The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans
Abstract
1. Introduction
2. Epidemiology of PRV
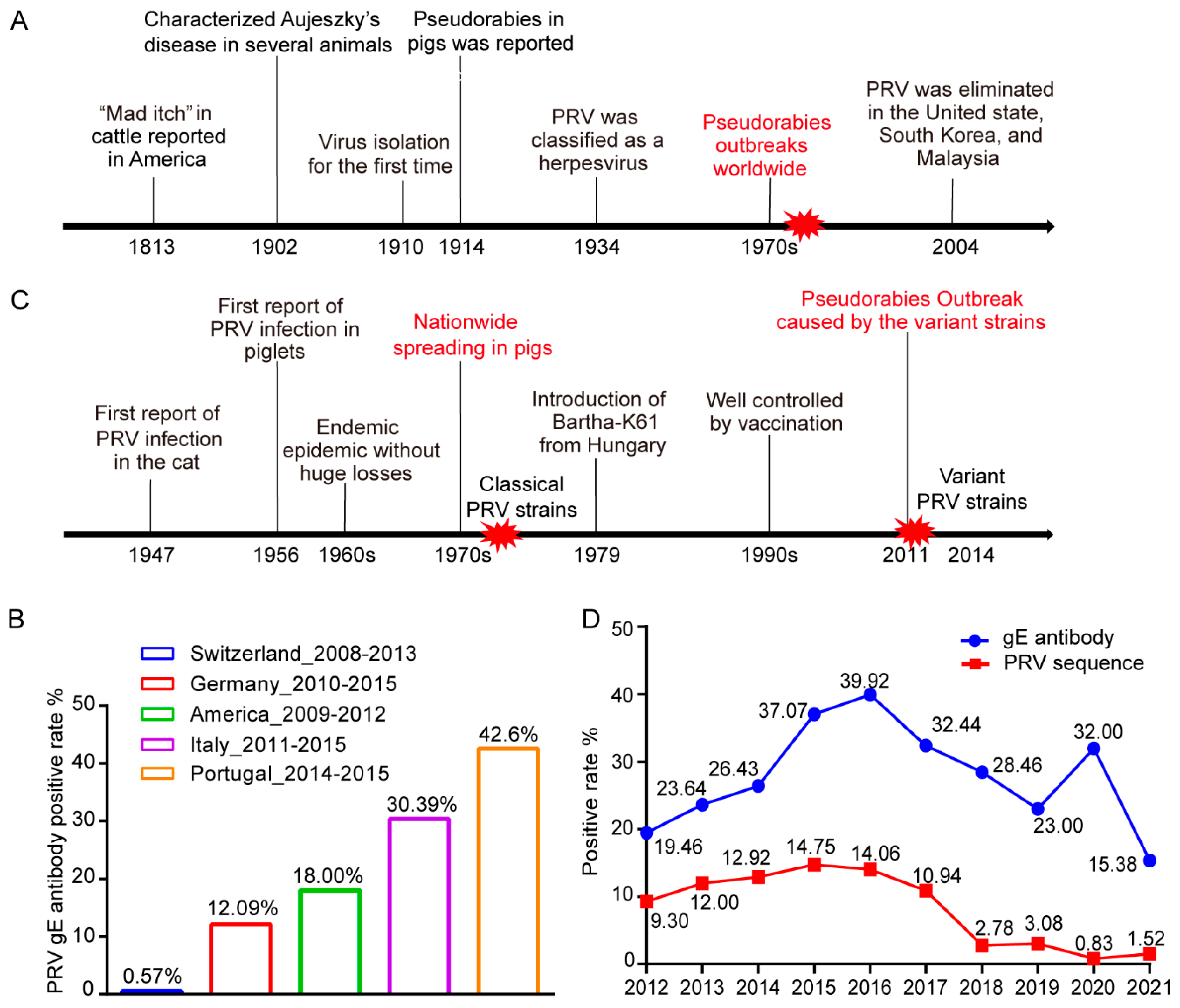
2.1. The Prevalence of PRV in the World
2.2. The Prevalence of PRV in China
3. Genotyping and Variation in PRV
3.1. Genotyping of PRV
3.2. The Evolution of PRV Based on Natural Mutation-Selection
3.3. Frequent Recombination between PRV Strains Significantly Contributes to Virus Evolution
4. PRV Infections in Animals and Potentially in Humans
4.1. PRV Infections in Pigs and Other Animals
4.2. Potiential PRV Infections in Humans
| Case | Year | Occupation | Contact History | Clinical Symptoms | Antibody Detection | Nucleotide | Outcome | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 2017 | Swineherder | Sewage spilled into eyes | Fever, headache, visually impaired, endophthalmitis | gB antibody | + | Survived | [74] |
| 2 | 2017 | Pork dealer | Cut hand by a meat cleaver | Fever, headache, consciousness disorders, seizures, retinitis, encephalitis | PRV antibody-positive in three patients | + | Survived | [71] |
| 3 | 2017 | Cook | / | Fever, headache, seizures, consciousness disorders | + | Died | ||
| 4 | 2017 | Pig butcher | / | Fever, headache, seizures, consciousness disorders | + | Survived | ||
| 5 | 2018 | Pig butcher | / | Fever, seizures, consciousness disorders, retinitis | + | Survived | ||
| 6 | 2018 | Veterinary | Hands were punctured by a knife used for the autopsy of dead swine | Fever, headache, seizures, respiratory failure, disturbance of consciousness, encephalitis | gB antibody gE antibody | + | Survived | [75] |
| 7 | 2018 | Swineherder | Needlestick injury | Fever, seizures, consciousness disorders, encephalitis | neutralizing antibody | + | Survived | [76] |
| 8 | 2018 | Pig butcher | Finger hurt by a pig | Fever, headache, visual disturbances, convulsions | / | + | Survived | [77] |
| 9 | 2018 | Pig butcher | Hand injury before hospitalization | Fever, memory loss, consciousness disorders, convulsions, respiratory failure | + | Survived | ||
| 10 | 2018 | Swineherder | Hand injury before hospitalization | Fever, extremity tremors, respiratory failure, vision loss | + | Survived | ||
| 11 | 2018 | Porker cutter | Hand injury at work | Fever, convulsions, respiratory failure | + | Survived | ||
| 12 | 2018 | Porker cutter | No injury | Fever, extremity tremors, respiratory failure, vision loss | + | Survived | ||
| 13 | 2011 | Pork dealer | / | Fever, psychotic behavior, seizures | Died | [78] | ||
| 14 | 2018 | Pig butcher | / | Fever, seizures, consciousness loss, retinal necrosis | + | Died | ||
| 15 | 2018 | Swineherder | / | Fever, seizures, cognitive decline, respiratory failure, blindness | + | Survived | ||
| 16 | 2018 | Driver | / | Fever, seizures, consciousness loss | + | Survived | ||
