An Updated Review on SARS-CoV-2 Infection in Animals
Abstract
:1. Introduction
2. SARS-CoV-2
3. SARS-CoV-2 Testing in Animals
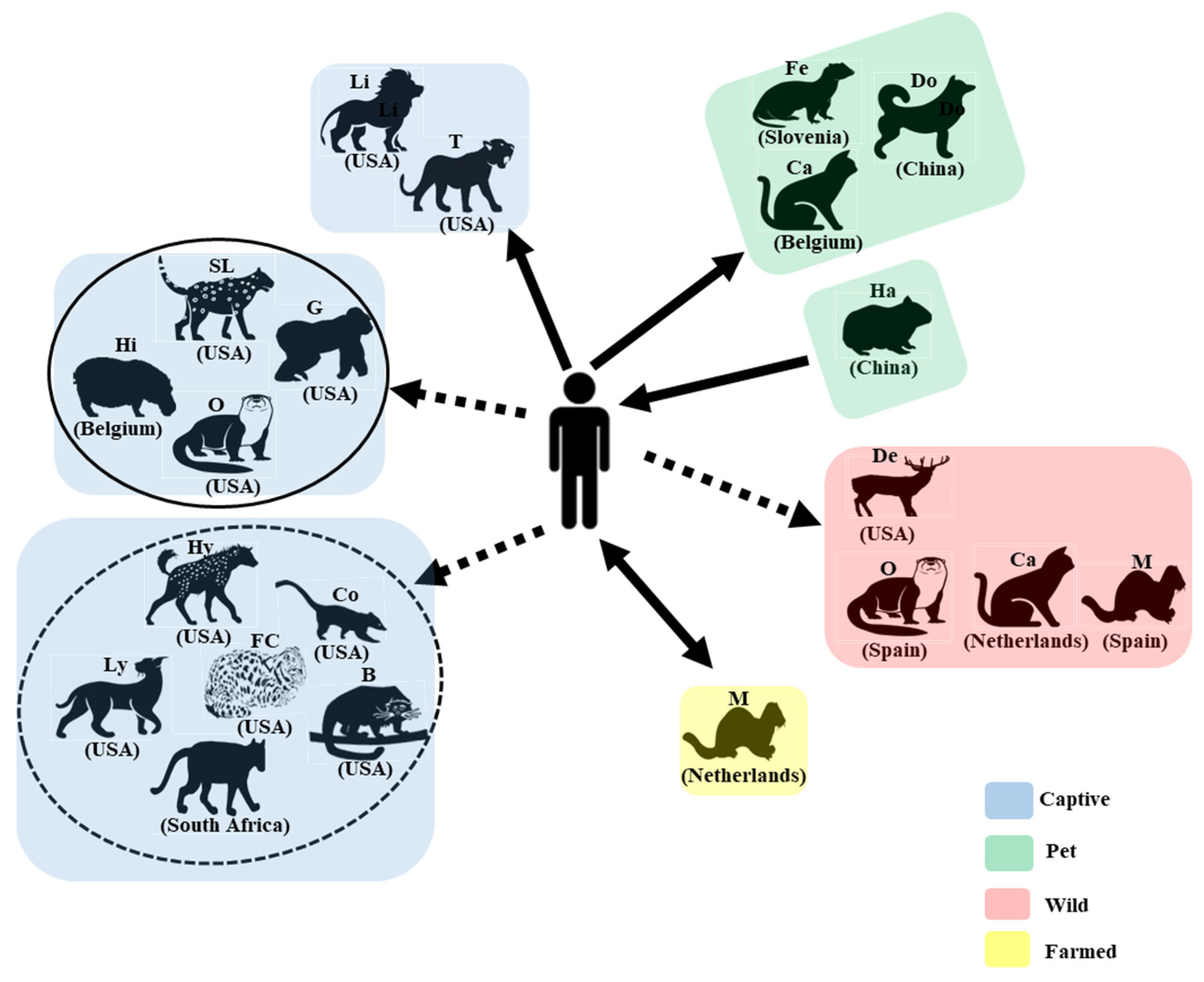
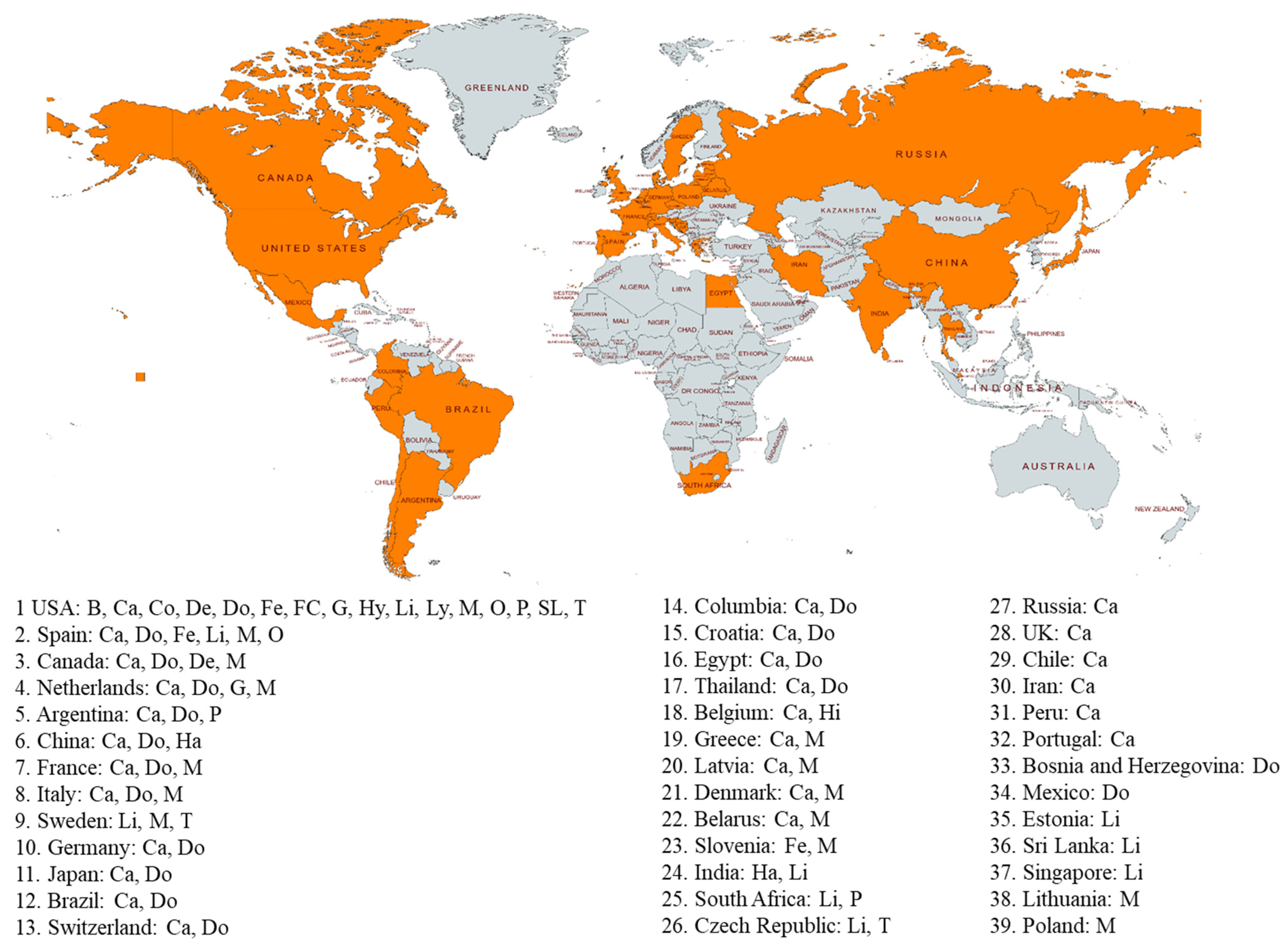
4. SARS-CoV-2 Infection in Animals
5. SARS-CoV-2 Strains in Animals
6. SARS-CoV-2 Transmission Routes
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schalk, A.F.; Hawn, M.C. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931, 78, 413–423. [Google Scholar]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Medina-Enriquez, M.M.; Lopez-Leon, S.; Carlos-Escalante, J.A.; Aponte-Torres, Z.; Cuapio, A.; Wegman-Ostrosky, T. ACE2: The molecular doorway to SARS-CoV-2. Cell Biosci. 2020, 10, 148. [Google Scholar] [CrossRef]
- Petersen, E.; Ntoumi, F.; Hui, D.S.; Abubakar, A.; Kramer, L.D.; Obiero, C.; Tambyah, P.A.; Blumberg, L.; Yapi, R.; Al-Abri, S.; et al. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B.1.1.529)—highlights Africa’s research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int. J. Infect. Dis. 2022, 114, 268–272. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220-20. [Google Scholar] [CrossRef]
- Fernandez-Bellon, H.; Rodon, J.; Fernandez-Bastit, L.; Almagro, V.; Padilla-Sole, P.; Lorca-Oro, C.; Valle, R.; Roca, N.; Grazioli, S.; Trogu, T.; et al. Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses. Viruses 2021, 13, 1683. [Google Scholar] [CrossRef]
- Garigliany, M.; Van Laere, A.S.; Clercx, C.; Giet, D.; Escriou, N.; Huon, C.; van der Werf, S.; Eloit, M.; Desmecht, D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. 2020, 26, 3069–3071. [Google Scholar] [CrossRef]
