Abstract
Classical swine fever can be controlled effectively by vaccination with C-strain vaccine. In this study, we developed a novel competitive enzyme-linked immunosorbent assay (cELISA) based on a C-strain Erns specific monoclonal antibody (mAb 1504), aiming to serologically measure immune responses to C-strain vaccine in pigs, and finally to make the C-strain become a DIVA-compatible vaccine. The cELISA system was established based on the strategy that mAb 1504 will compete with the C-strain induced antibodies in the pig serum to bind the C-strain Erns protein. The cELISA was optimized and was further evaluated by testing different categories of pig sera. It can efficiently differentiate C-strain immunized from wild-type CSFV-infected pigs and lacks cross-reaction with other common swine viruses and viruses in genus Pestivirus such as Bovine viral diarrhea virus (BVDV). The C-strain antibody can be tested in pigs 7–14 days post vaccination with this cELISA. The sensitivity and specificity of the established cELISA were 100% (95% confidence interval: 95.60 to 100%) and 100% (95% confidence interval: 98.30 to 100%), respectively. This novel cELISA is a reliable tool for specifically measuring and differentiating immune responses to C-strain vaccine in pigs. By combining with the wild-type CSFV-specific infection tests, it can make the C-strain have DIVA capability.
1. Introduction
Classical swine fever (CSF) is a highly contagious viral disease of domestic and wild pigs [1]. Despite the enormous control efforts, it continues to cause significant economic losses to the swine industry worldwide and represents a high-consequence threat to agriculture security and trade for CSF-free countries such as the United States [2,3,4]. The conventional Chinese vaccine (C-strain) is the most frequently used live attenuated vaccine for CSF control and prevention [5]. It was attenuated from a virulent strain over at least 480 passages in rabbits [6]. The immunity has been proven to persist for at least six to eleven months, probably even lifelong. It can cover all different genotypes, does not require adjuvants, and is suitable for the oral vaccination of wild boar populations [7,8,9]. The only drawback of the C-strain vaccine is the lack of a reliable accompanying diagnostic assay that allows the differentiation of infected from vaccinated animals (DIVA), which has hindered its application in the control and elimination of CSF outbreaks, especially in CSF-free countries.
CSF is caused by the classical swine fever virus (CSFV), a small enveloped single stranded positive-sense RNA virus that belongs to the genus Pestivirus in the family Flaviviridae [1]. CSFV exists as a single serotype and has evolved into three distinct genotypes and eleven sub-genotypes based on phylogenetic analysis with E2, 5′ UTR, or NS5B gene sequences [10,11,12]. The viral envelope glycoprotein Erns is one of the major targets for eliciting antibodies against CSFV in infected animals [13,14]. It has been shown that antibodies to Erns can be used as an indicator of CSFV infection in pigs. The Erns-based enzyme-linked immunosorbent assay (ELISA) can be used as a companion diagnostic test to identify CSFV-infected pigs vaccinated with the E2-based subunit or marker vaccines [15,16,17,18,19]. Here, we describe a competitive ELISA (cELISA) developed with a C-strain Erns specific monoclonal antibody (mAb), which can specifically measure and differentiate immune responses to C-strain vaccine in pigs.
2. Materials and Methods
2.1. Animals
Five female Balb/c mice (six weeks old) were purchased from Charles River Laboratories, Inc. Wilmington, MA, USA. The mice were fed with a standard commercial diet and housed in a clean facility at Kansas State University. Animal care and protocols were approved by Institutional Animal Care and Use Committee (IACUC#4490) at Kansas State University. All animal experiments were performed under strict adherence to the IACUC protocols.
2.2. Cell Lines and Media
Spodoptera frugiperda (Sf9; ATCC, Manassas, VA, USA) and High Five (ATCC, Manassas, VA, USA) insect cells were grown at 27 °C under an air atmosphere in Grace’s insect medium (Gibco, New York, NY, USA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA, USA) and 1% antibiotic-antimycotic solution (Gibco, New York, NY, USA), and Express Five SFM medium (Gibco, New York, NY, USA), respectively. Murine myeloma cell line Sp2/0Ag14 was purchased from the American Type Culture Collection (ATCC-CRL-1581, Rockville, MD, USA) and was maintained in RPMI-1640 (Gibco, New York, NY, USA) supplemented with 10% FBS at 37 °C with 5% CO2.
