Evaluation of Two Broadly Used Commercial Methods for Detection of Respiratory Viruses with a Recently Added New Target for Detection of SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
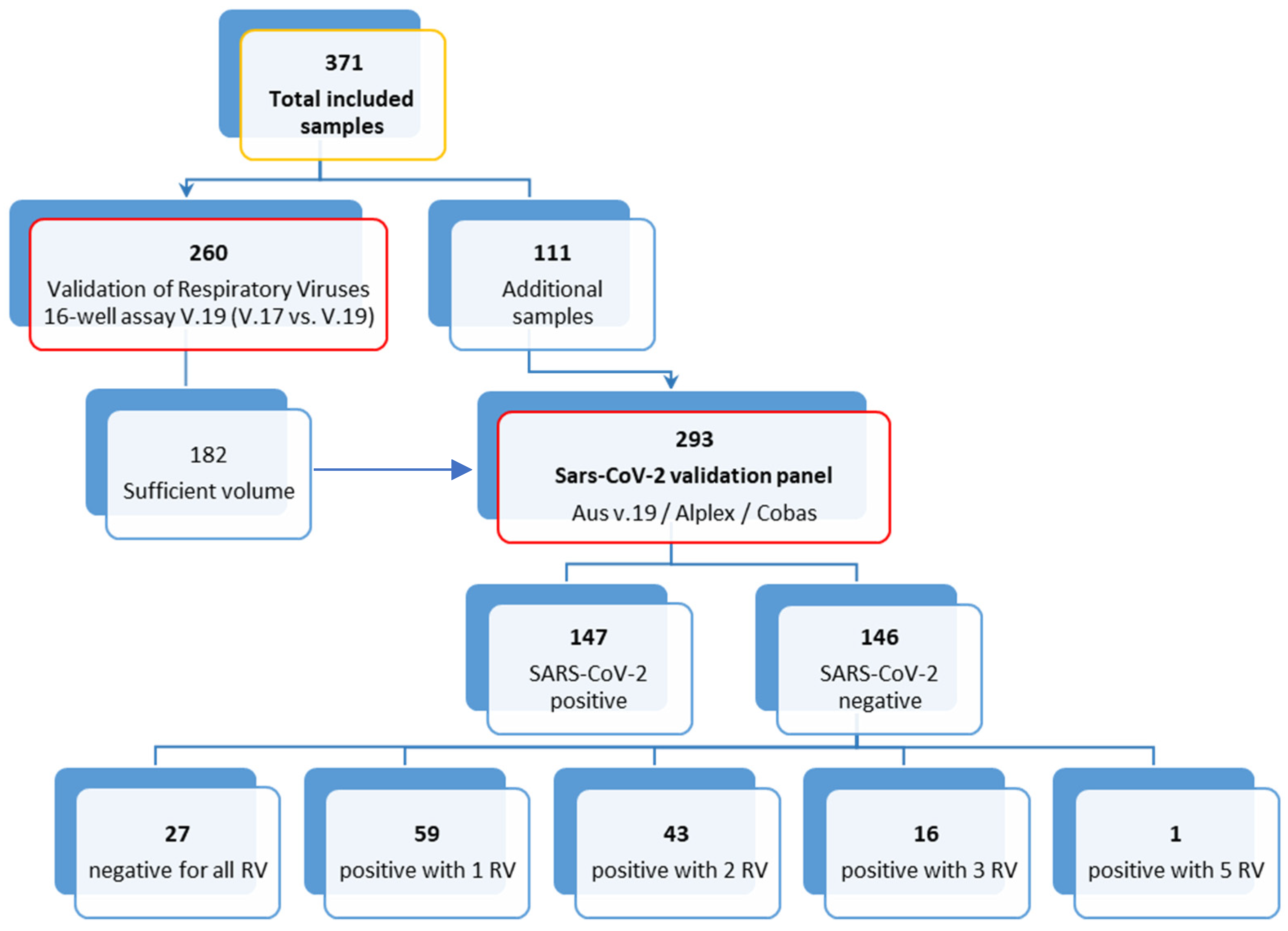
2.1. Specimen Selection, Collection and Routine Diagnostics
2.2. Respiratory Viruses’ Validation Panel
2.3. SARS-CoV-2 Validation Panel
2.4. Specificity Testing
2.5. Statistical Analysis
3. Results
3.1. Respiratory Viruses 16-Well Assay V.17 versus V.19
3.2. Respiratory Viruses 16-Well Assay V.19 and Allplex™-nCoV Assay SARS-CoV-2 Detection Performance for Detection of SARS-CoV-2
3.3. Comparison of the Respiratory Viruses 16-Well Assay V.19 and the Allplex™ RV Essential Assay
3.4. Cross Reactivity of SARS-CoV-2 with Seasonal HCoVs in the Respiratory Viruses 16-Well Assay V.19
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R. SARS-CoV-2 Omicron spike mediated immune escape and tropism shift. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Loeffelholz, M.J.; Tang, Y.W. Laboratory diagnosis of emerging human coronavirus infections—The state of the art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef]
- Chan, J.F.; Yip, C.C.; To, K.K.; Tang, T.H.; Wong, S.C.; Leung, K.H.; Fung, A.Y.; Ng, A.C.; Zou, Z.; Tsoi, H.W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, J.; Wang, M.X.; Ang, I.Y.H.; Tan, S.H.X.; Lewis, R.F.; Chen, J.I.; Gutierrez, R.A.; Gwee, S.X.W.; Chua, P.E.Y.; Yang, Q.; et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J. Clin. Med. 2020, 9, 623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poljak, M.; Korva, M.; Knap Gasper, N.; Fujs Komlos, K.; Sagadin, M.; Ursic, T.; Avsic Zupanc, T.; Petrovec, M. Clinical Evaluation of the cobas SARS-CoV-2 Test and a Diagnostic Platform Switch during 48 Hours in the Midst of the COVID-19 Pandemic. J. Clin. Microbiol. 2020, 58, e00599-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, S.J.; Azziz-Baumgartner, E.; Budd, A.P.; Brammer, L.; Sullivan, S.; Pineda, R.F.; Cohen, C.; Fry, A.M. Decreased Influenza Activity During the COVID-19 Pandemic—United States, Australia, Chile, and South Africa, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1305–1309. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.D.; Sordillo, E.M.; Gitman, M.R.; Paniz Mondolfi, A.E. Coinfection in SARS-CoV-2 infected patients: Where are influenza virus and rhinovirus/enterovirus? J. Med. Virol. 2020, 92, 1699–1700. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Maejima, M.; Shibusawa, M.; Amemiya, K.; Nagakubo, Y.; Hosaka, K.; Sueki, H.; Mochizuki, H.; Tsutsui, T.; Kakizaki, Y.; et al. Analysis of COVID-19 and non-COVID-19 viruses, including influenza viruses, to determine the influence of intensive preventive measures in Japan. J. Clin. Virol. 2020, 129, 104543. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Wang, C.Y.; Hsueh, P.R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect.=Wei Mian Yu Gan Ran Za Zhi 2020, 53, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, S.L. Coinfections in Hospitalized Children With Community-Acquired Pneumonia: What Does This Mean for the Clinician? J. Infect. Dis. 2018, 218, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Kogoj, R.; Korva, M.; Knap, N.; Resman Rus, K.; Pozvek, P.; Avsic-Zupanc, T.; Poljak, M. Comparative Evaluation of Six SARS-CoV-2 Real-Time RT-PCR Diagnostic Approaches Shows Substantial Genomic Variant-Dependent Intra- and Inter-Test Variability, Poor Interchangeability of Cycle Threshold and Complementary Turn-Around Times. Pathogens 2022, 11, 462. [Google Scholar] [CrossRef] [PubMed]
- Attwood, L.O.; Francis, M.J.; Hamblin, J.; Korman, T.M.; Druce, J.; Graham, M. Clinical evaluation of AusDiagnostics SARS-CoV-2 multiplex tandem PCR assay. J. Clin. Virol. 2020, 128, 104448. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.; Roberts, J.; Bond, K.; Tran, T.; Kostecki, R.; Yoga, Y.; Naughton, W.; Taiaroa, G.; Seemann, T.; et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020, 212, 459–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, H.; Carter, I.; Basile, K.; Donovan, L.; Kumar, S.; Tran, T.; Ko, D.; Alderson, S.; Sivaruban, T.; Eden, J.S.; et al. Interpret with caution: An evaluation of the commercial AusDiagnostics versus in-house developed assays for the detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 127, 104374. [Google Scholar] [CrossRef] [PubMed]
| V.19 for respiratory viruses | V.17 for Respiratory Viruses | % Agreement | Kappa | r | |||
| Pos | Neg | ||||||
| FluA | Pos | 22 | 0 | 100 (100–100) | 1 (1–1) | 0.95 | |
| Neg | 0 | 238 | |||||
| FluB | Pos | 24 | 0 | 100 (100–100) | 1 (1–1) | 0.70 | |
| Neg | 0 | 236 | |||||
| RSV | Pos | 35 | 1 | 99.6 (97.9–99.9) | 0.98 (0.95–1) | 0.70 | |
| Neg | 0 | 224 | |||||
| HRV | Pos | 52 | 0 | 100 (100–100) | 1 (1–1) | 0.98 | |
| Neg | 0 | 208 | |||||
| EV | Pos | 9 | 0 | 100 (100–100) | 1 (1–1) | 0.99 | |
| Neg | 0 | 251 | |||||
| Parecho | Pos | 8 | 0 | 100 (100–100) | 1 (1–1) | 0.98 | |
| Neg | 0 | 252 | |||||
| HBoV | Pos | 28 | 0 | 100 (100–100) | 1 (1–1) | 0.99 | |
| Neg | 0 | 232 | |||||
| PIV | Pos | 20 | 0 | 100 (100–100) | 1 (1–1) | 0.99 | |
| Neg | 0 | 240 | |||||
| AdV | Pos | 22 | 0 | 100 (100–100) | 1 (1–1) | 0.99 | |
| Neg | 0 | 238 | |||||
| HMPV | Pos | 22 | 0 | 100 (100–100) | 1 (1–1) | 0.99 | |
| Neg | 0 | 238 | |||||
| HCoV | Pos | 25 | 0 | 100 (100–100) | 1 (1–1) | 0.96 | |
| Neg | 0 | 235 | |||||
| Cobas 6800 | % Agreement | Kappa | |||
|---|---|---|---|---|---|
| Pos | Neg | ||||
| V.19 for respiratory viruses (AusDiagnostics) | Pos | 143 | 0 | 98.6 (96.5–99.5) | 0.97 (0.94–1.00) |
| Neg | 4 | 146 | |||