| 17 | 2019 | Pork dealer | Contact with pork with injured fingers | Fever, seizures, consciousness disorder, encephalitis | PRV antibody positive | + | Survived | [79] |
| 18 | 2018 | Veterinary | / | Fever, headache, memory loss, seizures, consciousness disorders | gB antibody gE antibody Neutralizing antibody | + | Survived | [70] |
| 19 | 2019 | Pig butcher | Hand injury | Fever, headache, respiratory failure, memory loss, seizures, consciousness disorders | + | Survived | ||
| 20 | 2019 | Pig butcher | Finger injury | Fever, headache, respiratory failure, memory loss, seizures, consciousness disorders | + | Survived | ||
| 21 | 2019 | Pig butcher | / | Fever, headache, consciousness loss, seizures, bilateral retinal detachment, encephalitis | / | + | Survived | [80] |
| 22 | 2020 | Swineherder | / | Fever, coma, endophthalmitis | / | + | Survived | [81] |
| 23 | 2021 | Housewife | / | Fever, headache, seizures, coma, respiratory failure | / | + | Survived | [82] |
| 24 | 2021 | Swineherder | / | / | + | Died | ||
| 25 | 2021 | Pig butcher | Hand injury at work | Fever, consciousness loss, seizures, respiratory failure | / | + | discharged with ventilator support | [83] |
5. Vaccines and Diagnosis Methods for Pseudorabies
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kolb, A.W.; Lewin, A.C.; Moeller Trane, R.; McLellan, G.J.; Brandt, C.R. Phylogenetic and recombination analysis of the herpesvirus genus varicellovirus. BMC Genom. 2017, 18, 887. [Google Scholar] [CrossRef]
- Szpara, M.L.; Kobiler, O.; Enquist, L.W. A common neuronal response to alphaherpesvirus infection. J. Neuroimmune Pharmacol. 2010, 5, 418–427. [Google Scholar] [CrossRef]
- Lu, J.J.; Yuan, W.Z.; Zhu, Y.P.; Hou, S.H.; Wang, X.J. Latent pseudorabies virus infection in medulla oblongata from quarantined pigs. Transbound. Emerg. Dis. 2021, 68, 543–551. [Google Scholar] [CrossRef]
- An, T.Q.; Peng, J.M.; Tian, Z.J.; Zhao, H.Y.; Li, N.; Liu, Y.M.; Chen, J.Z.; Leng, C.L.; Sun, Y.; Chang, D.; et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef]
- Freuling, C.M.; Muller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. MMBR 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Csabai, Z.; Tombacz, D.; Deim, Z.; Snyder, M.; Boldogkoi, Z. Analysis of the Complete Genome Sequence of a Novel, Pseudorabies Virus Strain Isolated in Southeast Europe. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 1806842. [Google Scholar] [CrossRef]
- Hanson, R.P. The history of pseudorabies in the United States. J. Am. Vet. Med. Assoc. 1954, 124, 259–261. [Google Scholar]
- Petrovskis, E.A.; Timmins, J.G.; Gierman, T.M.; Post, L.E. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J. Virol. 1986, 60, 1166–1169. [Google Scholar] [CrossRef]
- OIE. OIE World Animal Health Information System. Available online: https://wahis.oie.int/#/dashboards/country-or-disease-dashboard (accessed on 26 April 2022).