- Lenz, O.C.; Marques, A.D.; Kelly, B.J.; Rodino, K.G.; Cole, S.D.; Perera, R.; Weiss, S.R.; Bushman, F.D.; Lennon, E.M. SARS-CoV-2 Delta Variant (AY.3) in the Feces of a Domestic Cat. Viruses 2022, 14, 421. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Grome, H.N.; Meyer, B.; Read, E.; Buchanan, M.; Cushing, A.; Sawatzki, K.; Levinson, K.J.; Thomas, L.S.; Perry, Z.; Uehara, A.; et al. SARS-CoV-2 Outbreak among Malayan Tigers and Humans, Tennessee, USA, 2020. Emerg. Infect. Dis. 2022, 28, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, K.N.; Mendes, A.; Strydom, A.; Rotherham, L.; Mulumba, M.; Venter, M. SARS-CoV-2 Reverse Zoonoses to Pumas and Lions, South Africa. Viruses 2022, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Currie, D.W.; Shah, M.M.; Salvatore, P.P.; Ford, L.; Whaley, M.J.; Meece, J.; Ivacic, L.; Thornburg, N.J.; Tamin, A.; Harcourt, J.L.; et al. Relationship of SARS-CoV-2 Antigen and Reverse Transcription PCR Positivity for Viral Cultures. Emerg. Infect. Dis. 2022, 28, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Goletic, S.; Goletic, T.; Softic, A.; Zahirovic, A.; Rukavina, D.; Kavazovic, A.; Omeragic, J.; Umihanic, S.; Hukic, M. The Evidence of SARS-CoV-2 Human-to-Pets Transmission in Household Settings in Bosnia and Herzegovina. Front. Genet. 2022, 13, 839205. [Google Scholar]
- Zhao, S.; Schuurman, N.; Li, W.; Wang, C.; Smit, L.A.M.; Broens, E.M.; Wagenaar, J.A.; van Kuppeveld, F.J.M.; Bosch, B.J.; Egberink, H. Serologic Screening of Severe Acute Respiratory Syndrome Coronavirus 2 Infection in Cats and Dogs during First Coronavirus Disease Wave, the Netherlands. Emerg. Infect. Dis. 2021, 27, 1362–1370. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Racnik, J.; Kocevar, A.; Slavec, B.; Korva, M.; Rus, K.R.; Zakotnik, S.; Zorec, T.M.; Poljak, M.; Matko, M.; Rojs, O.Z.; et al. Transmission of SARS-CoV-2 from Human to Domestic Ferret. Emerg. Infect. Dis. 2021, 27, 2450–2453. [Google Scholar] [CrossRef]
