Pol, J.M. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J. Gen. Virol. 1994, 75 Pt 11, 3095–3106. [Google Scholar] [CrossRef]
- Sabó, A.; Rajcáni, J.; Blaskovic, D. Studies on the pathogenesis of Aujeszky’s disease. 3. The distribution of virulent virus in piglets after intranasal infection. Acta Virol. 1969, 13, 407–414. [Google Scholar]
- Nauwynck, H.J.; Pensaert, M.B. Interactions of Aujeszky’s disease virus and porcine blood mononuclear cells in vivo and in vitro. Acta Vet. Hung. 1994, 42, 301–308. [Google Scholar]
- Babic, N.; Mettenleiter, T.C.; Ugolini, G.; Flamand, A.; Coulon, P. Propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology 1994, 204, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Kramer, T.; Greco, T.M.; Taylor, M.P.; Ambrosini, A.E.; Cristea, I.M.; Enquist, L.W. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe 2012, 12, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Grinde, B. Herpesviruses: Latency and reactivation—Viral strategies and host response. J. Oral Microbiol. 2013, 5, 22766. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.G.; Osorio, F.A. Investigation of sites of pseudorabies virus latency, using polymerase chain reaction. Am. J. Vet. Res. 1991, 52, 1799–1803. [Google Scholar] [PubMed]
- van Oirschot, J.T.; Gielkens, A.L. In vivo and in vitro reactivation of latent pseudorabies virus in pigs born to vaccinated sows. Am. J. Vet. Res. 1984, 45, 567–571. [Google Scholar]
- Wigdahl, B.; Rong, B.L.; Kinney-Thomas, E. Varicella-zoster virus infection of human sensory neurons. Virology 1986, 152, 384–399. [Google Scholar] [CrossRef]
- Jones, C. Bovine Herpes Virus 1 (BHV-1) and Herpes Simplex Virus Type 1 (HSV-1) Promote Survival of Latently Infected Sensory Neurons, in Part by Inhibiting Apoptosis. J. Cell Death 2013, 6, 1–16. [Google Scholar] [CrossRef]
- Van de Walle, G.R.; Favoreel, H.W.; Nauwynck, H.J.; Mettenleiter, T.C.; Pensaert, M.B. Transmission of pseudorabies virus from immune-masked blood monocytes to endothelial cells. J. Gen. Virol. 2003, 84 Pt 3, 629–637. [Google Scholar] [CrossRef]
- Kluge, J.P.; Maré, C.J. Swine pseudorabies: Abortion, clinical disease, and lesions in pregnant gilts infected with pseudorabies virus (Aujeszky’s disease). Am. J. Vet. Res. 1974, 35, 991–995. [Google Scholar]
- Hsu, F.S.; Chu, R.M.; Lee, R.C.; Chu, S.H. Placental lesions caused by pseudorabies virus in pregnant sows. J. Am. Vet. Med. Assoc. 1980, 177, 636–641. [Google Scholar]
- Ka, H.; Seo, H.; Choi, Y.; Yoo, I.; Han, J. Endometrial response to conceptus-derived estrogen and interleukin-1β at the time of implantation in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Vélez, C.; Barbeito, C.; Koncurat, M. αvβ3 Integrin and fibronectin expressions and their relation to estrogen and progesterone during placentation in swine. Biotech. Histochem. 2018, 93, 15–24. [Google Scholar] [CrossRef]
- Bidarimath, M.; Tayade, C. Pregnancy and spontaneous fetal loss: A pig perspective. Mol. Reprod. Dev. 2017, 84, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Dieuzy, I.; Vannier, P.; Jestin, A. Effects of experimental pseudorabies virus infection on vaccinated pregnant sows. Ann. De Rech. Vet. Ann. Vet. Res. 1987, 18, 233–240. [Google Scholar]
- Iglesias, J.G.; Harkness, J.W. Studies of transplacental and perinatal infection with two clones of a single Aujeszky’s disease (pseudorabies) virus isolate. Vet. Microbiol. 1988, 16, 243–254. [Google Scholar] [CrossRef]
- Ceriatti, F.S.; Sabini, L.I.; Bettera, S.G.; Zanón, S.M.; Ramos, B.A. Experimental infection of pregnant gilts with Aujeszky’s disease virus strain RC/79. Rev. Argent. Microbiol. 1992, 24, 102–112. [Google Scholar] [PubMed]
- Wong, G.; Lu, J.; Zhang, W.; Gao, G.F. Pseudorabies virus: A neglected zoonotic pathogen in humans? Emerg. Microbes Infect. 2019, 8, 150–154. [Google Scholar] [CrossRef]
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef]
- Wang, X.; Wu, C.X.; Song, X.R.; Chen, H.C.; Liu, Z.F. Comparison of pseudorabies virus China reference strain with emerging variants reveals independent virus evolution within specific geographic regions. Virology 2017, 506, 92–98. [Google Scholar] [CrossRef]
- Zhai, X.; Zhao, W.; Li, K.; Zhang, C.; Wang, C.; Su, S.; Zhou, J.; Lei, J.; Xing, G.; Sun, H.; et al. Genome Characteristics and Evolution of Pseudorabies Virus Strains in Eastern China from 2017 to 2019. Virol. Sin. 2019, 34, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhu, L.; Zhao, J.; Yin, X.; Feng, Y.; Wang, X.; Sun, X.; Zhou, Y.; Xu, Z. Genetic evolution analysis of novel recombinant pseudorabies virus strain in Sichuan, China. Transbound. Emerg. Dis. 2020, 67, 1428–1432. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Tan, L.; Wang, C.; He, S.; Fang, L.; Wang, Z.; Zhong, Y.; Zhang, K.; Liu, D.; Yang, Q.; et al. Serological Investigation and Genetic Characteristics of Pseudorabies Virus in Hunan Province of China From 2016 to 2020. Front. Vet. Sci. 2021, 8, 762326. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, C.; Li, X. Novel Chinese pseudorabies virus variants undergo extensive recombination and rapid interspecies transmission. Transbound. Emerg. Dis. 2020, 67, 2274–2276. [Google Scholar] [CrossRef]
- Ye, C.; Zhang, Q.; Tian, Z.; Zheng, H.; Zhao, K.; Liu, F.; Guo, J.; Tong, W.; Jiang, C.; Wang, S.; et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 2015, 483, 32–43. [Google Scholar] [CrossRef]