- Caruso, C.; Vitale, N.; Prato, R.; Radaelli, M.C.; Zoppi, S.; Possidente, R.; Dondo, A.; Chiavacci, L.; Moreno Martin, A.M.; Masoero, L. Pseudorabies virus in North-West Italian wild boar (Sus scrofa) populations: Prevalence and risk factors to support a territorial risk-based surveillance. Vet. Ital. 2018, 54, 337–341. [Google Scholar] [CrossRef]
- Denzin, N.; Conraths, F.J.; Mettenleiter, T.C.; Freuling, C.M.; Muller, T. Monitoring of Pseudorabies in Wild Boar of Germany-A Spatiotemporal Analysis. Pathogens 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.K.; Ruiz-Fons, F.; Ryser-Degiorgis, M.P. A picture of trends in Aujeszky’s disease virus exposure in wild boar in the Swiss and European contexts. BMC Vet. Res. 2015, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, K.; Bevins, S.N.; Baroch, J.A.; Cumbee, J.C., Jr.; Chandler, S.C.; Woodruff, B.S.; Bigelow, T.T.; DeLiberto, T.J. Pseudorabies in feral swine in the United States, 2009–2012. J. Wildl. Dis. 2013, 49, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Jia, Z.; Zhang, D.; Liu, X.; Liu, D.; Guo, Y.; Liu, L.; Sun, Y.; Liu, Y.; Chen, H. Serological investigation of pseudorabies in large-scale pig farms in Beijing. Gansu Anim. Husb. Vet. 2017, 47, 65–68. (In Chinese) [Google Scholar] [CrossRef]
- Song, Q.; Liu, G.; Lin, W.; Wang, K.; Liu, Y.; Zhang, Y. Big data analysis of pseudorabies immune detection in the medium and large pig farms nationwide. Vet. Orientat. 2020, 17, 12–14. (In Chinese) [Google Scholar]
- Chen, C. Analysis on Surveillance of Pig Pseudorabies Antibody in Chongqing from 2015 to 2019. Master’s Thesis, Southwest University, Chongqing, China, 2020. (In Chinese). [Google Scholar]
- Lin, R.; Xia, L.; Yin, H.; Fei, S.; Fan, K.; Huang, S.; Li, X.; Yang, X.; Dai, A. Epidemiological survey of the wild pseudorabies virus infection in some large scale pig farms of southwest Fujian from 2013 to 2018 in Chinese. J. Anim. Infect. Dis. 2021, 29, 85–90. (In Chinese) [Google Scholar]
- Liu, X. Investigation and Control of Pseudorabies Infection in Large-Scale Pig Farms in Guizhou Province. Ph.D. Dissertation, Gansu Agricultural University, Lanzhou, China, 2019. (In Chinese). [Google Scholar]
- He, H.; Hu, S.; Li, J.; Feng, S.; Zhong, S.; Liu, F.; Chen, Z.; Pan, Y. Seroepidemiological investigation of the major viral diseases on Large-scale Pig Farms in Guangxi in 2015–2017. Heilongjiang Anim. Sci. Vet. Med. 2019, 14, 83–86+175. (In Chinese) [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, S.; Ran, X.; Yan, R. Surveillance of gE antibody against pseudorabies virus in pigs in Henan Province from 2017 to 2019. Anim. Breed. Feed. 2020, 19, 15–17. (In Chinese) [Google Scholar] [CrossRef]
- Zuo, Y.; Wang, B.; Han, L.; Wang, J.; Yuan, G.; Zhang, J.; Fan, J.; Zhong, F. Investigation of pseudorabies virus infection in Hebei province and phylogenetic analysis of its gE gene. Chin. J. Vet. Sci. 2021, 41, 224–230. (In Chinese) [Google Scholar]
- Zhou, H.; Pan, Y.; Liu, M.; Han, Z. Prevalence of Porcine Pseudorabies Virus and Its Coinfection Rate in Heilongjiang Province in China from 2013 to 2018. Viral Immunol. 2020, 33, 550–554. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.; Du, L.; Peng, M.; Liu, B.; Qiu, M. Investigation on the prevalence of main viral diseases in pig farms in some areas of Hunan Province in recent five years. Hunan J. Anim. Sci. Vet. Med. 2018, 4, 35–37. (In Chinese) [Google Scholar]
- Li, H.; Kang, Z.; Tan, M.; Zeng, Y.; Ji, H. Seroepidemiological survey of swine pseudorabies on large-scale pig farms in Jiangxi Province from 2016 to 2017. Acta Agric. Univ. Jiangxiensis 2018, 40, 1037–1041. (In Chinese) [Google Scholar] [CrossRef]
- Cui, W.; Fu, Y.; Wang, Y.; Lin, Y.; Li, X.; Zhang, Y.; Ying, L. Spotted surveillance and analysis of swine pseudorabies in Qinghai Province in 2016 and 2017. Shandong J. Anim. Sci. Vet. Med. 2019, 40, 13–14. (In Chinese) [Google Scholar]
- Ma, Z.; Han, Z.; Liu, Z.; Meng, F.; Wang, H.; Cao, L.; Li, Y.; Jiao, Q.; Liu, S.; Liu, M. Epidemiological investigation of porcine pseudorabies virus and its coinfection rate in Shandong Province in China from 2015 to 2018. J. Vet. Sci. 2020, 21, e36. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, W.; Chi, J.; Lu, C.; Li, X.; Li, C.; Jiang, S.; Tian, X.; Li, F.; Wang, L.; et al. Epidemiology of Porcine Pseudorabies from 2010 to 2018 in Tianjin, China. Viral Immunol. 2021, 34, 714–721. [Google Scholar] [CrossRef]
- Christensen, L.S.; Soerensen, K.J.; Lei, J.C. Restriction fragment pattern (RFP) analysis of genomes from Danish isolates of suid herpesvirus 1 (Aujezsky’s disease virus). Arch. Virol. 1987, 97, 215–224. [Google Scholar] [CrossRef]