- Kok, K.H.; Wong, S.C.; Chan, W.M.; Wen, L.; Chu, A.W.; Ip, J.D.; Lee, L.K.; Wong, I.T.; Lo, H.W.; Cheng, V.C.; et al. Co-circulation of two SARS-CoV-2 variant strains within imported pet hamsters in Hong Kong. Emerg. Microbes Infect. 2022, 11, 689–698. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Aguilo-Gisbert, J.; Padilla-Blanco, M.; Lizana, V.; Maiques, E.; Munoz-Baquero, M.; Chillida-Martinez, E.; Cardells, J.; Rubio-Guerri, C. First Description of SARS-CoV-2 Infection in Two Feral American Mink (Neovison vison) Caught in the Wild. Animals 2021, 11, 1422. [Google Scholar] [CrossRef] [PubMed]
- Van Aart, A.E.; Velkers, F.C.; Fischer, E.A.J.; Broens, E.M.; Egberink, H.; Zhao, S.; Engelsma, M.; Hakze-van der Honing, R.W.; Harders, F.; de Rooij, M.M.T.; et al. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Padilla-Blanco, M.; Aguilo-Gisbert, J.; Rubio, V.; Lizana, V.; Chillida-Martinez, E.; Cardells, J.; Maiques, E.; Rubio-Guerri, C. The Finding of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) in a Wild Eurasian River Otter (Lutra lutra) Highlights the Need for Viral Surveillance in Wild Mustelids. Front. Vet. Sci. 2022, 9, 826991. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Rivero, R.; Garay, E.; Botero, Y.; Serrano-Coll, H.; Gastelbondo, B.; Munoz, M.; Ballesteros, N.; Castaneda, S.; Patino, L.H.; Ramirez, J.D.; et al. Human-to-dog transmission of SARS-CoV-2, Colombia. Sci. Rep. 2022, 12, 7880. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Yen, H.L.; Sit, T.H.C.; Brackman, C.J.; Chuk, S.S.Y.; Gu, H.; Tam, K.W.S.; Law, P.Y.T.; Leung, G.M.; Peiris, M.; Poon, L.L.M.; et al. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: A case study. Lancet 2022, 399, 1070–1078. [Google Scholar] [CrossRef]
- Chavda, V.P.; Feehan, J.; Apostolopoulos, V. A Veterinary Vaccine for SARS-CoV-2: The First COVID-19 Vaccine for Animals. Vaccines 2021, 9, 631. [Google Scholar] [CrossRef]