- Fan, J.; Zeng, X.; Zhang, G.; Wu, Q.; Niu, J.; Sun, B.; Xie, Q.; Ma, J. Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013-2014. J. Vet. Sci. 2016, 17, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Ren, H.; Gu, J.; Wang, J.; Jiang, L.; Gao, S. The Epidemiological Analysis of Pseudorabies Virus and Pathogenicity of the Variant Strain in Shandong Province. Front. Vet. Sci. 2022, 9, 806824. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, W.; Liu, Q.; Zhao, T.; Zhu, H.; Hua, L.; Peng, Z.; Tang, X.; Stratton, C.; Zhou, D.; et al. Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. PeerJ 2018, 6, e5785. [Google Scholar] [CrossRef]
- Tan, L.; Yao, J.; Yang, Y.; Luo, W.; Yuan, X.; Yang, L.; Wang, A. Current Status and Challenge of Pseudorabies Virus Infection in China. Virol. Sin. 2021, 36, 588–607. [Google Scholar] [CrossRef]
- Liu, H.; Shi, Z.; Liu, C.; Wang, P.; Wang, M.; Wang, S.; Liu, Z.; Wei, L.; Sun, Z.; He, X.; et al. Implication of the Identification of an Earlier Pseudorabies Virus (PRV) Strain HLJ-2013 to the Evolution of Chinese PRVs. Front. Microbiol. 2020, 11, 612474. [Google Scholar] [CrossRef]
- Tan, L.; Yao, J.; Lei, L.; Xu, K.; Liao, F.; Yang, S.; Yang, L.; Shu, X.; Duan, D.; Wang, A. Emergence of a Novel Recombinant Pseudorabies Virus Derived From the Field Virus and Its Attenuated Vaccine in China. Front. Vet. Sci. 2022, 9, 872002. [Google Scholar] [CrossRef]
- Ma, Z.; Han, Z.; Liu, Z.; Meng, F.; Liu, M. Epidemiological investigation of porcine pseudorabies virus and its coinfection rate in Shandong Province in China from 2015 to 2018. J. Vet. Sci. 2020, 21, e36. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Gao, F.; Guo, H.; Liu, J. Establishment of Indirect ELISA Antibody Detection Kit for gE Protein of Porcine Pseudorabies Virus. China Anim. Health Insp. 2018, 35, 91–94. [Google Scholar]
- Morenkov, O.S.; Sobko, Y.A.; Panchenko, O.A. Glycoprotein gE blocking ELISAs to differentiate between Aujeszky’s disease-vaccinated and infected animals. J. Virol. Methods 1997, 65, 83–94. [Google Scholar] [CrossRef]
- Xu, L.; Peng, Z.; Zhao, T.T.; Xu, S.; Chen, H.C.; Bin, W.U. Development and preliminary application of a direct immunofluorescence method for the detection of pseudorabies virus. Chin. J. Prev. Vet. Med. 2017, 39, 993–997. [Google Scholar]
- Zhang, P.; Lv, L.; Sun, H.; Li, S.; Fan, H.; Wang, X.; Bai, J.; Jiang, P. Identification of linear B cell epitope on gB, gC, and gE proteins of porcine pseudorabies virus using monoclonal antibodies. Vet. Microbiol. 2019, 234, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, Y.; Yang, S.; Wang, Y.; Yang, J.; Liu, Y.; Jin, Q.; Li, X.; Guo, C.; Zhang, G. Development of an immunochromatographic strip for antibody detection of pseudorabies virus in swine. J. Vet. Diagn. Investig. 2015, 27, 739–742. [Google Scholar] [CrossRef]
- Wang, Y.B.; Li, Y.H.; Li, Q.M.; Xie, W.T.; Guo, C.L.; Guo, J.Q.; Deng, R.G.; Zhang, G.P. Development of a blocking immunoperoxidase monolayer assay for differentiation between pseudorabies virus-infected and vaccinated animalss. Pol. J. Vet. Sci. 2019, 22, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Kinker, D.R.; Swenson, S.L.; Wu, L.L.; Zimmerman, J.J. Evaluation of serological tests for the detection of pseudorabies gE antibodies during early infection. Vet. Microbiol. 1997, 55, 99–106. [Google Scholar] [CrossRef]
- Panyasing, Y.; Kedkovid, R.; Kittawornrat, A.; Ji, J.; Zimmerman, J.; Thanawongnuwech, R. Detection of Aujeszky’s disease virus DNA and antibody in swine oral fluid specimens. Transbound. Emerg. Dis. 2018, 65, 1828–1835. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, Z.; Lv, L.; Xiao, Y.; Qu, Y.; Ma, H.; Niu, Y.; Wang, G.; Liu, S. Outbreak of variant pseudorabies virus in Bartha-K61-vaccinated piglets in central Shandong Province, China. J. Vet. Diagn. Investig. 2015, 27, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, S.; Xu, Q.; Wu, J.; Zhai, X.; Li, S.; Wang, J.; Ni, J.; Yuan, L.; Song, X.; et al. Investigation on pseudorabies prevalence in Chinese swine breeding farms in 2013–2016. Trop. Anim. Health Prod. 2018, 50, 1279–1285. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, H.; Dong, H.; Mehmood, K.; Chang, Z.; Li, K.; Liu, S.; Rehman, M.U.; Nabi, F.; Javed, M.T.; et al. Seroprevalence and risk factors associated with Pseudorabies virus infection in Tibetan pigs in Tibet. BMC Vet. Res. 2018, 14, 25. [Google Scholar] [CrossRef]
- Xia, L.; Sun, Q.; Wang, J.; Chen, Q.; Liu, P.; Shen, C.; Sun, J.; Tu, Y.; Shen, S.; Zhu, J.; et al. Epidemiology of pseudorabies in intensive pig farms in Shanghai, China: Herd-level prevalence and risk factors. Prev. Vet. Med. 2018, 159, 51–56. [Google Scholar] [CrossRef]
- Ji, C.; Wei, Y.; Wang, J.; Zeng, Y.; Pan, H.; Liang, G.; Ma, J.; Gong, L.; Zhang, W.; Zhang, G.; et al. Development of a Dual Fluorescent Microsphere Immunological Assay for Detection of Pseudorabies Virus gE and gB IgG Antibodies. Viruses 2020, 12, 912. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.L.; Xia, S.L.; Wang, Y.; Du, M.; Xiang, G.T.; Cong, X.; Luo, Y.; Li, L.F.; Zhang, L.; Yu, J.; et al. Safety and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant expressing the E2 protein of classical swine fever virus in pigs. Immunol. Lett. 2016, 174, 63–71. [Google Scholar] [CrossRef]
- Aytogu, G.; Toker, E.B.; Yavas, O.; Kadiroglu, B.; Ates, O.; Ozyigit, M.O.; Yesilbag, K. First isolation and molecular characterization of pseudorabies virus detected in Turkey. Mol. Biol. Rep. 2022, 49, 1679–1686. [Google Scholar] [CrossRef]