- Herrmann, S.-C.; Heppner, B.; Ludwig, H. Pseudorabies Viruses from Clinical Outbreaks and Latent Infections Grouped into Four Major Genome Types. In Proceedings of the Latent Herpes Virus Infections in Veterinary Medicine: A Seminar in the CEC Programme of Coordination of Research on Animal Pathology, Tübingen, Germany, 21–24 September 1982; Wittmann, G., Gaskell, R.M., Rziha, H.J., Eds.; Springer: Dordrecht, The Netherlands, 1984; pp. 387–401. [Google Scholar]
- Christensen, L.S. The population biology of suid herpesvirus 1. Apmis Suppl. 1995, 48, 1–48. [Google Scholar]
- Muller, T.; Klupp, B.G.; Freuling, C.; Hoffmann, B.; Mojcicz, M.; Capua, I.; Palfi, V.; Toma, B.; Lutz, W.; Ruiz-Fon, F.; et al. Characterization of pseudorabies virus of wild boar origin from Europe. Epidemiol. Infect. 2010, 138, 1590–1600. [Google Scholar] [CrossRef][Green Version]
- Fonseca, A.A., Jr.; Magalhaes, C.G.; Sales, E.B.; D’Ambros, R.M.; Ciacci-Zanella, J.; Heinemann, M.B.; Leite, R.C.; Dos Reis, J.K. Genotyping of the pseudorabies virus by multiplex PCR followed by restriction enzyme analysis. ISRN Microbiol. 2011, 2011, 458294. [Google Scholar] [CrossRef]
- Deblanc, C.; Oger, A.; Simon, G.; Le Potier, M.F. Genetic Diversity among Pseudorabies Viruses Isolated from Dogs in France from 2006 to 2018. Pathogens 2019, 8, 266. [Google Scholar] [CrossRef]
- Ye, C.; Zhang, Q.Z.; Tian, Z.J.; Zheng, H.; Zhao, K.; Liu, F.; Guo, J.C.; Tong, W.; Jiang, C.G.; Wang, S.J.; et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 2015, 483, 32–43. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Szpara, M.L.; Tafuri, Y.R.; Parsons, L.; Shamim, S.R.; Verstrepen, K.J.; Legendre, M.; Enquist, L.W. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 2011, 7, e1002282. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Zhao, W.; Li, K.; Zhang, C.; Wang, C.; Su, S.; Zhou, J.; Lei, J.; Xing, G.; Sun, H.; et al. Genome Characteristics and Evolution of Pseudorabies Virus Strains in Eastern China from 2017 to 2019. Virol. Sin. 2019, 34, 601–609. [Google Scholar] [CrossRef]
- Hu, R.; Wang, L.; Liu, Q.; Hua, L.; Huang, X.; Zhang, Y.; Fan, J.; Chen, H.; Song, W.; Liang, W.; et al. Whole-Genome Sequence Analysis of Pseudorabies Virus Clinical Isolates from Pigs in China between 2012 and 2017 in China. Viruses 2021, 13, 1322. [Google Scholar] [CrossRef] [PubMed]
- Thiry, E.; Meurens, F.; Muylkens, B.; McVoy, M.; Gogev, S.; Thiry, J.; Vanderplasschen, A.; Epstein, A.; Keil, G.; Schynts, F. Recombination in alphaherpesviruses. Rev. Med. Virol. 2005, 15, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Henderson, L.M.; Katz, J.B.; Erickson, G.A.; Mayfield, J.E. In vivo and in vitro genetic recombination between conventional and gene-deleted vaccine strains of pseudorabies virus. Am. J. Vet. Res. 1990, 51, 1656–1662. [Google Scholar] [PubMed]
- Christensen, L.S.; Lomniczi, B. High frequency intergenomic recombination of suid herpesvirus 1 (SHV-1, Aujeszky’s disease virus). Arch. Virol. 1993, 132, 37–50. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, L.; Zhao, J.; Yin, X.; Feng, Y.; Wang, X.; Sun, X.; Zhou, Y.; Xu, Z. Genetic evolution analysis of novel recombinant pseudorabies virus strain in Sichuan, China. Transbound. Emerg. Dis. 2020, 67, 1428–1432. [Google Scholar] [CrossRef] [PubMed]
- Bo, Z.; Miao, Y.; Xi, R.; Gao, X.; Miao, D.; Chen, H.; Jung, Y.S.; Qian, Y.; Dai, J. Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound. Emerg. Dis. 2021, 68, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Guo, J.C.; Gao, J.C.; Wang, T.Y.; Zhao, K.; Chang, X.B.; Wang, Q.; Peng, J.M.; Tian, Z.J.; Cai, X.H.; et al. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 2016, 491, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Nauwynck, H.J.; Pensaert, M.B. Abortion induced by cell-associated pseudorabies virus in vaccinated sows. Am. J. Vet. Res. 1992, 53, 489–493. [Google Scholar]
- Salogni, C.; Lazzaro, M.; Giacomini, E.; Giovannini, S.; Zanoni, M.; Giuliani, M.; Ruggeri, J.; Pozzi, P.; Pasquali, P.; Boniotti, M.B.; et al. Infectious agents identified in aborted swine fetuses in a high-density breeding area: A three-year study. J. Vet. Diagn. Investig. 2016, 28, 550–554. [Google Scholar] [CrossRef]
- Sehl, J.; Teifke, J.P. Comparative Pathology of Pseudorabies in Different Naturally and Experimentally Infected Species—A Review. Pathogens 2020, 9, 633. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Kong, Z.; Liu, P.; Fu, Z.; Zhang, J.; Liu, M.; Shang, Y. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound. Emerg. Dis. 2020, 67, 518–522. [Google Scholar] [CrossRef]