2.3. Serum Samples
The negative control sera from phosphate-buffered saline (PBS) inoculated pigs (n = 159), C-strain/ C-strain Erns immunized pig sera (n = 45), C-strain E2 immunized pig sera (n = 151), CSFV Alford (Genotype 1.1)-infected pig sera (n = 223), CSFV Honduras/1997 (Genotype 1.3)-infected pig sera (n = 59), porcine reproductive and respiratory syndrome virus (PRRSV)-infected pig sera (n = 107) are kept in our laboratory at Kansas State University. Pseudorabies virus (PRV)-infected pig serum sample (n = 1) and BVDV-infected pig serum samples (n = 2) were purchased from National Veterinary Services Laboratories (NVSL, Ames, IA, USA). The existence of CSFV Erns antibodies in these serum samples were tested by ELISA as we described previously [20]. Briefly, 62.5 ng/mL of purified Erns was used as coating antigen on 96-well flat-bottomed microtiter plates (Corning®, ARI, Corning, NY, USA). Diluted sera (1:2000) were added to plates and incubated for 1 h (hr) at room temperature. Then, horseradish peroxidase (HRP)-conjugated goat anti-porcine IgG was used as secondary antibody (Southern Biotech, Birmingham, AL, USA). The ELISA plates were developed using 3,3,5,5 tetramethylbenzidine (TMB) stabilized chromogen (Thermo Scientific, Rockford, IL, USA), and the reactions were stopped with 2 N sulfuric acid. Relative antibody concentrations were determined using optical spectrophotometer readings at 450 nm using a SpectraMAX microplate reader.
2.4. Expression of C-Strain Erns Protein and Generation of Monoclonal Antibodies
Expression and purification of C-strain Erns protein using a baculovirus expression system were performed as we previously described [20]. Briefly, PCR amplified Erns gene from hog cholera lapinized virus C-strain (HCLV, Genotype 1.1) was cloned into pFastBacTM 1 plasmid and transformed into DH10BacTM E. coli host strain (Thermo Scientific, IL, USA). To generate recombinant baculovirus stock for Erns expression, Sf9 insect cells were transfected with the bacmid DNA extracted from the DH10BacTM E. coli and passaged three times to amplify the Erns bearing recombinant baculovirus. At passage 3, Sf9 cell culture supernatant was collected and clarified by centrifugation at 500× g for 5 min to obtain the baculovirus stock that was used to infect High Five™ Cells for Erns expression. Erns protein was purified using Ni-NTA Agarose (Thermo Scientific, IL, USA) as described by the manufacturer. The purified C-strain Erns protein was concentrated using Amicon Ultra Centrifugal Filters 30,000 NMWL (Millipore, Billerica, MA, USA) and measured using BCA assay kit (Thermo Scientific, IL, USA) according to the manufacture’s recommendations.
Generation of monoclonal antibodies against C-strain Erns were performed as we previously described [21]. Briefly, 50 µL (1 µg/µL) purified Erns protein plus equal volume of Alhydrogel 2% (InvivoGen, San Diego, CA, USA) was used as an immunogen to inject each of five female Balb/c mice (purchased from Charles River Laboratories, Inc. Wilmington, MA, USA) via intraperitoneal injection for mAb production. Three booster immunizations with same dose were conducted at two-week intervals. Three days after the final booster injection, the mice were euthanized and spleen cells were fused with the mouse myeloma partner SP2/0-Ag14 (ATCC, Gaithersburg, MD, USA) by using polyethylene glycol 1500 (Roche Diagnostics, Indianapolis, IN, USA) at a ratio of 10:1. The hybridoma cells were maintained in RPMI1640 medium (Gibco, New York, NY, USA) with 20% FBS. Supernatants from growing hybridomas were screened by an ELISA for reactivity to Erns protein. The positive hybridoma clones were subcloned three times by limiting dilution until monoclonals were obtained. Isotypes of the generated mAbs were determined with an antibody-isotyping kit (Roche Diagnostics, Indianapolis, IN, USA).
2.5. SDS-PAGE and Western Blotting
For analyzing the insect cell expressed C-strain Erns protein, the purified protein was treated without (Native) or with β-mercaptoethanol (Reduced), separated by SDS-PAGE in a Mini-Protean TGX Gel (Bio-Rad, Hercules, CA, USA) and stained with PageBlue Protein Staining Solution (Thermo Scientific, IL, USA) according to the manufacture’s recommendations.
For analyzing if the monoclonal antibody recognizes linear or conformational epitope, Erns proteins (native or reduced) were separated by electrophoresis in 10% polyacrylamide gels, and transferred to PVDF membranes (Millipore, Burlington, MA, USA). Membranes were blocked with 5% milk and then incubated with monoclonal antibodies. Incubation with horseradish peroxidase (HRP) conjugated secondary antibody, and detection and imaging were performed as we described previously [20].
2.6. Competitive Enzyme-Linked Immunosorbent Assay (cELISA)
The mAb1504 was purified by HiTrap™ Protein G column (GE Healthcare Life Sciences, Pittsburgh, PA, USA) followed by conjugating with horseradish peroxidase (HRP) using EZ-Link™ Plus Activated Peroxidase (Thermo Scientific, Bridgewater, NJ, USA) according to the manufacturer’s instruction. The HRP-1504 was dialyzed with Slide-A-Lyzer Dialysis Cassettes (Thermo Scientific, Bridgewater, NJ, USA) against PBS and stored in Pierce™ Peroxidase Conjugate Stabilizer (Thermo Scientific, Bridgewater, NJ, USA).