- Moreira-Soto, A.; Walzer, C.; Czirjak, G.A.; Richter, M.H.; Marino, S.F.; Posautz, A.; De Yebra Rodo, P.; McEwen, G.K.; Drexler, J.F.; Greenwood, A.D. Serological Evidence That SARS-CoV-2 Has Not Emerged in Deer in Germany or Austria during the COVID-19 Pandemic. Microorganisms 2022, 10, 748. [Google Scholar] [CrossRef] [PubMed]
| Species | Total | Alpha | Delta | Gamma | Omicron | Lambda | MU GH | Non-Var |
|---|---|---|---|---|---|---|---|---|
| Cat | 140 | 11 | 27 | 1 | 5 | 3 | 93 | |
| Deer | 159 | 3 | 38 | 1 | 4 | 113 | ||
| Mink | 1366 | 6 | 60 | 2 | 1298 | |||
| Dog | 84 | 5 | 24 | 7 | 48 | |||
| Lion | 74 | 3 | 42 | 29 | ||||
| Tiger | 43 | 3 | 27 | 13 | ||||
| Gorilla | 15 | 2 | 12 | 1 | ||||
| Otter | 8 | 5 | 3 | |||||
| Hamster | 24 | 12 | 12 | |||||
| Snow Leopard | 9 | 4 | 4 | 1 | ||||
| Binturong | 1 | 1 | ||||||
| Hyena | 1 | 1 | ||||||
| Ferret | 1 | 1 | ||||||
| Hippo | 1 | 1 | ||||||
| Fishing Cat | 1 | 1 | ||||||
| Lynx | NA | |||||||
| Cougar | NA | |||||||
| Coatimundi | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, S.; Liu, Y.; Zhao, J.; Peng, X.; Lu, G.; Shi, W.; Pan, Y.; Zhang, D.; Yang, P.; Wang, Q. An Updated Review on SARS-CoV-2 Infection in Animals. Viruses 2022, 14, 1527. https://doi.org/10.3390/v14071527
Cui S, Liu Y, Zhao J, Peng X, Lu G, Shi W, Pan Y, Zhang D, Yang P, Wang Q. An Updated Review on SARS-CoV-2 Infection in Animals. Viruses. 2022; 14(7):1527. https://doi.org/10.3390/v14071527
Chicago/Turabian StyleCui, Shujuan, Yimeng Liu, Jiachen Zhao, Xiaomin Peng, Guilan Lu, Weixian Shi, Yang Pan, Daitao Zhang, Peng Yang, and Quanyi Wang. 2022. "An Updated Review on SARS-CoV-2 Infection in Animals" Viruses 14, no. 7: 1527. https://doi.org/10.3390/v14071527
APA StyleCui, S., Liu, Y., Zhao, J., Peng, X., Lu, G., Shi, W., Pan, Y., Zhang, D., Yang, P., & Wang, Q. (2022). An Updated Review on SARS-CoV-2 Infection in Animals. Viruses, 14(7), 1527. https://doi.org/10.3390/v14071527