- Ciarello, F.P.; Capucchio, M.T.; Ippolito, D.; Colombino, E.; Gibelli, L.R.M.; Fiasconaro, M.; Moreno Martin, A.M.; Di Marco Lo Presti, V. First Report of a Severe Outbreak of Aujeszky’s Disease in Cattle in Sicily (Italy). Pathogens 2020, 9, 954. [Google Scholar] [CrossRef]
- McFerran, J.B.; Dow, C. Virus Studies on Experimental Aujeszky’s Disease In Calves. J. Comp. Pathol. 1964, 74, 173–179. [Google Scholar] [CrossRef]
- Henderson, J.P.; Graham, D.A.; Stewart, D. An outbreak of Aujeszky’s disease in sheep in Northern Ireland. Vet. Rec. 1995, 136, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Mocsari, E.; Toth, C.; Meder, M.; Saghy, E.; Glavits, R. Aujeszky’s disease of sheep: Experimental studies on the excretion and horizontal transmission of the virus. Vet. Microbiol. 1987, 13, 353–359. [Google Scholar] [CrossRef]
- Cramer, S.D.; Campbell, G.A.; Njaa, B.L.; Morgan, S.E.; Smith, S.K., 2nd; McLin, W.R.T.; Brodersen, B.W.; Wise, A.G.; Scherba, G.; Langohr, I.M.; et al. Pseudorabies virus infection in Oklahoma hunting dogs. J. Vet. Diagn. Investig. 2011, 23, 915–923. [Google Scholar] [CrossRef]
- Pedersen, K.; Turnage, C.T.; Gaston, W.D.; Arruda, P.; Alls, S.A.; Gidlewski, T. Pseudorabies detected in hunting dogs in Alabama and Arkansas after close contact with feral swine (Sus scrofa). BMC Vet. Res. 2018, 14, 388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhong, C.; Wang, J.; Lu, Z.; Liu, L.; Yang, W.; Lyu, Y. Pathogenesis of natural and experimental Pseudorabies virus infections in dogs. Virol. J. 2015, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Hahn, E.C.; Tottewitz, F.; Kramer, M.; Klupp, B.G.; Mettenleiter, T.C.; Freuling, C. Pseudorabies virus in wild swine: A global perspective. Arch. Virol. 2011, 156, 1691–1705. [Google Scholar] [CrossRef]
- Caruso, C.; Dondo, A.; Cerutti, F.; Masoero, L.; Rosamilia, A.; Zoppi, S.; D’Errico, V.; Grattarola, C.; Acutis, P.L.; Peletto, S. Aujeszky’s disease in red fox (Vulpes vulpes): Phylogenetic analysis unravels an unexpected epidemiologic link. J. Wildl. Dis. 2014, 50, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Lian, K.; Zhang, M.; Zhou, L.; Song, Y.; Wang, G.; Wang, S. First report of a pseudorabies-virus-infected wolf (Canis lupus) in China. Arch. Virol. 2020, 165, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Zanin, E.; Capua, I.; Casaccia, C.; Zuin, A.; Moresco, A. Isolation and characterization of Aujeszky’s disease virus in captive brown bears from Italy. J. Wildl. Dis. 1997, 33, 632–634. [Google Scholar] [CrossRef]
- Schultze, A.E.; Maes, R.K.; Taylor, D.C. Pseudorabies and volvulus in a black bear. J. Am. Vet. Med. Assoc. 1986, 189, 1165–1166. [Google Scholar] [PubMed]
- Glass, C.M.; McLean, R.G.; Katz, J.B.; Maehr, D.S.; Cropp, C.B.; Kirk, L.J.; McKeirnan, A.J.; Evermann, J.F. Isolation of pseudorabies (Aujeszky’s disease) virus from a Florida panther. J. Wildl. Dis. 1994, 30, 180–184. [Google Scholar] [CrossRef]
- Masot, A.J.; Gil, M.; Risco, D.; Jimenez, O.M.; Nunez, J.I.; Redondo, E. Pseudorabies virus infection (Aujeszky’s disease) in an Iberian lynx (Lynx pardinus) in Spain: A case report. BMC Vet. Res. 2017, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, C.M.; Kanitz, C.L.; McCrocklin, S.M. Possible role of wild mammals in transmission of pseudorabies to swine. J. Wildl. Dis. 1980, 16, 601–614. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.T.; Hu, B.; Deng, X.Y.; Zhang, L.; Lian, S.Z.; Zhang, H.L.; Lv, S.; Xue, X.H.; Lu, R.G.; et al. Outbreak of severe pseudorabies virus infection in pig-offal-fed farmed mink in Liaoning Province, China. Arch. Virol. 2017, 162, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Skinner, G.R.; Ahmad, A.; Davies, J.A. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp. Immunol. Microbiol. Infect. Dis. 2001, 24, 255–269. [Google Scholar] [CrossRef]
- Mravak, S.; Bienzle, U.; Feldmeier, H.; Hampl, H.; Habermehl, K.O. Pseudorabies in man. Lancet 1987, 1, 501–502. [Google Scholar] [CrossRef] [PubMed]
- Schükrü-Aksel, I.; Tunman, Z. Aujeskysche Erkrankung in der Türkei bei Mensch und Tier. Z. Gesamte Neurol. Psychiatr. 1940, 169, 598–606. [Google Scholar] [CrossRef]