Optimization of the concentration of capture antigen and HRP-1504, the dilution of serum, and the blocking solution was performed as we described previously [21]. Briefly, Corning® 96 Well Clear Flat Bottom Polystyrene High Bind Microplates (Corning, NY, USA) were coated overnight with C-strain Erns (0.31 µg/mL, 100 µL/well) in PBS (without calcium and magnesium, pH7.4, Thermo Scientific, Bridgewater, NJ, USA) at 4 °C. After blocking, 50 µL of diluted serum samples and 50 µL of diluted HRP-1504 (0.625 µg/mL) were added to each well and mixed well by pipetting. After adding the TMB Stabilized Chromogen (Invitrogen, Carlsbad, CA, USA) and 2N Sulfuric Acid (Ricca Chemical Company, Arlington, TX, USA), the optical density at 450 nm (OD450) were obtained using SpectraMAX microplate reader (Molecular Devices, San Jose, CA, USA). The OD450 of the samples were converted to a percent inhibition (PI) value using the following formulation: PI (%) = (OD450 value of negative controls − OD450 value of sample)/OD450 value of negative controls × 100%.
2.7. Reproducibility and Statistical Analysis
Inter-assay and intra-assay reproducibility for the established cELISA was evaluated by testing C-strain Erns antibody negative (n = 20) and C-strain Erns positive (n = 20) pig serum samples. For the intra-assay reproducibility, each serum sample (in duplicate) was detected by the same batch of pre-coated ELISA plates. For the inter-assay reproducibility, each serum sample was detected by three batches of pre-coated ELISA plates. Sensitivity and specificity analysis were carried out by the web-based MedCalc statistical software (https://www.medcalc.org/calc/diagnostic_test.php, accessed on 30 May 2022). Statistical analysis of reproducibility was carried out by calculating the mean PI value and coefficient of variation (CV) of replications of each test.
3. Results
3.1. Generation of Capture Antigen and Suitable Competitive Monoclonal Antibody
The envelope glycoprotein Erns of C-strain was successfully expressed in insect cells by using Bac-to-Bac® Baculovirus Expression System. The purified C-strain Erns protein exists as homodimer and monomer under non-reducing condition with a molecular weight of ~74 kDa and ~37 kDa, respectively (Figure 1A).
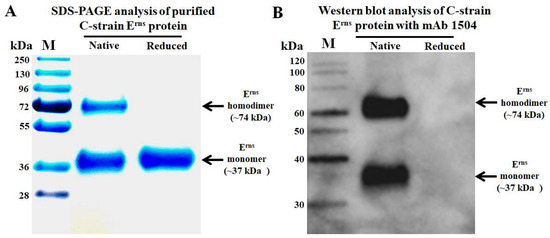
Figure 1.
Analysis of purified C-strain Erns protein and monoclonal antibody 1504. (A) Purified C-strain Erns protein exists as homodimer and monomer under native condition. (B) mAb 1504 recognizes the conformational epitope as it only reacts with the native C-strain Erns protein. The left lines are protein molecular weight size marker.
To generate suitable competitive mAb, the purified C-strain Erns protein was used as an immunogen in Balb/c mice. After three injections and the fusion of spleen cells with myeloma cells, we generated one panel of more than ten hybridomas which secreting mAbs against C-strain Erns protein. After further evaluation by Western blot, mAb 1504 (IgG1 and kappa chain) which recognizes the conformational epitope of C-strain Erns (Figure 1B) showed the best of capability to differentiate C-strain from wild-type CSFVs. It can only recognize insect cell expressed C-strain Erns, but not react with insect cell expressed Erns of wild-type CSFVs which are prevalent in Europe (sub-genotypes 2.1 and 2.3) [22], Asia (sub-genotypes 1.1 and 2.1–2.3) [23], and other separated geographic regions (sub-genotypes 3.1, 3.2 and 3.4) [22,23,24]. Indirect fluorescent antibody assay (IFA) testing further confirmed that mAb 1504 lacks reaction to CSFV field isolates (genotypes 1.1, 2.1a, 2.1b, 2.1c, 2.1g, 2.1h, 2.1i, 2.1j, 2.2 and 2.3). Detailed data are described in our article for characterization of mAbs that specifically differentiate field isolates from vaccine strains of CSFV [25].