- Tian, R.B.; Jin, Y.; Xu, T.; Zhao, Y.; Wang, Z.Y.; Chen, H.Y. Development of a SYBR green I-based duplex real-time PCR assay for detection of pseudorabies virus and porcine circovirus 3. Mol. Cell. Probes 2020, 53, 101593. [Google Scholar] [CrossRef]
- Song, C.; Gao, L.; Bai, W.; Zha, X.; Yin, G.; Shu, X. Molecular epidemiology of pseudorabies virus in Yunnan and the sequence analysis of its gD gene. Virus Genes 2017, 53, 392–399. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, C.; Shen, H.; Sun, J.; Zhang, J. Duplex fluorescence melting curve analysis as a new tool for rapid detection and differentiation of genotype I, II and Bartha-K61 vaccine strains of pseudorabies virus. BMC Vet. Res. 2018, 14, 372. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.H.; Bai, Y.L.; Xu, T.; Zheng, L.L.; Li, X.S.; Chen, H.Y.; Wang, Z.Y. Isolation and Phylogenetic Analysis of Reemerging Pseudorabies Virus Within Pig Populations in Central China During 2012 to 2019. Front. Vet. Sci. 2021, 8, 764982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.F.; Cui, S.J.; Zhu, C. Loop-mediated isothermal amplification for rapid detection and differentiation of wild-type pseudorabies and gene-deleted virus vaccines. J. Virol. Methods 2010, 169, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nan, W.; Qin, L.; Gong, M.; Wu, F.; Hu, S.; Chen, Y. A NanoPCR Assay for Detection of Pseudorabies Virus. China Anim. Health Insp. 2017, 34, 102–105. [Google Scholar]
- Ren, M.; Lin, H.; Chen, S.; Yang, M.; An, W.; Wang, Y.; Xue, C.; Sun, Y.; Yan, Y.; Hu, J. Detection of pseudorabies virus by duplex droplet digital PCR assay. J. Vet. Diagn. Investig. 2018, 30, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Tu, F.; Zhang, Y.; Xu, S.; Yang, X.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H. Detection of pseudorabies virus with a real-time recombinase-aided amplification assay. Transbound. Emerg. Dis. 2022, 69, 2266–2274. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, H.; Liu, W.; Li, S.; Guo, D. Diagnosis and gI antibody dynamics of pseudorabies virus in an intensive pig farm in Hei Longjiang Province. J. Vet. Sci. 2021, 22, e23. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Y.; Luo, Y.; Liu, Y.; Shao, L.; Sun, Y.; Li, Y.; Li, S.; Ji, S.; Qiu, H.J. A triplex real-time PCR for differential detection of classical, variant and Bartha-K61 vaccine strains of pseudorabies virus. Arch. Virol. 2016, 161, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liang, L.; Zhou, L.; Zhao, K.; Cui, S. Concurrent infections of pseudorabies virus and porcine bocavirus in China detected by duplex nanoPCR. J. Virol. Methods 2015, 219, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.Y.; Henao-Diaz, A.; Poonsuk, K.; Buckley, A.; van Geelen, A.; Lager, K.; Harmon, K.; Gauger, P.; Wang, C.; Ambagala, A.; et al. Pseudorabies (Aujeszky’s disease) virus DNA detection in swine nasal swab and oral fluid specimens using a gB-based real-time quantitative PCR. Prev. Vet. Med. 2021, 189, 105308. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Zhang, W.; Li, Z.; Zhang, S.; Li, Y.; Zhang, Z. Development of an isothermal recombinase polymerase amplification assay for rapid detection of pseudorabies virus. Mol. Cell. Probes 2017, 33, 32–35. [Google Scholar] [CrossRef]
- Yang, H.; Guo, Y.; Li, S.; Lan, G.; Jiang, Q.; Yang, X.; Fan, J.; Ali, Z.; Tang, Y.; Mou, X.; et al. Magnetic beads-based chemiluminescent assay for ultrasensitive detection of pseudorabies virus. J. Nanosci. Nanotechnol. 2014, 14, 3337–3342. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, X.; Zhang, X.; Xu, H.; Zhao, X.; Zhou, S.; Huang, S.; Liu, X. Establishment of a multiplex RT-PCR assay for identification of atmospheric virus contamination in pig farms. Environ. Pollut. (Barking Essex 1987) 2019, 253, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Sunaga, F.; Tsuchiaka, S.; Kishimoto, M.; Aoki, H.; Kakinoki, M.; Kure, K.; Okumura, H.; Okumura, M.; Okumura, A.; Nagai, M.; et al. Development of a one-run real-time PCR detection system for pathogens associated with porcine respiratory diseases. J. Vet. Med. Sci. 2020, 82, 217–223. [Google Scholar] [CrossRef]
- Lee, C.-S.; Moon, H.-J.; Yang, J.-S.; Park, S.-J.; Song, D.-S.; Kang, B.-K.; Park, B.-K. Multiplex PCR for the simultaneous detection of pseudorabies virus, porcine cytomegalovirus, and porcine circovirus in pigs. J. Virol. Methods 2007, 139, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Pérez, L.J.; Perera, C.L.; Frías, M.T.; Núñez, J.I.; Ganges, L.; de Arce, H.D. A multiple SYBR Green I-based real-time PCR system for the simultaneous detection of porcine circovirus type 2, porcine parvovirus, pseudorabies virus and Torque teno sus virus 1 and 2 in pigs. J. Virol. Methods 2012, 179, 233–241. [Google Scholar] [CrossRef]
- Huang, C.; Hung, J.J.; Wu, C.Y.; Chien, M.S. Multiplex PCR for rapid detection of pseudorabies virus, porcine parvovirus and porcine circoviruses. Vet. Microbiol. 2004, 101, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Tombácz, D.; Sharon, D.; Szűcs, A.; Moldován, N.; Snyder, M.; Boldogkői, Z. Transcriptome-wide survey of pseudorabies virus using next- and third-generation sequencing platforms. Sci. Data 2018, 5, 180119. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, X.T.; Hu, B.; Deng, X.Y.; Zhang, L.; Lian, S.Z.; Zhang, H.L.; Lv, S.; Xue, X.H.; Lu, R.G.; et al. Outbreak of severe pseudorabies virus infection in pig-offal-fed farmed mink in Liaoning Province, China. Arch. Virol. 2017, 162, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Lian, K.; Zhang, M.; Zhou, L.; Song, Y.; Wang, G.; Wang, S. First report of a pseudorabies-virus-infected wolf (Canis lupus) in China. Arch. Virol. 2020, 165, 459–462. [Google Scholar] [CrossRef]