- Anusz, Z.; Szweda, W.; Popko, J.; Trybala, E. Is Aujeszky’s disease a zoonosis? Prz. Epidemiol. 1992, 46, 181–186. [Google Scholar]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.L.; Wu, Y.H.; Li, H.F.; Li, S.Y.; Fan, S.Y.; Wu, H.L.; Li, Y.J.; Lu, Y.L.; Han, J.; Zhang, W.C.; et al. Clinical experience and next-generation sequencing analysis of encephalitis caused by pseudorabies virus. Zhonghua Yi Xue Za Zhi 2018, 98, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, X.; Chen, H.; Yan, R.; Li, W.; Wang, X. Reply to Kitaura and Okamoto. Clin. Infect. Dis. 2021, 72, e693–e694. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Fu, S.H.; Chen, L.Y.; Li, F.; Deng, J.H.; Lu, X.C.; Wang, H.Y.; Tian, K.G. Detection of Pseudorabies Virus Antibodies in Human Encephalitis Cases. Biomed. Environ. Sci. BES 2020, 33, 444–447. [Google Scholar] [CrossRef]
- Ai, J.W.; Weng, S.S.; Cheng, Q.; Cui, P.; Li, Y.J.; Wu, H.L.; Zhu, Y.M.; Xu, B.; Zhang, W.H. Human Endophthalmitis Caused by Pseudorabies Virus Infection, China, 2017. Emerg. Infect. Dis. 2018, 24, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Han, H.; Wang, H.; Cui, Y.; Liu, H.; Ding, S. A Case of Human Viral Encephalitis Caused by Pseudorabies Virus Infection in China. Front. Neurol. 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, X.; Yuan, D.; Li, R.; Lu, J.; Li, X.; Tian, K.; Dai, E. Dynamic cerebrospinal fluid analyses of severe pseudorabies encephalitis. Transbound. Emerg. Dis. 2019, 66, 2562–2565. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guan, H.; Li, C.; Li, Y.; Wang, S.; Zhao, X.; Zhao, Y.; Liu, Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int. J. Infect. Dis. IJID 2019, 87, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Yuan, H.; Liu, L.; Li, H.; Wang, S.; Zhao, W.; Wu, Y.; Wang, P.; Hu, Y.; Han, J.; et al. Pseudorabies virus encephalitis in humans: A case series study. J. Neurovirol. 2020, 26, 556–564. [Google Scholar] [CrossRef]
- Wang, D.; Tao, X.; Fei, M.; Chen, J.; Guo, W.; Li, P.; Wang, J. Human encephalitis caused by pseudorabies virus infection: A case report. J. Neurovirol. 2020, 26, 442–448. [Google Scholar] [CrossRef]
- Hu, F.; Wang, J.; Peng, X.Y. Bilateral Necrotizing Retinitis following Encephalitis Caused by the Pseudorabies Virus Confirmed by Next-Generation Sequencing. Ocul. Immunol. Inflamm. 2021, 29, 922–925. [Google Scholar] [CrossRef]
- Ying, M.; Hu, X.; Wang, M.; Cheng, X.; Zhao, B.; Tao, Y. Vitritis and retinal vasculitis caused by pseudorabies virus. J. Int. Med. Res. 2021, 49, 3000605211058990. [Google Scholar] [CrossRef]
- Zhou, Y.; Nie, C.; Wen, H.; Long, Y.; Zhou, M.; Xie, Z.; Hong, D. Human viral encephalitis associated with suid herpesvirus 1. Neurol. Sci. 2021, 43, 2681–2692. [Google Scholar] [CrossRef]
- Yan, W.; Hu, Z.; Zhang, Y.; Wu, X.; Zhang, H. Case Report: Metagenomic Next-Generation Sequencing for Diagnosis of Human Encephalitis and Endophthalmitis Caused by Pseudorabies Virus. Front. Med. 2021, 8, 753988. [Google Scholar] [CrossRef]
- Luo, Y.; Li, N.; Cong, X.; Wang, C.H.; Du, M.; Li, L.; Zhao, B.; Yuan, J.; Liu, D.D.; Li, S.; et al. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet. Microbiol. 2014, 174, 107–115. [Google Scholar] [CrossRef]
- Yu, T.; Chen, F.; Ku, X.; Fan, J.; Zhu, Y.; Ma, H.; Li, S.; Wu, B.; He, Q. Growth characteristics and complete genomic sequence analysis of a novel pseudorabies virus in China. Virus Genes 2016, 52, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Gu, Z.; Jin, L.; Lv, L.; Wang, J.; Sun, T.; Bai, J.; Sun, H.; Wang, X.; Jiang, P. Polymorphisms affecting the gE and gI proteins partly contribute to the virulence of a newly-emergent highly virulent Chinese pseudorabies virus. Virology 2018, 519, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Husak, P.J.; Kuo, T.; Enquist, L.W. Pseudorabies virus membrane proteins gI and gE facilitate anterograde spread of infection in projection-specific neurons in the rat. J. Virol. 2000, 74, 10975–10983. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heldwein, E.E.; Krummenacher, C. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. CMLS 2008, 65, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.Q.; Tong, W.; Zheng, H.; Li, L.W.; Li, G.X.; Gao, F.; Wang, T.; Liang, C.; Ye, C.; Wu, J.Q.; et al. Variations in glycoprotein B contribute to immunogenic difference between PRV variant JS-2012 and Bartha-K61. Vet. Microbiol. 2017, 208, 97–105. [Google Scholar] [CrossRef]
- Sozzi, E.; Moreno, A.; Lelli, D.; Cinotti, S.; Alborali, G.L.; Nigrelli, A.; Luppi, A.; Bresaola, M.; Catella, A.; Cordioli, P. Genomic characterization of pseudorabies virus strains isolated in Italy. Transbound. Emerg. Dis. 2014, 61, 334–340. [Google Scholar] [CrossRef]
- Spear, P.G.; Eisenberg, R.J.; Cohen, G.H. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 2000, 275, 1–8. [Google Scholar] [CrossRef]
- Menotti, L.; Lopez, M.; Avitabile, E.; Stefan, A.; Cocchi, F.; Adelaide, J.; Lecocq, E.; Dubreuil, P.; Campadelli-Fiume, G. The murine homolog of human Nectin1delta serves as a species nonspecific mediator for entry of human and animal alpha herpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. USA 2000, 97, 4867–4872. [Google Scholar] [CrossRef]