3.2. Establishment of Competitive ELISA System with C-Strain Erns Protein and mAb 1504
The development and optimization of the cELISA system were performed as described in our previous publication [21]. A concentration of 0.31 µg/mL of C-strain Erns protein and a concentration of 0.625 µg/mL of HRP-1504 were chosen for optimal concentrations of capture protein and competitive mAb, because they consistently produced an OD450 value around 2.2 and at a point in the linear range in the standard curve (Figure 2A); 2% FBS in PBST was chosen as the optimal blocking buffer. The preferred dilution of serum samples is 1:5 as it is the lowest dilution with the highest differential ability between negative pig sera and C-strain-vaccinated pig sera (Figure 2B). These conditions were used in all subsequent cELISA experiments.
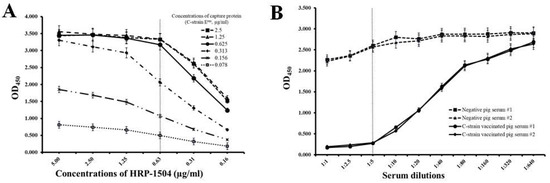
Figure 2.
Determination of concentrations of capture antigen and competitive antibody, and dilution of serum. (A) Determination of optimal concentrations of capture protein (C-strain Erns) and competitive antibody (HRP-1504). (B) Determination of an optimal dilution of serum. Data are expressed as the mean ± standard deviation from independently repeated experiments.
3.3. Standardization of the Cut-Off Value of the Established cELISA
A total of 549 pig serum samples were used to standardize the cut-off value of the established cELISA. Among them, 504 samples were from unvaccinated negative control pigs and 45 samples were from C-strain or C-strain Erns protein immunized pigs (14–56 days post vaccination, DPV). Distributions of the cELISA PI values showing the frequency of positive and negative samples are calculated and shown in Figure 3. The mean PI value (x-axis) of the negative sera detected by cELISA is −0.12%. When the mean PI value of negative sera plus two standard deviation (SD) (i.e., 10.63%) was used as threshold, the sensitivity and specificity of the cELISA were 100% (95% confidence interval: 95.60 to 100%) and 97.90% (95% confidence interval: 95.70 to 99.10%), respectively. When mean PI value of negative sera plus three SD (i.e., 15.99%) was used as threshold, the sensitivity and specificity of the cELISA were 100% (95% confidence interval: 95.60 to 100%) and 100% (95% confidence interval: 98.30 to 100%), respectively.
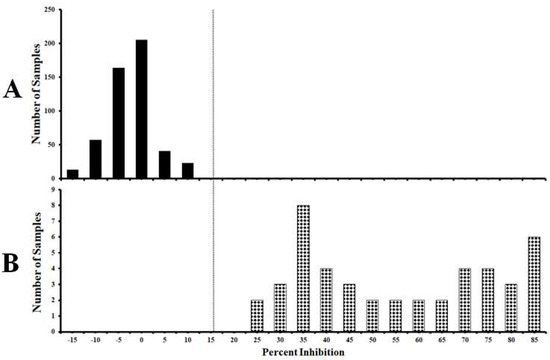
Figure 3.
Standardization of the cut-off value of mAb1504-based cELISA. (A) Negative serum samples from unvaccinated pigs (n = 504). (B) C-strain antibody positive serum samples from C-strain or C-strain Erns protein immunized pigs (14–56 DPV) (n = 45). The dotted line represents cut-off value of 15.99% inhibition when using the mean PI of negative sera plus three SD as the threshold.
3.4. The Established cELISA Can Efficiently Differentiate C-Strain/C-Strain Erns Immunized Pigs from Wild-Type CSFV and Other Common Swine Virus-Infected Pigs
For validating the specificity of the established cELISA, we tested eight categories of serum samples including negative control sera from PBS inoculated pigs (n = 159), C-strain/C-strain Erns immunized pig sera (n = 37, 14–56 DPV), C-strain E2 immunized pig sera (n = 151, 0–52 DPV), CSFV Alford (Genotype 1.1)-infected pig sera (n = 223, 0–15 days post infection, DPI), CSFV Honduras/1997 (Genotype 1.3)-infected pig sera (n = 59, 0–15 DPI), PRRSV-infected pig sera (n = 107, 0–38 DPI), PRV-infected pig sera (n = 1) and BVDV-infected pig sera (n = 2). The negative control and CSFV E2 immunized pig sera showed PI value from −19.59 % to 8.26%. The wild-type CSFVs Alford and Honduras/1997-infected pig sera showed PI value from −19.12% to 8.10%. The other common swine viruses-infected pig sera showed PI value from −14.93% to 3.69%. However, all C-strain/C-strain Erns immunized pig sera showed PI value from 23.82% to 92.03% (Figure 4). By using mean PI value (−0.32%) of negative sera plus three SD (i.e., 16.39%) as a threshold, this cELISA can efficiently differentiate C-strain/C-strain Erns immunized pig sera from the other seven pig sera categories.