- Guo, X.; Hou, J.; Yuan, Z.; Li, H.; Sang, S. A novel paper biosensor based on Fe(3)O(4)@SiO(2)-NH(2)and MWCNTs for rapid detection of pseudorabies virus. Nanotechnology 2021, 32, 355102. [Google Scholar] [CrossRef] [PubMed]
- McFerran, J.B.; Dow, C. Experimental Aujeszky’s disease (pseudorabies) in rats. Br. Vet. J. 1970, 126, 173–179. [Google Scholar] [CrossRef]
- Su, D.; Wu, S.; Guo, J.; Wu, X.; Yang, Q.; Xiong, X. Protective effect of resveratrol against pseudorabies virus-induced reproductive failure in a mouse model. Food Sci. Biotechnol. 2016, 25, 103–106. [Google Scholar] [CrossRef]
- Lomniczi, B.; Watanabe, S.; Ben-Porat, T.; Kaplan, A.S. Genetic basis of the neurovirulence of pseudorabies virus. J. Virol. 1984, 52, 198–205. [Google Scholar] [CrossRef]
- Yin, H.; Li, Z.; Zhang, J.; Huang, J.; Kang, H.; Tian, J.; Qu, L. Construction of a US7/US8/UL23/US3-deleted recombinant pseudorabies virus and evaluation of its pathogenicity in dogs. Vet. Microbiol. 2020, 240, 108543. [Google Scholar] [CrossRef]
- Kit, S.; Sheppard, M.; Ichimura, H.; Kit, M. Second-generation pseudorabies virus vaccine with deletions in thymidine kinase and glycoprotein genes. Am. J. Vet. Res. 1987, 48, 780–793. [Google Scholar]
- Moormann, R.J.; de Rover, T.; Briaire, J.; Peeters, B.P.; Gielkens, A.L.; van Oirschot, J.T. Inactivation of the thymidine kinase gene of a gI deletion mutant of pseudorabies virus generates a safe but still highly immunogenic vaccine strain. J. Gen. Virol. 1990, 71 Pt 7, 1591–1595. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Weiland, F.; Visser, N. Characterization of a quadruple glycoprotein-deleted pseudorabies virus mutant for use as a biologically safe live virus vaccine. J. Gen. Virol. 1994, 75 Pt 7, 1723–1733. [Google Scholar] [CrossRef]
- Peeters, B.; Bouma, A.; de Bruin, T.; Moormann, R.; Gielkens, A.; Kimman, T. Non-transmissible pseudorabies virus gp50 mutants: A new generation of safe live vaccines. Vaccine 1994, 12, 375–380. [Google Scholar] [CrossRef]
- Ling, Z.; Wan-Zhu, G.; Zhi-Wen, X.U. Fluctuant Rule of Colostral Antibodies and the Date of Initial Immunization for the Piglet from Sows Inoculated with Pseudorabies Virus Gene-deleted Vaccine SA215. Chin. J. Vet. 2004, 24, 320–322. [Google Scholar]
- Xu, X.J.; Xu, G.Y.; Chen, H.C.; Liu, Z.F.; He, Q.G. Construction and characterization of a pseudorabies virus TK-/gG- mutant. China J. Biotechnol. 2004, 20, 532–535. [Google Scholar]
- Ye, C.; Guo, J.C.; Gao, J.C.; Wang, T.Y.; Zhao, K.; Chang, X.B.; Wang, Q.; Peng, J.M.; Tian, Z.J.; Cai, X.H.; et al. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 2016, 491, 56–63. [Google Scholar] [CrossRef]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.J.; Jiang, Y.F.; Shan, T.L.; Gao, F.; Li, G.X.; Tong, G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, Y.; Wang, C.H.; Yuan, J.; Li, N.; Song, K.; Qiu, H.J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016, 183, 119–124. [Google Scholar] [CrossRef]
- Wang, T.; Xiao, Y.; Yang, Q.; Wang, Y.; Sun, Z.; Zhang, C.; Yan, S.; Wang, J.; Guo, L.; Yan, H.; et al. Construction of a gE-Deleted Pseudorabies Virus and Its Efficacy to the New-Emerging Variant PRV Challenge in the Form of Killed Vaccine. BioMed Res. Int. 2015, 2015, 684945. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.; Wang, L.Q.; Zheng, H.H.; Yang, Y.R.; Liu, F.; Zheng, L.L.; Jin, Y.; Chen, H.Y. Construction and immunogenicity of a gE/gI/TK-deleted PRV based on porcine pseudorabies virus variant. Mol. Cell. Probes 2020, 53, 101605. [Google Scholar] [CrossRef]
- Lv, L.; Liu, X.; Jiang, C.; Wang, X.; Cao, M.; Bai, J.; Jiang, P. Pathogenicity and immunogenicity of a gI/gE/TK/UL13-gene-deleted variant pseudorabies virus strain in swine. Vet. Microbiol. 2021, 258, 109104. [Google Scholar] [CrossRef]
- Gu, Z.; Dong, J.; Wang, J.; Hou, C.; Sun, H.; Yang, W.; Bai, J.; Jiang, P. A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 2015, 195, 57–63. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, L.; Jia, X.; Wang, T.; Wang, J.; Sun, Z.; Wang, L.; Li, X.; Tan, F.; Tian, K. Construction of a triple gene-deleted Chinese Pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine 2015, 33, 2432–2437. [Google Scholar] [CrossRef]
- Wang, J.; Cui, X.; Wang, X.; Wang, W.; Gao, S.; Liu, X.; Kai, Y.; Chen, C. Efficacy of the Bartha-K61 vaccine and a gE(-)/gI(-)/TK(-) prototype vaccine against variant porcine pseudorabies virus (vPRV) in piglets with sublethal challenge of vPRV. Res. Vet. Sci. 2020, 128, 16–23. [Google Scholar] [CrossRef]
- Dong, J.; Bai, J.; Sun, T.; Gu, Z.; Wang, J.; Sun, H.; Jiang, P. Comparative pathogenicity and immunogenicity of triple and double gene-deletion pseudorabies virus vaccine candidates. Res. Vet. Sci. 2017, 115, 17–23. [Google Scholar] [CrossRef]
- Sun, L.; Tang, Y.; Yan, K.; Zhang, H. Construction of a quadruple gene-deleted vaccine confers complete protective immunity against emerging PRV variant challenge in piglets. Virol. J. 2022, 19, 19. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Chen, S.; Qiao, Y.; Guo, M.; Zheng, Y.; Xu, M.; Wang, Z.; Hou, J.; Wang, J. A gD&gC-substituted pseudorabies virus vaccine strain provides complete clinical protection and is helpful to prevent virus shedding against challenge by a Chinese pseudorabies variant. BMC Vet. Res. 2019, 15, 2. [Google Scholar] [CrossRef]