- Milne, R.S.; Connolly, S.A.; Krummenacher, C.; Eisenberg, R.J.; Cohen, G.H. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 2001, 281, 315–328. [Google Scholar] [CrossRef]
- Li, A.; Lu, G.; Qi, J.; Wu, L.; Tian, K.; Luo, T.; Shi, Y.; Yan, J.; Gao, G.F. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. PLoS Pathog. 2017, 13, e1006314. [Google Scholar] [CrossRef]
- Nixdorf, R.; Schmidt, J.; Karger, A.; Mettenleiter, T.C. Infection of Chinese hamster ovary cells by pseudorabies virus. J. Virol. 1999, 73, 8019–8026. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.; Klupp, B.G.; Karger, A.; Mettenleiter, T.C. Adaptability in herpesviruses: Glycoprotein D-independent infectivity of pseudorabies virus. J. Virol. 1997, 71, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Holper, J.E.; Grey, F.; Baillie, J.K.; Regan, T.; Parkinson, N.J.; Hoper, D.; Thamamongood, T.; Schwemmle, M.; Pannhorst, K.; Wendt, L.; et al. A Genome-Wide CRISPR/Cas9 Screen Reveals the Requirement of Host Sphingomyelin Synthase 1 for Infection with Pseudorabies Virus Mutant gD(-)Pass. Viruses 2021, 13, 1574. [Google Scholar] [CrossRef]
- Tiwari, V.; Oh, M.J.; Kovacs, M.; Shukla, S.Y.; Valyi-Nagy, T.; Shukla, D. Role for nectin-1 in herpes simplex virus 1 entry and spread in human retinal pigment epithelial cells. FEBS J. 2008, 275, 5272–5285. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Shukla, S.Y.; Yue, B.; Shukla, D. Herpes simplex virus type 2 entry into cultured human corneal fibroblasts is mediated by herpesvirus entry mediator. J. Gen. Virol. 2007, 88, 2106–2110. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Clement, C.; Scanlan, P.M.; Kowlessur, D.; Yue, B.Y.; Shukla, D. A role for herpesvirus entry mediator as the receptor for herpes simplex virus 1 entry into primary human trabecular meshwork cells. J. Virol. 2005, 79, 13173–13179. [Google Scholar] [CrossRef]
- Akhtar, J.; Tiwari, V.; Oh, M.J.; Kovacs, M.; Jani, A.; Kovacs, S.K.; Valyi-Nagy, T.; Shukla, D. HVEM and nectin-1 are the major mediators of herpes simplex virus 1 (HSV-1) entry into human conjunctival epithelium. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4026–4035. [Google Scholar] [CrossRef]
- Shah, A.; Farooq, A.V.; Tiwari, V.; Kim, M.J.; Shukla, D. HSV-1 infection of human corneal epithelial cells: Receptor-mediated entry and trends of re-infection. Mol. Vis. 2010, 16, 2476–2486. [Google Scholar]
- Karaba, A.H.; Kopp, S.J.; Longnecker, R. Herpesvirus entry mediator and nectin-1 mediate herpes simplex virus 1 infection of the murine cornea. J. Virol. 2011, 85, 10041–10047. [Google Scholar] [CrossRef]
- Edwards, R.G.; Kopp, S.J.; Karaba, A.H.; Wilcox, D.R.; Longnecker, R. Herpesvirus entry mediator on radiation-resistant cell lineages promotes ocular herpes simplex virus 1 pathogenesis in an entry-independent manner. mBio 2015, 6, e01532-15. [Google Scholar] [CrossRef]
- Tang, Y.D.; Liu, J.T.; Wang, T.Y.; An, T.Q.; Sun, M.X.; Wang, S.J.; Fang, Q.Q.; Hou, L.L.; Tian, Z.J.; Cai, X.H. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus Res. 2016, 225, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Li, G.; Liang, C.; Liu, F.; Tian, Q.; Cao, Y.; Li, L.; Zheng, X.; Zheng, H.; Tong, G. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antivir. Res. 2016, 130, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.M.; Zhou, Q.; Song, W.B.; Sun, E.C.; Zhang, M.M.; He, Q.G.; Chen, H.C.; Wu, B.; Liu, Z.F. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine 2015, 33, 5733–5740. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yao, J.; Yang, Y.; Luo, W.; Yuan, X.; Yang, L.; Wang, A. Current Status and Challenge of Pseudorabies Virus Infection in China. Virol. Sin. 2021, 36, 588–607. [Google Scholar] [CrossRef]
- Cong, X.; Lei, J.L.; Xia, S.L.; Wang, Y.M.; Li, Y.; Li, S.; Luo, Y.; Sun, Y.; Qiu, H.J. Pathogenicity and immunogenicity of a gE/gI/TK gene-deleted pseudorabies virus variant in susceptible animals. Vet. Microbiol. 2016, 182, 170–177. [Google Scholar] [CrossRef]
- Lv, L.; Liu, X.; Jiang, C.; Wang, X.; Cao, M.; Bai, J.; Jiang, P. Pathogenicity and immunogenicity of a gI/gE/TK/UL13-gene-deleted variant pseudorabies virus strain in swine. Vet. Microbiol. 2021, 258, 109104. [Google Scholar] [CrossRef]
- Yan, S.; Huang, B.; Bai, X.; Zhou, Y.; Guo, L.; Wang, T.; Shan, Y.; Wang, Y.; Tan, F.; Tian, K. Construction and Immunogenicity of a Recombinant Pseudorabies Virus Variant With TK/gI/gE/11k/28k Deletion. Front. Vet. Sci. 2021, 8, 797611. [Google Scholar] [CrossRef]