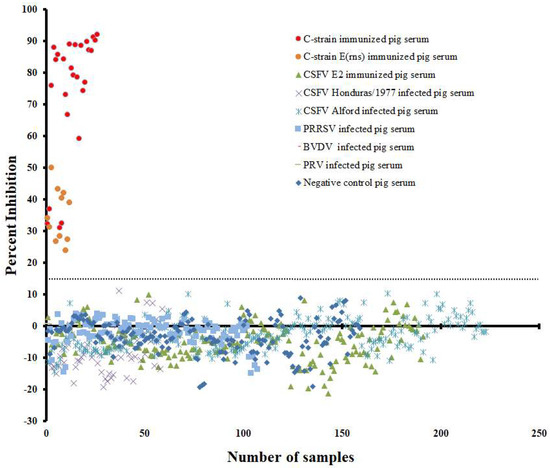
Figure 4.
Validating the diagnostic specificity of the mAb1504-based cELISA. The dotted line represents cut-off value of 16.39% inhibition when using the mean PI of negative sera plus three SD as the threshold.
3.5. Reproducibility of the cELISA
The reproducibility of the cELISA was determined by calculating the coefficient of variation (CV) of PI values by testing 20 C-strain Erns negative pig serum samples and 20 C-strain Erns positive pig serum samples. The intra-assay CVs of the C-strain Erns positive samples ranged from 0.05 to 1.37%. The inter-assay CVs of those same samples ranged from 0.86 to 8.50%. The intra-assay and inter-assay of the C-strain Erns negative samples also exhibited excellent repeatability, showing 0.10–0.45% and 0.56–5.45%, respectively (Table 1).

Table 1.
Coefficient values of the samples tested by mAb1504-based cELSIA.
3.6. Kinetics of Erns Antibody Response of C-Strain-Vaccinated Pigs at Different Time Intervals
Serial serum samples from 0 to 56 DPV at every 7 days were derived from three pigs vaccinated with C-strain and were tested by the established cELISA. The C-strain Erns antibody could be detected in all three pigs between 7 to 14 DPV. Significant increase of inhibition was observed between 7 DPV and 21 DPV. The levels of antibody titer kept relatively stable from 28 DPV to 56 DPV with inhibition values ranging from 62.81% to 84.14% (Figure 5).
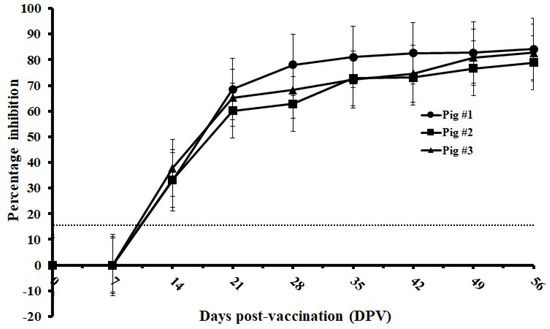
Figure 5.
Kinetics of Erns antibody response of C-strain-vaccinated pigs tested by mAb1504-based cELISA. Serum samples were derived from C-strain-vaccinated pigs (n = 3) at every 7 days. The dotted line represents cut-off value: 16%.
4. Discussion
Vaccines with DIVA capability with their accompanying diagnostic tests are essential to control and to enabling a country’s rapid return to free status following a CSF outbreak. Up to now, only one CSF DIVA vaccine was licensed by the European Medicines Agency: “CP7_E2alf” (Alfort/187 E2-based “CP7_E2alf” chimeric pestivirus, Suvaxyn CSF Marker, Zoetis) [26]. Its DIVA concept is based on the fact that field-virus-infected animals react positive in an Erns-specific antibody ELISA, while vaccinated animals only develop a CSFV E2-specific antibody response. Two ELISAs are commercially available that could work with CP7_E2alf DIVA vaccine. One is the PrioCHECK CSFV Erns (Thermofisher, former Prionics), and the other is the pigtype CSF Marker (Qiagen, Germantown, MD, USA). However, both of them showed limited specificity and sensitivity [18,19,27]. Therefore, novel reliable DIVA diagnostic assays, which can accompany current and/or future CSF vaccines, are critically needed.
C-strain is the most frequently used vaccine all over the world and showed outstanding efficacy and safety from many years of use [2,3,4,5,6,7,8,9]. A single vaccination with a C-strain-based vaccine can provide lifelong immunity with an onset as early as 2–3 days after vaccination [5]. The only disadvantage of the C-strain vaccine is that it does not allow for DIVA, which can lead to trade restrictions with significant economic consequences. As alternatives, virus replicon particles (VRP) were developed with deletions of Erns, E2, or partial E2 sequences of C-strain to make it compatible with a DIVA approach to serology [28,29]. However, it has been reported that the use of this type of vaccine is limited depending on the replicon type and administration route. This caused differences in the level of protection of pigs against the challenge of virulent CSFVs [9,28,29].