| Allplex™-nCoV Assay (automatic calling) | Pos | 143 | 0 | 98.6 (96.5–99.5) | 0.97 (0.94–1.00) |
| Neg | 4 | 146 | |||
| Allplex™-nCoV Assay (all 3 genes for pos) | Pos | 133 | 0 | 95.2 (92.1–97.1) | 0.90 (0.85–0.95) |
| Neg | 14 | 146 | |||
| Allplex™-nCoV Assay (at least 2 genes for pos) | Pos | 135 | 2 | 95.9 (93.0–97.6) | 0.92 (0.87–0.96) |
| Neg | 10 | 146 | |||
| V.19 for Respiratory Viruses (AusDiagnostics) | % Agreement | Kappa | |||
|---|---|---|---|---|---|
| Pos | Neg | ||||
| Allplex™-nCoV Assay (automatic calling) | Pos | 141 | 2 | 98.6 (96.5–99.5) | 0.97 (0.95–1.00) |
| Neg | 2 | 148 | |||
| Allplex™-nCoV Assay (all 3 genes for pos) | Pos | 133 | 0 | 96.6 (93.8–98.1) | 0.93 (0.89–0.97) |
| Neg | 10 | 150 | |||
| Allplex™-nCoV Assay (at least 2 genes for pos) | Pos | 135 | 2 | 96.6 (93.8–98.1) | 0.93 (0.89–0.97) |
| Neg | 8 | 148 | |||
| V.19 for Respiratory Viruses (AusDiagnostics) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FluA | FluB | PIV | RSV | MPV | AdV | HRV | |||||||||
| Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | ||
| Seegene | Pos | 14 | 0 | 14 | 1 | 7 | 0 | 24 | 2 | 12 | 2 | 22 | 3 | 32 | 2 |
| Neg | 1 | 278 | 0 | 278 | 3 | 283 | 1 | 266 | 1 | 278 | 0 | 268 | 11 | 248 | |
| % agreement | 99.7 (98.1–99.9) | 99.7 (98.1–99.9) | 99.0 (97.0–99.7) | 99.0 (97.0–99.7) | 99.0 (97.0–99.7) | 99.0 (97.0–99.7) | 95.6 (92.6–97.4) | ||||||||
| Kappa | 0.96 (0.89–1.00) | 0.96 (0.89–1.00) | 0.82 (0.62–1.00) | 0.94 (0.86–1.00) | 0.88 (0.75–1.00) | 0.93 (0.85–1.00) | 0.81 (0.70–0.91) | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jevšnik Virant, M.; Uršič, T.; Kogoj, R.; Korva, M.; Petrovec, M.; Avšič-Županc, T. Evaluation of Two Broadly Used Commercial Methods for Detection of Respiratory Viruses with a Recently Added New Target for Detection of SARS-CoV-2. Viruses 2022, 14, 1530. https://doi.org/10.3390/v14071530
Jevšnik Virant M, Uršič T, Kogoj R, Korva M, Petrovec M, Avšič-Županc T. Evaluation of Two Broadly Used Commercial Methods for Detection of Respiratory Viruses with a Recently Added New Target for Detection of SARS-CoV-2. Viruses. 2022; 14(7):1530. https://doi.org/10.3390/v14071530
Chicago/Turabian StyleJevšnik Virant, Monika, Tina Uršič, Rok Kogoj, Miša Korva, Miroslav Petrovec, and Tatjana Avšič-Županc. 2022. "Evaluation of Two Broadly Used Commercial Methods for Detection of Respiratory Viruses with a Recently Added New Target for Detection of SARS-CoV-2" Viruses 14, no. 7: 1530. https://doi.org/10.3390/v14071530
APA StyleJevšnik Virant, M., Uršič, T., Kogoj, R., Korva, M., Petrovec, M., & Avšič-Županc, T. (2022). Evaluation of Two Broadly Used Commercial Methods for Detection of Respiratory Viruses with a Recently Added New Target for Detection of SARS-CoV-2. Viruses, 14(7), 1530. https://doi.org/10.3390/v14071530