- Yin, Y.; Xu, Z.; Liu, X.; Li, P.; Yang, F.; Zhao, J.; Fan, Y.; Sun, X.; Zhu, L. A live gI/gE-deleted pseudorabies virus (PRV) protects weaned piglets against lethal variant PRV challenge. Virus Genes 2017, 53, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Huang, B.; Bai, X.; Zhou, Y.; Guo, L.; Wang, T.; Shan, Y.; Wang, Y.; Tan, F.; Tian, K. Construction and Immunogenicity of a Recombinant Pseudorabies Virus Variant With TK/gI/gE/11k/28k Deletion. Front. Vet. Sci. 2021, 8, 797611. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fang, K.; Rong, Z.; Li, X.; Ren, X.; Ma, H.; Chen, H.; Li, X.; Qian, P. Comparison of gE/gI- and TK/gE/gI-Gene-Deleted Pseudorabies Virus Vaccines Mediated by CRISPR/Cas9 and Cre/Lox Systems. Viruses 2020, 12, 369. [Google Scholar] [CrossRef]
- Quint, W.; Gielkens, A.; Van Oirschot, J.; Berns, A.; Cuypers, H.T. Construction and characterization of deletion mutants of pseudorabies virus: A new generation of ‘live’ vaccines. J. Gen. Virol. 1987, 68 Pt 2, 523–534. [Google Scholar] [CrossRef]
- Wang, J.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Gu, Y.; Liu, C.; Hou, J. An inactivated gE-deleted pseudorabies vaccine provides complete clinical protection and reduces virus shedding against challenge by a Chinese pseudorabies variant. BMC Vet. Res. 2016, 12, 277. [Google Scholar] [CrossRef][Green Version]
- Visser, N.; Luetticken, D. Experiences with a gI-/TK- modified live pseudorabies virus vaccine: Strain Begonia. Curr. Top. Vet. Med. 1989, 48, 37–44. [Google Scholar]
- Wardley, R.C.; Post, L.E. The use of the gX deleted vaccine PRV.TK.gX-1 in the control of Aujeszky’s disease. In Proceedings of the CEC Seminar on Vaccination and Control of Aujeszky’s Disease, Brussels, Belgium, 5–6 July 1988; Van Oirschot, J.T., Ed.; Kluwer Academic: Dordrecht, The Netherlands, 1989; pp. 13–25. [Google Scholar]
- Yang, D.K.; Kim, H.H.; Choi, S.S.; Hyun, B.H.; Song, J.Y. An oral Aujeszky’s disease vaccine (YS-400) induces neutralizing antibody in pigs. Clin. Exp. Vaccine Res. 2016, 5, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Li, G.; Liang, C.; Liu, F.; Tian, Q.; Cao, Y.; Li, L.; Zheng, X.; Zheng, H.; Tong, G. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antivir. Res. 2016, 130, 110–117. [Google Scholar] [CrossRef]
- Wu, C.Y.; Liao, C.M.; Chi, J.N.; Chien, M.S.; Huang, C. Growth properties and vaccine efficacy of recombinant pseudorabies virus defective in glycoprotein E and thymidine kinase genes. J. Biotechnol. 2016, 229, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, Z.; Ge, A.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Zheng, Y.; Fan, H.; et al. Safety and immunogenicity of an attenuated Chinese pseudorabies variant by dual deletion of TK&gE genes. BMC Vet. Res. 2018, 14, 287. [Google Scholar] [CrossRef]
- Ferrari, M.; Brack, A.; Romanelli, M.G.; Mettenleiter, T.C.; Corradi, A.; Dal Mas, N.; Losio, M.N.; Silini, R.; Pinoni, C.; Pratelli, A. A study of the ability of a TK-negative and gI/gE-negative pseudorabies virus (PRV) mutant inoculated by different routes to protect pigs against PRV infection. J. Vet. Med. B 2000, 47, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yi, Y.; Xu, Z.; Cheng, L.; Tang, S.; Guo, W. Growth, physicochemical properties, and morphogenesis of Chinese wild-type PRV Fa and its gene-deleted mutant strain PRV SA215. Virol. J. 2011, 8, 272. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.M.; Zhou, Q.; Song, W.B.; Sun, E.C.; Zhang, M.M.; He, Q.G.; Chen, H.C.; Wu, B.; Liu, Z.F. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine 2015, 33, 5733–5740. [Google Scholar] [CrossRef]
- Tang, Y.D.; Liu, J.T.; Wang, T.Y.; An, T.Q.; Sun, M.X.; Wang, S.J.; Fang, Q.Q.; Hou, L.L.; Tian, Z.J.; Cai, X.H. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus Res. 2016, 225, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, Z.; Feng, Z.; Fang, Z.; Chen, J.; Chen, W.; Liang, W.; Chen, Q. Pseudorabies virus (PRV) strain with defects in gE, gC, and TK genes protects piglets against an emerging PRV variant. J. Vet. Med. Sci. 2020, 82, 846–855. [Google Scholar] [CrossRef]
- Gao, J.F.; Lai, Z.; Shu, Y.H.; Qi, S.; Ma, J.; Wu, B.; Gong, J.P. Isolation and identification of porcine pseudorabies virus(PRV)C strain. Acta Agric. Shanghai 2015, 31, 32–36. [Google Scholar]
- Kimman, T.G.; de Wind, N.; Oei-Lie, N.; Pol, J.M.; Berns, A.J.; Gielkens, A.L. Contribution of single genes within the unique short region of Aujeszky’s disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J. Gen. Virol. 1992, 73 Pt 2, 243–251. [Google Scholar] [CrossRef]
- Dong, B.; Zarlenga, D.S.; Ren, X. An overview of live attenuated recombinant pseudorabies viruses for use as novel vaccines. J. Immunol. Res. 2014, 2014, 824630. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, W.; Xu, Z.; Yan, Q.; Luo, Y.; Shi, Q.; Chen, D.; Zhu, L.; Wang, X. A novel recombinant pseudorabies virus expressing parvovirus VP2 gene: Immunogenicity and protective efficacy in swine. Virol. J. 2011, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Zheng, H.; Li, G.X.; Gao, F.; Shan, T.L.; Zhou, Y.J.; Yu, H.; Jiang, Y.F.; Yu, L.X.; Li, L.W.; et al. Recombinant pseudorabies virus expressing E2 of classical swine fever virus (CSFV) protects against both virulent pseudorabies virus and CSFV. Antivir. Res. 2020, 173, 104652. [Google Scholar] [CrossRef]