In this study, we successfully generated a C-strain-specific mAb 1504, which can only recognize Erns protein of C-strain, but not react with all prevalent CSFV strains in different genotypes. This unique character combined with lacking cross-reaction with other antigenically closely related pestiviruses make mAb 1504 an ideal competitive antibody to develop a C-strain-specific cELISA. The hypothesis of the C-strain-specific cELISA is that mAb 1504 can compete with the C-strain induced antibodies in the pig serum to bind C-strain Erns protein. cELISA assays have been shown to increase specificity and reduce the cross-reactivity [30,31]. Indeed, after optimizing, our established cELISA can efficiently differentiate C-strain immunized from wild-type CSFVs or other common swine virus-infected pigs and can test the C-strain antibody during 7–14 days post vaccination. Both sensitivity and specificity can reach 100% when using mean PI value of negative sera plus three SD as threshold. By combining this C-strain-specific cELISA with the wild-type CSFV specific infection tests, such as the genetic detection systems developed to specifically detect wild-type CSFVs [32,33,34,35,36,37], it can differentiate the infected (C-strain-specific C-ELISA “-”, wild-type CSFV specific infection tests “+”) from C-strain-vaccinated pigs (C-strain-specific C-ELISA “+”, wild-type CSFV specific infection tests “-”), and it can also be used to detect the infected pigs that are incompletely protected by C-strain vaccination (C-strain-specific C-ELISA “+”, wild-type CSFV specific infection tests “+”) as well. This will play a critical role in making decisions for a control strategy that employs vaccination with C-strain.
5. Conclusions
The presented mAb 1504-based cELISA showed excellent specificity and sensitivity. This cELISA is a reliable and cost-effective tool for specifically measuring and differentiating immune responses to the C-strain vaccine in pigs. By combining with the wild-type CSFV-specific infection tests, it renders the C-strain vaccine DIVA compatible.
Author Contributions
Conceptualization, J.S. and L.W.; methodology, J.S., L.W., S.M., W.G. and C.T.; formal analysis, J.S. and L.W.; investigation, L.W., S.M., W.G., R.M. and Y.L.; resources, J.S., L.W., W.G. and C.T.; writing—original draft preparation, L.W.; writing —review and editing, J.S., L.W., W.G. and C.T.; supervision, J.S.; funding acquisition, J.S. and L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by awards from the National Bio and Agro-Defense Facility Transition Fund, the USDA National Institute of Food and Agriculture, Hatch-Multistate project, grant number [1021491]; USDA ARS Non-Assistance Cooperative Agreements, grant numbers [58-8064-8-011, 58-8064-9-007, 58-3020-9-020, 59-0208-9-222]; USDA NIFA Award #2022-67015-36516 and USDA NIFA Subaward #25-6226-0633-002; National Pork Board Grant, grant number [18-059].
Institutional Review Board Statement
The study was conducted according to the guidelines and approved protocols by the Institutional Animal Care and Use Committee (IACUC) at Kansas State University (KSU) (protocol code 4490, date of approval 18 December 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets for the current study are available from the corresponding author on reasonable request.
Acknowledgments
We thank the Comparative Medicine staff and staff of Biosecurity Research Institute at Kansas State University for their technical help.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moennig, V. Introduction to classical swine fever: Virus, disease and control policy. Vet. Microbiol. 2000, 73, 93–102. [Google Scholar] [CrossRef]
- Ganges, L.; Crooke, H.R.; Bohórquez, J.A.; Postel, A.; Sakoda, Y.; Becher, P.; Ruggli, N. Classical swine fever virus: The past, present and future. Virus Res. 2020, 289, 198151. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Madera, R.; Li, Y.; McVey, D.S.; Drolet, B.S.; Shi, J. Recent Advances in the Diagnosis of Classical Swine Fever and Future Perspectives. Pathogens 2020, 9, 658. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, L.; McVey, D.S. Of pigs and men: The best-laid plans for prevention and control of swine fevers. Anim. Front. 2021, 11, 6–13. [Google Scholar] [CrossRef]