| Region | gE Positive Rate (gE Positive Samples/Total Samples) | Reference | |||
|---|---|---|---|---|---|
| 2016 | 2017 | 2018 | 2019 | ||
| Beijing | 33.66% (662/1966) | / | 20% (4/20) | / | [15,16] |
| Chongqing | 1.6% (11/702) | 9.4% (60/637) | 7.5% (60/798) | 11.5% (53/460) | [17] |
| Fujian | 37.37% (111/297) | 26.11% (53/203) | 27.32% (50/183) | / | [18] |
| Guizhou | 1.89% (27/1480) | 16.85% (538/3192) | 16.85% (538/3192) | 8.5% (92/1078) | [19] |
| Guangdong | / | / | 33.60% (1084/3226) | / | [16] |
| Guangxi | 22.87% (854/3734) | 23.71% (996/4200) | 20.60% (766/3718) | / | [16,20] |
| Henan | 26.21% (3513/13,404) | 28.82% (4755/16,497) | 25.31% (3000/11,854) | 26.69% (3460/12,963) | [21] |
| Hebei | / | / | 62.74% (367/585) | 50.05% (5245/10,479) | [16,22] |
| Heilongjiang | 15.36% (474/3086) | 15.50% (539/3478) | 11.64% (318/2731) | / | [23] |
| Hubei | / | / | 13.21% (123/931) | / | [16] |
| Hunan | 24.4% (344/1410) | 23.2% (349/1504) | 44.64% (1011/2265) | / | [24] |
| Jiangxi | 40.1% (362/902) | 34.6% (318/919) | 27.41% (1769/6455) | / | [16,25] |
| Qinghai | 28.17% (131/465) | 19.75% (157/794) | / | / | [26] |
| Shandong | 57.8% (2909/5033) | 50.4% (2476/4915) | 55.2% (2072/3753) | / | [27] |
| Sichuan | / | / | 32.49% (952/2930) | / | [16] |
| Yunnan | / | / | 17.07% (306/1793) | / | |
| Tianjin | 40.43% (970/2399) | 37.02% (2219/3793) | 51.59% (1957/3793) | / | [28] |
| Strain | Isolation Country | Recombination Pattern | Recombination Site | Reference |
|---|---|---|---|---|
| Yangsan | South Korean | genotype I and genotype II | UL21 | [35] |
| FJ-W2, FJ-ZXF | Fujian, China | genotype I and genotype II | gB | [38] |
| FJ62 | Sichuan, China | genotype I (Wild boar) and genotype II | gB | [43] |
| JSY13 | Jiangsu, China | genotype I (Bartha) and genotype II (JSY7) | UL42, UL19, UL18, UL10 | [44] |
| SC | China | genotype I (Bartha) and genotype II | gC | [45] |
| HeN1, Qihe547 | China | genotype I and genotype II (vaccine strains) | / | [36] |
| SC, LA | China | genotype I and genotype II (early strains) | / | |
| ZJ01 | China | genotype I and genotype II | / |
| Case | Year | Occupation | Contact History | Clinical Symptoms | Antibody Detection | Pathogen Detection | Outcome | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 1914 | Lab technician | A laboratory cat with pseudorabies | Swelling, reddening, and intense itching of the wound and the surrounding area | / | / | Survived | [66] |
| 2 | 1914 | Lab technician | / | / | Survived | |||
| 3 | 1940 | Lab technician | Got injured during contacting with a dog infected with PRV | Pruritus, erythema, pain, and aphthous stomatitis | / | / | Survived | [68] |
| 4 | 1940 | Lab technician | / | / | Survived | |||
| 5 | 1963 | Animal handler | A dog infected with PRV following an outbreak of pseudorabies on a pig farm | Severe throat pain and weakness in the legs | / | / | Survived | [66] |
| 6 | 1963 | Animal handler | / | / | Survived | |||
| 7 | 1963 | Veterinary | / | / | Survived | |||
| 8 | 1963 | Nightwatchman | / | / | Survived | |||
| 9 | 1983 | Tourist in Denmark | Indirect contact with a sick cat | Anorexia, weight loss, headache, arthralgia | Neutralizing antibody Titer: 1:8–1:16 | / | Survived | [67] |
| 10 | 1986 | Tourist in France | Close contact with cats and other domestic animals | Dysphagia, experienced strange smells and taste | / | |||
| 11 | 1986 | Tourist in France | / | |||||
| 12–17 | 1992 | Six workers on a cattle farm | Direct contact with PRV infected cattle | Pruritus of the palms that spread onto the arms and shoulders and lasted for several days | / | / | Survived | [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Kuang, Y.; Li, Y.; Guo, H.; Zhou, C.; Guo, S.; Tan, C.; Wu, B.; Chen, H.; Wang, X. The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans. Viruses 2022, 14, 1463. https://doi.org/10.3390/v14071463
Liu Q, Kuang Y, Li Y, Guo H, Zhou C, Guo S, Tan C, Wu B, Chen H, Wang X. The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans. Viruses. 2022; 14(7):1463. https://doi.org/10.3390/v14071463
Chicago/Turabian StyleLiu, Qingyun, Yan Kuang, Yafei Li, Huihui Guo, Chuyue Zhou, Shibang Guo, Chen Tan, Bin Wu, Huanchun Chen, and Xiangru Wang. 2022. "The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans" Viruses 14, no. 7: 1463. https://doi.org/10.3390/v14071463
APA StyleLiu, Q., Kuang, Y., Li, Y., Guo, H., Zhou, C., Guo, S., Tan, C., Wu, B., Chen, H., & Wang, X. (2022). The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans. Viruses, 14(7), 1463. https://doi.org/10.3390/v14071463