- Zheng, H.H.; Wang, L.Q.; Fu, P.F.; Zheng, L.L.; Chen, H.Y.; Liu, F. Characterization of a recombinant pseudorabies virus expressing porcine parvovirus VP2 protein and porcine IL-6. Virol. J. 2020, 17, 19. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, J.; Cong, X.; Qin, H.Y.; Wang, C.H.; Li, Y.; Li, S.; Luo, Y.; Sun, Y.; Qiu, H.J. Generation and Efficacy Evaluation of a Recombinant Pseudorabies Virus Variant Expressing the E2 Protein of Classical Swine Fever Virus in Pigs. Clin. Vaccine Immunol. 2015, 22, 1121–1129. [Google Scholar] [CrossRef]
- Yao, L.; Wu, C.X.; Zheng, K.; Xu, X.J.; Zhang, H.; Chen, C.F.; Liu, Z.F. Immunogenic response to a recombinant pseudorabies virus carrying bp26 gene of Brucella melitensis in mice. Res. Vet. Sci. 2015, 100, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Chen, J.; Liang, W.; Chen, W.; Li, Z.; Chen, Q.; Cai, S. The recombinant pseudorabies virus expressing African swine fever virus CD2v protein is safe and effective in mice. Virol. J. 2020, 17, 180. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, X.; Shao, G.; Shao, Y.; Hu, Z.; Feng, K.; Xie, Z.; Li, H.; Chen, W.; Lin, W.; et al. Construction and Evaluation of Recombinant Pseudorabies Virus Expressing African Swine Fever Virus Antigen Genes. Front. Vet. Sci. 2022, 9, 832255. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhang, E.; Zhou, M.; Lin, J.; Yang, Q. Intranasal administration with recombinant Bacillus subtilis induces strong mucosal immune responses against pseudorabies. Microb. Cell Factories 2019, 18, 103. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, A.; Feng, H.; Wei, Q.; Zhou, E.; Zhang, G. A single dose glycoprotein D-based subunit vaccine against pseudorabies virus infection. Vaccine 2020, 38, 6153–6161. [Google Scholar] [CrossRef]
- Kong, H.; Zhang, K.; Liu, Y.; Shang, Y.; Wu, B.; Liu, X. Attenuated live vaccine (Bartha-K16) caused pseudorabies (Aujeszky’s disease) in sheep. Vet. Res. Commun. 2013, 37, 329–332. [Google Scholar] [CrossRef]
- Lin, W.; Shao, Y.; Tan, C.; Shen, Y.; Zhang, X.; Xiao, J.; Wu, Y.; He, L.; Shao, G.; Han, M.; et al. Commercial vaccine against pseudorabies virus: A hidden health risk for dogs. Vet. Microbiol. 2019, 233, 102–112. [Google Scholar] [CrossRef]
- Moreno, A.; Chiapponi, C.; Sozzi, E.; Morelli, A.; Silenzi, V.; Gobbi, M.; Lavazza, A.; Paniccià, M. Detection of a gE-deleted Pseudorabies virus strain in an Italian red fox. Vet. Microbiol. 2020, 244, 108666. [Google Scholar] [CrossRef]
- Cheng, C.; Yu, X. Research Progress in Chinese Herbal Medicines for Treatment of Sepsis: Pharmacological Action, Phytochemistry, and Pharmacokinetics. Int. J. Mol. Sci. 2021, 22, 11078. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhang, J.; Lai, R.; Li, Q.; Ju, J.; Xu, H. Chinese Herbal Medicines and Active Metabolites: Potential Antioxidant Treatments for Atherosclerosis. Front. Pharmacol. 2021, 12, 675999. [Google Scholar] [CrossRef]
- Sun, J.; Ren, J.; Hu, X.; Hou, Y.; Yang, Y. Therapeutic effects of Chinese herbal medicines and their extracts on diabetes. Biomed. Pharmacother. 2021, 142, 111977. [Google Scholar] [CrossRef]
- Hsiao, Y.H.; Lin, C.W.; Wang, P.H.; Hsin, M.C.; Yang, S.F. The Potential of Chinese Herbal Medicines in the Treatment of Cervical Cancer. Integr. Cancer Ther. 2019, 18, 1534735419861693. [Google Scholar] [CrossRef]
- Wang, Z.; Qi, F.; Cui, Y.; Zhao, L.; Sun, X.; Tang, W.; Cai, P. An update on Chinese herbal medicines as adjuvant treatment of anticancer therapeutics. Biosci. Trends 2018, 12, 220–239. [Google Scholar] [CrossRef]
- Zhao, M.; Yu, Y.; Wang, R.; Chang, M.; Ma, S.; Qu, H.; Zhang, Y. Mechanisms and Efficacy of Chinese Herbal Medicines in Chronic Kidney Disease. Front. Pharmacol. 2020, 11, 619201. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L. Chinese herbal medicine: Fighting SARS-CoV-2 infection on all fronts. J. Ethnopharmacol. 2021, 270, 113869. [Google Scholar] [CrossRef]
- Yang, H.; Jiang, C.; Chen, X.; He, K.; Hu, Y. Protective effects of sinomenine against LPS-induced inflammation in piglets. Microb. Pathog. 2017, 110, 573–577. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Zhang, X.L.; Liang, X.Q.; Gu, H.G.; Zhu, P.T. Effects of different Chinese herbal medicines on biochemical parameters in guinea-pig with pigment gallstones. J. Chin. Integr. Med. 2008, 6, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Horng, C.T.; Tsai, S.C.; Lee, Y.L.; Hsu, S.C.; Tsai, Y.J.; Tsai, F.J.; Chiang, J.H.; Kuo, D.H.; Yang, J.S. Relaxant and vasoprotective effects of ginger extracts on porcine coronary arteries. Int. J. Mol. Med. 2018, 41, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Duan, J.; Mu, F.; Bian, H.; Zhao, M.; Zhou, M.; Li, Y.; Wen, A.; Yang, Y.; Xi, M. Cardioprotective effects and underlying mechanism of Radix Salvia miltiorrhiza and Lignum Dalbergia odorifera in a pig chronic myocardial ischemia model. Int. J. Mol. Med. 2018, 42, 2628–2640. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, X.; Song, X.; Muhammad, A.; Jia, R.; Zou, Y.; Yin, L.; Li, L.; He, C.; Ye, G.; et al. The immune-adjuvant activity and the mechanism of resveratrol on pseudorabies virus vaccine in a mouse model. Int. Immunopharmacol. 2019, 76, 105876. [Google Scholar] [CrossRef]
- He, W.; Zhai, X.; Su, J.; Ye, R.; Zheng, Y.; Su, S. Antiviral Activity of Germacrone against Pseudorabies Virus in Vitro. Pathogens 2019, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Chen, Z.; Liu, F.; Qiao, Y.; Chen, T.; Wang, X. Antiviral activities of Radix isatidis polysaccharide against pseudorabies virus in swine testicle cells. BMC Complementary Med. Ther. 2020, 20, 48. [Google Scholar] [CrossRef]
- Lv, C.; Liu, W.; Wang, B.; Dang, R.; Qiu, L.; Ren, J.; Yan, C.; Yang, Z.; Wang, X. Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antivir. Res. 2018, 159, 55–62. [Google Scholar] [CrossRef]