- van Oirschot, J.T. Vaccinology of classical swine fever: From lab to field. Vet. Microbiol. 2003, 96, 367–384. [Google Scholar] [CrossRef]
- Luo, Y.; Li, S.; Sun, Y.; Qiu, H.J. Classical swine fever in China: A minireview. Vet. Microbiol. 2014, 172, 1–6. [Google Scholar] [CrossRef]
- Blome, S.; Moß, C.; Reimann, I.; König, P.; Beer, M. Classical swine fever vaccines: State-of-the-art. Vet. Microbiol. 2017, 206, 10–20. [Google Scholar] [CrossRef]
- Postel, A.; Austermann-Busch, S.; Petrov, A.; Moennig, V.; Becher, P. Epidemiology, diagnosis and control of classical swine fever: Recent developments and future challenges. Transbound. Emerg. Dis. 2018, 65, 248–261. [Google Scholar] [CrossRef]
- Coronado, L.; Perera, C.L.; Rios, L.; Frías, M.T.; Pérez, L.J. A Critical Review about Different Vaccines against Classical Swine Fever Virus and Their Repercussions in Endemic Regions. Vaccines 2021, 9, 154. [Google Scholar] [CrossRef]
- Paton, D.J.; McGoldrick, A.; Greiser-Wilke, I.; Parchariyanon, S.; Song, J.Y.; Liou, P.P.; Stadejek, T.; Lowings, J.P.; Björklund, H.; Belák, S. Genetic typing of classical swine fever virus. Vet. Microbiol. 2000, 73, 137–157. [Google Scholar] [CrossRef]
- Postel, A.; Schmeiser, S.P.; Perera, C.L.; Rodríguez, L.J.; Frias-Lepoureau, M.T.; Becher, P. Classical swine fever virus isolates from Cuba form a new subgenotype 1.4. Vet. Microbiol. 2013, 161, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Lv, L.; Gu, J.; Chen, T.; Xiao, Y.; Liu, S. Genetic diversity and positive selection analysis of classical swine fever virus envelope protein gene E2 in east China under C-strain vaccination. Front. Microbiol. 2016, 7, 85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- König, M.; Lengsfeld, T.; Pauly, T.; Stark, R.; Thiel, H.J. Classical swine fever virus: Independent induction of protective immunity by two structural glycoproteins. J. Virol. 1995, 69, 6479–6486. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Bai, Y.; Song, Y.; Liu, Y.; Yu, W.; Sun, Y.; Wang, L.; Deng, R.; Xing, G.; Zhang, G. Generation and immunogenicity analysis of recombinant classical swine fever virus glycoprotein E2 and Erns expressed in baculovirus expression system. Virol. J. 2021, 18, 44. [Google Scholar] [CrossRef]
- Moormann, R.J.; Bouma, A.; Kramps, J.A.; Terpstra, C.; De Smit, H.J. Development of a classical swine fever subunit marker vaccine and companion diagnostic test. Vet. Microbiol. 2000, 73, 209–219. [Google Scholar] [CrossRef]
- Langedijk, J.P.; Middel, W.G.; Meloen, R.H.; Kramps, J.A.; de Smit, J.A. Enzyme-linked immunosorbent assay using a virus type-specific peptide based on a subdomain of envelope protein Erns for serologic diagnosis of pestivirus infections in swine. J. Clin. Microbiol. 2001, 39, 906–912. [Google Scholar] [CrossRef]
- Lin, M.; Trottier, E.; Pasick, J. Antibody responses of pigs to defined Erns fragments after infection with classical swine fever virus. Clin. Diagn. Lab. Immunol. 2005, 12, 180–186. [Google Scholar]
- Schroeder, S.; von Rosen, T.; Blome, S.; Loeffen, W.; Haegeman, A.; Koenen, F.; Uttenthal, A. Evaluation of classical swine fever virus antibody detection assays with an emphasis on the differentiation of infected from vaccinated animals. Rev. Sci. Tech. 2012, 31, 997–1010. [Google Scholar] [CrossRef]
- Pannhorst, K.; Fröhlich, A.; Staubach, C.; Meyer, D.; Blome, S.; Becher, P. Evaluation of an Erns-based enzyme-linked immunosorbent assay to distinguish Classical swine fever virus-infected pigs from pigs vaccinated with CP7_E2alf. J. Vet. Diagn. Investig. 2015, 27, 449–460. [Google Scholar] [CrossRef]
- Madera, R.; Gong, W.; Wang, L.; Burakova, Y.; Lleellish, K.; Galliher-Beckley, A.; Nietfeld, J.; Henningson, J.; Jia, K.; Li, P.; et al. Pigs immunized with a novel E2 subunit vaccine are protected from subgenotype heterologous classical swine fever virus challenge. BMC Vet. Res. 2016, 12, 197. [Google Scholar] [CrossRef]
- Wang, L.; Mi, S.; Madera, R.; Ganges, L.; Borca, M.V.; Ren, J.; Cunningham, C.; Cino-Ozuna, A.G.; Li, H.; Tu, C.; et al. A neutralizing monoclonal antibody-based competitive ELISA for classical swine fever C-strain post-vaccination monitoring. BMC Vet. Res. 2020, 16, 14. [Google Scholar]