- Chen, X.; Song, X.; Li, L.; Chen, Y.; Jia, R.; Zou, Y.; Wan, H.; Zhao, L.; Tang, H.; Lv, C.; et al. Resveratrol Inhibits Pseudorabies Virus Replication by Targeting IE180 Protein. Front. Microbiol. 2022, 13, 891978. [Google Scholar] [CrossRef]
- Pezzuto, J.M. Resveratrol: Twenty Years of Growth, Development and Controversy. Biomol. Ther. 2019, 27, 1–14. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, Q.; Fu, Q.; Song, X.; Jia, R.; Yang, Y.; Zou, Y.; Li, L.; He, C.; Liang, X.; et al. Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Sci. Rep. 2017, 7, 8782. [Google Scholar] [CrossRef]
- Galindo, I.; Hernáez, B.; Berná, J.; Fenoll, J.; Cenis, J.L.; Escribano, J.M.; Alonso, C. Comparative inhibitory activity of the stilbenes resveratrol and oxyresveratrol on African swine fever virus replication. Antivir. Res. 2011, 91, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, R.; Hu, H.; Chen, X.; Yin, Z.; Liang, X.; He, C.; Yin, L.; Ye, G.; Zou, Y.; et al. The antiviral activity of kaempferol against pseudorabies virus in mice. BMC Vet. Res. 2021, 17, 247. [Google Scholar] [CrossRef]
- Huan, C.; Zhou, Z.; Yao, J.; Ni, B.; Gao, S. The Antiviral Effect of Panax Notoginseng Polysaccharides by Inhibiting PRV Adsorption and Replication In Vitro. Molecules 2022, 27, 1254. [Google Scholar] [CrossRef]
- Huan, C.; Zhang, W.; Xu, Y.; Ni, B.; Gao, S. Plantago asiaticaAntiviral Activity of Polysaccharide against Pseudorabies Virus. Oxidative Med. Cell. Longev. 2022, 2022, 3570475. [Google Scholar] [CrossRef]
- Sun, Y.; Li, C.; Li, Z.; Shangguan, A.; Jiang, J.; Zeng, W.; Zhang, S.; He, Q. Quercetin as an antiviral agent inhibits the Pseudorabies virus in vitro and in vivo. Virus Res. 2021, 305, 198556. [Google Scholar] [CrossRef]
- Hsuan, S.L.; Chang, S.C.; Wang, S.Y.; Liao, T.L.; Jong, T.T.; Chien, M.S.; Lee, W.C.; Chen, S.S.; Liao, J.W. The cytotoxicity to leukemia cells and antiviral effects of Isatis indigotica extracts on pseudorabies virus. J. Ethnopharmacol. 2009, 123, 61–67. [Google Scholar] [CrossRef]
- Zhu, W.; Zou, M.; Sun, M.; Liu, J.; Wang, Y. The anti-PRV and anti-PRRSV effects of Marine Bacillus S-12 low temperature lysozyme in vitro. China J. Mar. Drugs 2013, 32, 26–32. [Google Scholar] [CrossRef]
- Sui, X.; Yin, J.; Ren, X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antivir. Res. 2010, 85, 346–353. [Google Scholar] [CrossRef]
- Liu, J.; Tu, C.; Zhang, M.; Hou, S.; Wang, E. Activity of vanadium-substituted heteropolytungstate to antipseudorabies virus. China J. Appl. Chem. 1998, 15, 41–44. [Google Scholar]
- Ren, X.; Li, G.; Sui, X. Antiviral activities of phosphonoformate sodium to pseudorabies herpesvirus infection in vitro. Pharm. Biol. 2011, 49, 608–613. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, X.; Chen, Q.; Yi, S.; Chen, J.; Xiao, Q.; Yu, M.; Wei, Y.; Hu, T. Effects of Quercitrin on PRV-Induced Secretion of Reactive Oxygen Species and Prediction of lncRNA Regulatory Targets in 3D4/2 Cells. Antioxidants 2022, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Shao, K.; Li, Z.; Guo, N.; Zuo, Y.; Li, Q.; Lu, Z.; Chen, L.; He, Q.; Han, H. Antiviral Activity of Graphene Oxide: How Sharp Edged Structure and Charge Matter. ACS Appl. Mater. Interfaces 2015, 7, 21571–21579. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Reviews. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.R.; Abd-Aziz, N.; Affendi, S.; Poh, C.L. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020, 27, 4. [Google Scholar] [CrossRef]
- Tan, L.; Yuan, X.; Liu, Y.; Cai, X.; Guo, S.; Wang, A. Non-muscle Myosin II: Role in Microbial Infection and Its Potential as a Therapeutic Target. Front. Microbiol. 2019, 10, 401. [Google Scholar] [CrossRef]
- Wang, Y.P.; Huang, L.P.; Du, W.J.; Wei, Y.W.; Wu, H.L.; Feng, L.; Liu, C.M. Targeting the pseudorabies virus DNA polymerase processivity factor UL42 by RNA interference efficiently inhibits viral replication. Antivir. Res. 2016, 132, 219–224. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science (New York, NY, USA) 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Stagakis, E.; Bertsias, G.; Verginis, P.; Nakou, M.; Hatziapostolou, M.; Kritikos, H.; Iliopoulos, D.; Boumpas, D.T. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann. Rheum. Dis. 2011, 70, 1496–1506. [Google Scholar] [CrossRef]
- Huang, J.; Ma, G.; Fu, L.; Jia, H.; Zhu, M.; Li, X.; Zhao, S. Pseudorabies viral replication is inhibited by a novel target of miR-21. Virology 2014, 456, 319–328. [Google Scholar] [CrossRef]
- Liu, H.; Yang, L.; Shi, Z.; Lv, R.; Yang, X.; Wang, C.; Chen, L.; Chang, H. Functional analysis of prv-miR-LLT11a encoded by pseudorabies virus. J. Vet. Sci. 2019, 20, e68. [Google Scholar] [CrossRef]
- Laval, K.; Enquist, L. The Neuropathic Itch Caused by Pseudorabies Virus. Pathogens 2020, 9, 254. [Google Scholar] [CrossRef] [PubMed]

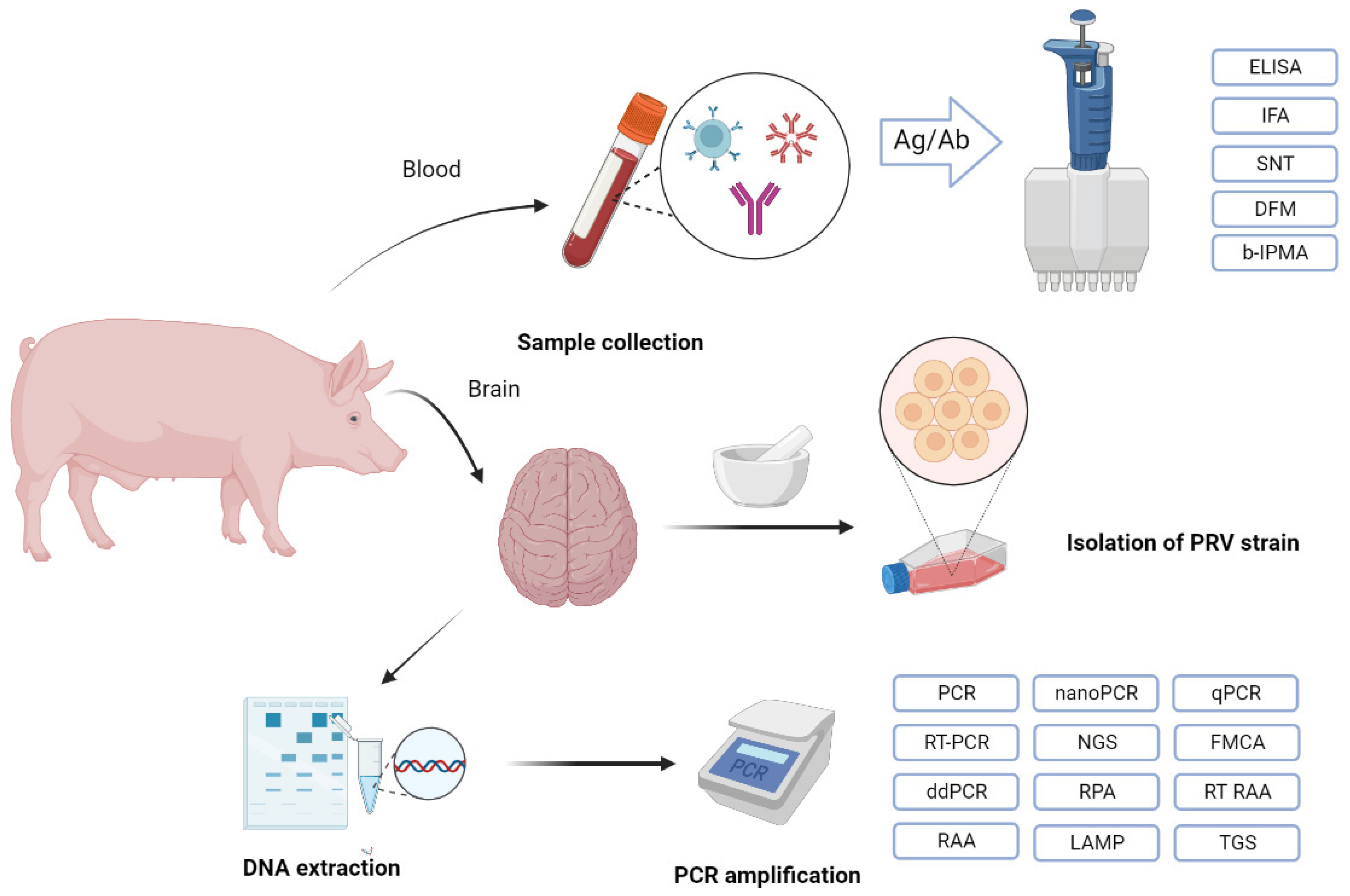
| Molecular | Serology | |
|---|---|---|
| Test type | Viral | Antibody |
| Description | Nucleic acid amplification test to detect viral DNA | Detects the presence of IgA, IgM/IgG antibodies against PRV |
| Platform technology | PCR, RT-PCR, LAMP, qPCR, ddPCR, FMCA | ELISA, SNT, IFA, IPMA, DFM |
| Sample type | Brains, Hearts, livers, spleens, lungs, kidneys and lymph nodes | Plasma, serum, whole blood |
| Result turnaround time | <5 h | 15–30 min |
| Name of Diagnostic Assay | Sensitivity | Target Gene | Turnaround Time | Samples Used | References |
|---|---|---|---|---|---|
| Conventional polymerase chain reaction (PCR) | —— | gE gene | Result in <5 h | Various tissue | [119,121] |
| Duplex droplet digital PCR (ddPCR) assary | 4.75 copies/µL | Both gE and gB genes | Result in <2 h | Lung, brain, liver and spleen | [124] |
| SYBR green I-based duplex real-time PCR assay | 37.8 copies/μL | gE gene | Result within 50 min | Hearts, livers, spleens, lungs, kidneys, brains and lymph nodes | [118] |
| Real-time recombinase-aided amplification assay (RAA) | Three 50% TCID50 | gE gene | Result in 75 min | Lung, lymph node, tonsil and spleen | [125] |
| Triplex real-time PCR | 0.5 TCID50 for classical strains, 0.2 TCID50 for variant strains and 0.05 TCID50 for vaccine strains | gE and gI genes | Result within 1 h | PRV strains | [127] |
| Probe-based fluorescence melting curve analysis (FMCA) | 1 × 100 copies per reaction | gC and gE genes | Result in <2 h | PRV strains | [120] |
| Loop-mediated isothermal amplification (LAMP) assay | 10 copies per sample | gE and gG genes | Result in <2 h | PRV strains and clinical tissue samples | [122] |
| Duplex nanoparticle-assisted polymerase chain reaction (nanoPCR) | 6 copies/μL | gE gene | Result in 80 min | The recombinant plasmids pET30a-PRV-gE and pUC57-PBoVNS1 | [128] |
| Real-time quantitative PCR (RT-qPCR) | Oral fluid of 53% and nasal swab of 70% | gB gene | Result in <1 h | Oral fluid and nasal swab | [129] |
| Metagenomic next-and third-generation sequencing (mNGS/TGS) | —— | Short- and long-read sequencing | —— | Brains | [137] |
| Real-time fluorescent detection (real-time RPA assay) | 100 copies per reaction | gD gene | Result within 20 min | Tissue | [130] |
| Lateral flow dipstick (RPA LFD assay) | 160 copies per reaction | gD gene | Result within 20 min | Tissue | [130] |
| Magnetic beads-based chemiluminescent assay | 100 μmol/5 pM | —— | Result in 20 min | Serum samples | [131] |
| Gene-Deleted Vaccines | Vaccine Strains | Progenitor Strains | Deleted Gene | Technology Used | Authorization | References |
|---|---|---|---|---|---|---|
| Single gene-deleted vaccine | Omnivac | BUK | TK gene | Natural losses | Licensed | [27] |
| 2.4N3A | NIA-3 (field strain) | gE gene | HR | Licensed | [166] | |
| PRV(LA-AB) | AH02LA (field strain) | gE gene | BCA | Not available | [167] | |
| HN1201ΔgE (inactivated) | HN1201 (field strain) | gE gene | HR | Licensed | [154] | |
| rPRVTJ-delgE | TJ (field strain) | gE gene | HR | Not available | [11] | |
| Double gene-deleted vaccine | Omnimark | Omnivac (BUK) | TK and gIII genes | Natural losses | Licensed | [145] |
| Begonia | 2.4N3A (NIA-3) | TK and gE genes | Natural losses | Licensed | [168] | |
| NIA3-783 | 2.4N3A (NIA-3) | TK and gE genes | HR | Licensed | [146] | |
| Tolvi | field strain | TK and gpX genes | HR | Licensed | [169] | |
| D1200/D560 | NIA-3 | gD and gI genes | HR | Not available | [148] | |
| AD-YS400 | Yangsan (field strain) | TK and gE genes | HR | Not available | [170] | |
| JS-2012-ΔgE/gI | JS-2012 (field strain) | gE and gI genes | HR | Not available | [171] | |
| gE-TK-PRV | TNL (field strain) | TK and gE genes | HR | Not available | [172] | |
| vZJ01ΔgE/gI (inactivated) | ZJ01 (field strain) | gE and gI genes | BCA | Not available | [157] | |
| PRV (PRVΔTK&gE-AH02) | AH02LA (field strain) | TK and gE genes | HR | Not available | [173] | |
| Triple gene-deleted vaccine | 6C2 | Field strain | TK, gE and gI genes | HR | Not available | [174] |
| SA215 | Fa (classical strain) | gE, gI and TK genes | HR | Licensed | [175] | |
| rSMXΔgI/gEΔTK | Field strain | TK, gE and gI genes | HR | Not available | [176] | |
| rPRVTJ-delgE/gI/TK- | rPRVTJ-delgE (TJ strain) | TK, gE and gI genes | HR | Not available | [29] | |
| vPRV HN1201 | HN1201 (field strain) | TK, gE and gI genes | HR | Not available | [158] | |
| gE-/gI-/TK- PRV | HeN1 (field strain) | TK, gE and gI genes | CRISPR/Cas9 | Not available | [177] | |
| rPRV NY-gE−/gI−/TK− | NY (field strain) | TK, gE and gI genes | HR and CRISPR/Cas9 | Not available | [155] | |
| 201715 (field strain) | gE, gC and TK genes | CRISPR/Cas9 | Not available | [178] | ||
| rPRV/XJ5-gE−/gI−/TK− | XJ5 (field strain) | gE, gI and TK genes | HR | Not available | [159] | |
| rGXΔTK/gE/gI | GX (field strain) | TK, gE and gI genes | Not available | [165] | ||
| Four gene-deleted vaccine | PrV (376) | PrV (376) | gD, gG, gI and gE genes | Not available | [147] | |
| —— | C (field strain) | gI, gE, Us9 and Us2 genes | Natura losses | Licensed | [179] | |
| PRV GDFS-delgI/gE/US9/US2 | GDFS (field strain) | gI, gE, Us9 and Us2 genes | CRISPR/Cas9 | Not available | [161] | |
| rZJ01-ΔgI/gE/TK/UL13 | ZJ01 | gI, gE, TK and UL13 genes | CRISPR/Cas9 | Not available | [29] | |
| Five gene-deleted vaccine | PRV rHN1201TK−/gE−/gI−/11k−/28k− | HN1201 (field strain)) | TK, gI, gE, 11k and 28k genes | BCA | Not available | [164] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, H.-H.; Fu, P.-F.; Chen, H.-Y.; Wang, Z.-Y. Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses 2022, 14, 1638. https://doi.org/10.3390/v14081638
Zheng H-H, Fu P-F, Chen H-Y, Wang Z-Y. Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses. 2022; 14(8):1638. https://doi.org/10.3390/v14081638
Chicago/Turabian StyleZheng, Hui-Hua, Peng-Fei Fu, Hong-Ying Chen, and Zhen-Ya Wang. 2022. "Pseudorabies Virus: From Pathogenesis to Prevention Strategies" Viruses 14, no. 8: 1638. https://doi.org/10.3390/v14081638
APA StyleZheng, H.-H., Fu, P.-F., Chen, H.-Y., & Wang, Z.-Y. (2022). Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses, 14(8), 1638. https://doi.org/10.3390/v14081638