- Postel, A.; Moennig, V.; Becher, P. Classical swine fever in Europe—The current situation. Berl. Munch. Tierarztl. Wochenschr. 2013, 126, 468–475. [Google Scholar] [PubMed]
- Postel, A.; Nishi, T.; Kameyama, K.I.; Meyer, D.; Suckstorff, O.; Fukai, K.; Becher, P. Reemergence of Classical Swine Fever, Japan, 2018. Emerg. Infect. Dis. 2019, 25, 1228–1231. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Villamandos, J.C.; Carrasco, L.; Bautista, M.J.; Sierra, M.A.; Quezada, M.; Hervas, J.; Chacón, M.; Ruiz-Villamor, E.; Salguero, F.J.; Sónchez-Cordón, P.J.; et al. African swine fever and classical swine fever: A review of the pathogenesis. Dtsch. Tierarztl. Wochenschr. 2003, 110, 165–169. [Google Scholar]
- Mi, S.; Wang, L.; Li, H.; Fei, B.; Madera, R.; Shi, X.; Zhang, L.; Mao, Y.; Yan, X.; Xia, X.; et al. Characterization of monoclonal antibodies that specifically differentiate field isolates from vaccine strains of classical swine fever virus. Front. Immunol. 2022; accepted. [Google Scholar]
- Blome, S.; Wernike, K.; Reimann, I.; König, P.; Moß, C.; Beer, M. A decade of research into classical swine fever marker vaccine CP7_E2alf (Suvaxyn® CSF Marker): A review of vaccine properties. Vet. Res. 2017, 48, 51. [Google Scholar] [CrossRef] [PubMed]
- Jelsma, T.; Post, J.; Born, E.; Segers, R.; Kortekaas, J. Assessing the Protective Dose of a Candidate DIVA Vaccine against Classical Swine Fever. Vaccines 2021, 9, 483. [Google Scholar] [CrossRef]
- Widjojoatmodjo, M.N.; van Gennip, H.G.; Bouma, A.; van Rijn, P.A.; Moormann, R.J. Classical swine fever virus Erns deletion mutants: Trans-complementation and potential use as nontransmissible, modified, live-attenuated marker vaccines. J. Virol. 2000, 74, 2973–2980. [Google Scholar] [CrossRef][Green Version]
- van Gennip, H.G.; Bouma, A.; van Rijn, P.A.; Widjojoatmodjo, M.N.; Moormann, R.J. Experimental non-transmissible marker vaccines for classical swine fever (CSF) by trans-complementation of Erns or E2 of CSFV. Vaccine 2002, 20, 1544–1556. [Google Scholar] [CrossRef]
- Greiser-Wilke, I.; Blome, S.; Moennig, V. Diagnostic methods for detection of Classical swine fever virus-status quo and new developments. Vaccine 2007, 25, 5524–5530. [Google Scholar] [CrossRef]
- OIE Terrestrial Manual Chapter 2.2.1. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access (accessed on 11 February 2022).
- Li, Y.; Zhao, J.J.; Li, N.; Shi, Z.; Cheng, D.; Zhu, Q.H.; Tu, C.; Tong, G.Z.; Qiu, H.J. A multiplex nested RT-PCR for the detection and differentiation of wild-type viruses from C-strain vaccine of classical swine fever virus. J. Virol. Methods 2007, 143, 16–22. [Google Scholar] [CrossRef]
- Leifer, I.; Depner, K.; Blome, S.; Le Potier, M.F.; Le Dimna, M.; Beer, M.; Hoffmann, B. Differentiation of C-strain “Riems” or CP7_E2alf vaccinated animals from animals infected by classical swine fever virus field strains using real-time RT-PCR. J. Virol. Methods 2009, 158, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xia, H.; Everett, H.; Sosan, O.; Crooke, H.; Meindl-Böhmer, A.; Qiu, H.J.; Moennig, V.; Belák, S.; Widén, F. A generic real-time TaqMan assay for specific detection of lapinized Chinese vaccines against classical swine fever. J. Virol. Methods 2011, 175, 170–174. [Google Scholar] [CrossRef]
- Zhang, X.J.; Han, Q.Y.; Sun, Y.; Zhang, X.; Qiu, H.J. Development of a triplex TaqMan real-time RT-PCR assay for differential detection of wild-type and HCLV vaccine strains of classical swine fever virus and bovine viral diarrhea virus 1. Res. Vet. Sci. 2012, 92, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Everett, H.E.; Crudgington, B.S.; Sosan-Soulé, O.; Crooke, H.R. Differential detection of classical swine fever virus challenge strains in C-strain vaccinated pigs. BMC Vet. Res. 2014, 10, 281. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Widén, F.; Everett, H.; Blome, S.; Fernandez Pinero, J.; Uttenthal, A.; Cortey, M.; von Rosen, T.; Tignon, M.; Liu, L. Comparison of two real-time RT-PCR assays for differentiation of C-strain vaccinated from classical swine fever infected pigs and wild boars. Res. Vet. Sci. 2014, 97, 455–457. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).