Biological Characteristics of Infectious Laryngotracheitis Viruses Isolated in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Sequencing
2.3. Sequence Analysis
2.4. Proliferation Test of Three ILTVs
2.5. Chicken Experiment
2.6. Quantitative Analysis by Real-Time PCR
2.7. Antibodies Detected in Chickens
2.8. Statistical Analysis
3. Results
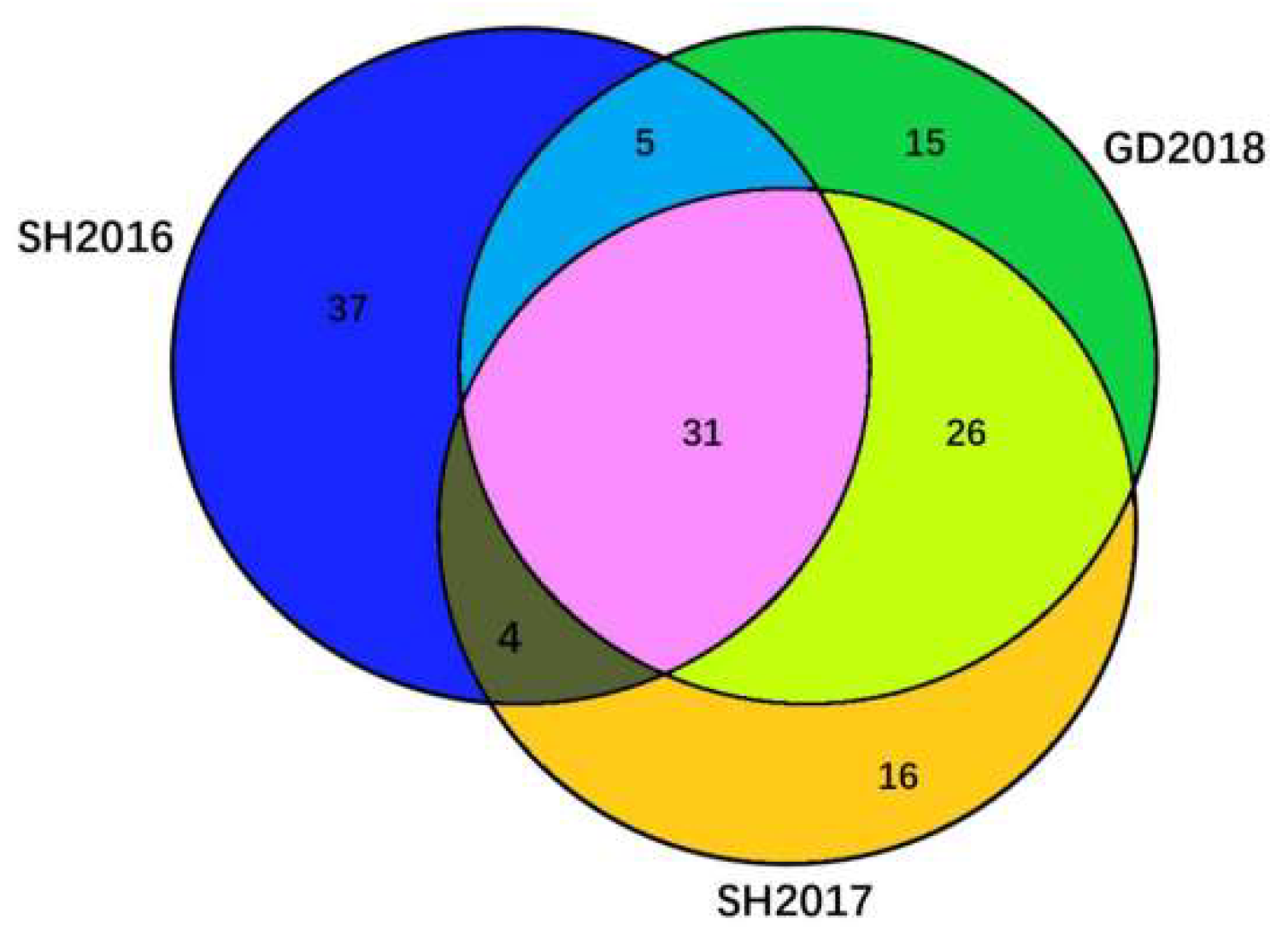
3.1. Genomic Characteristics of Three ILTVs
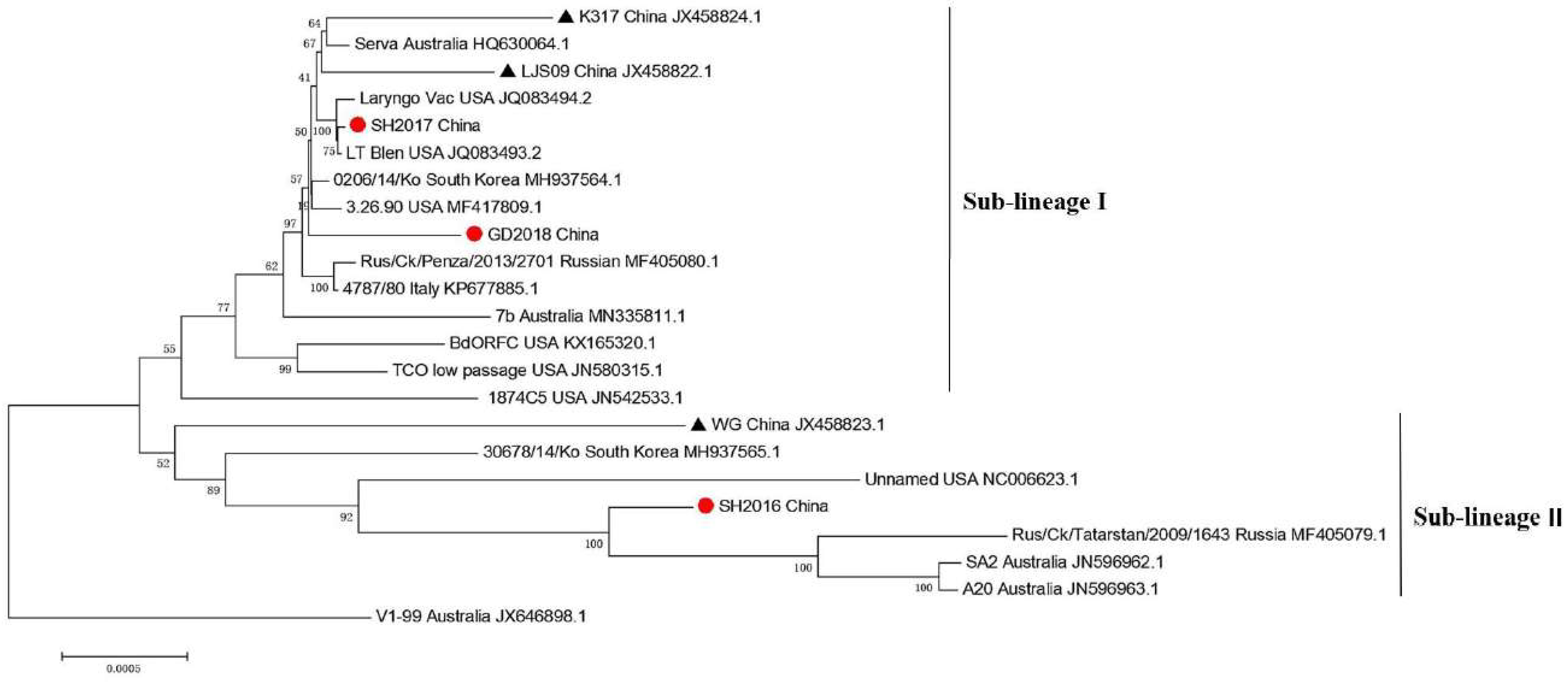
3.2. Phylogenetics of Three ILTVs
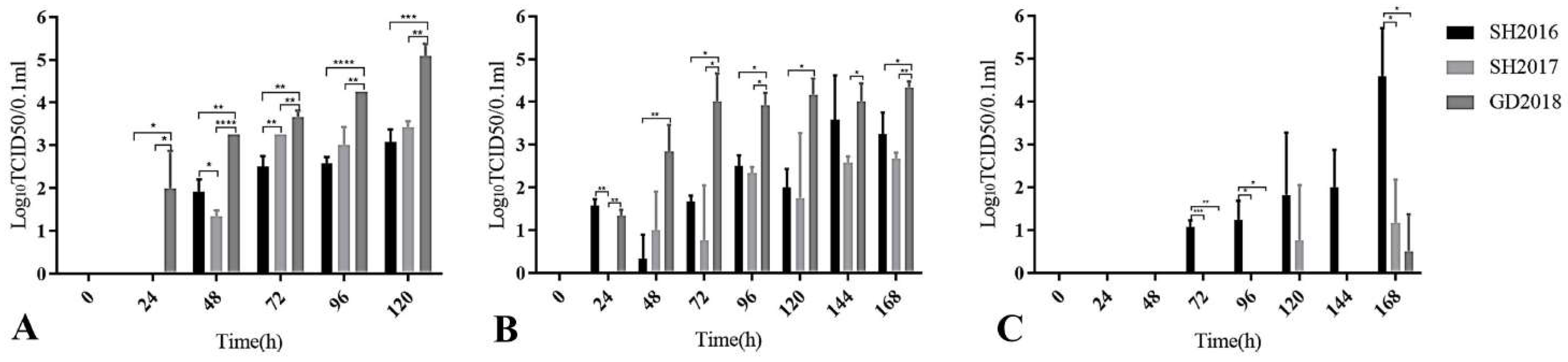
3.3. Proliferation of Three ILTVs
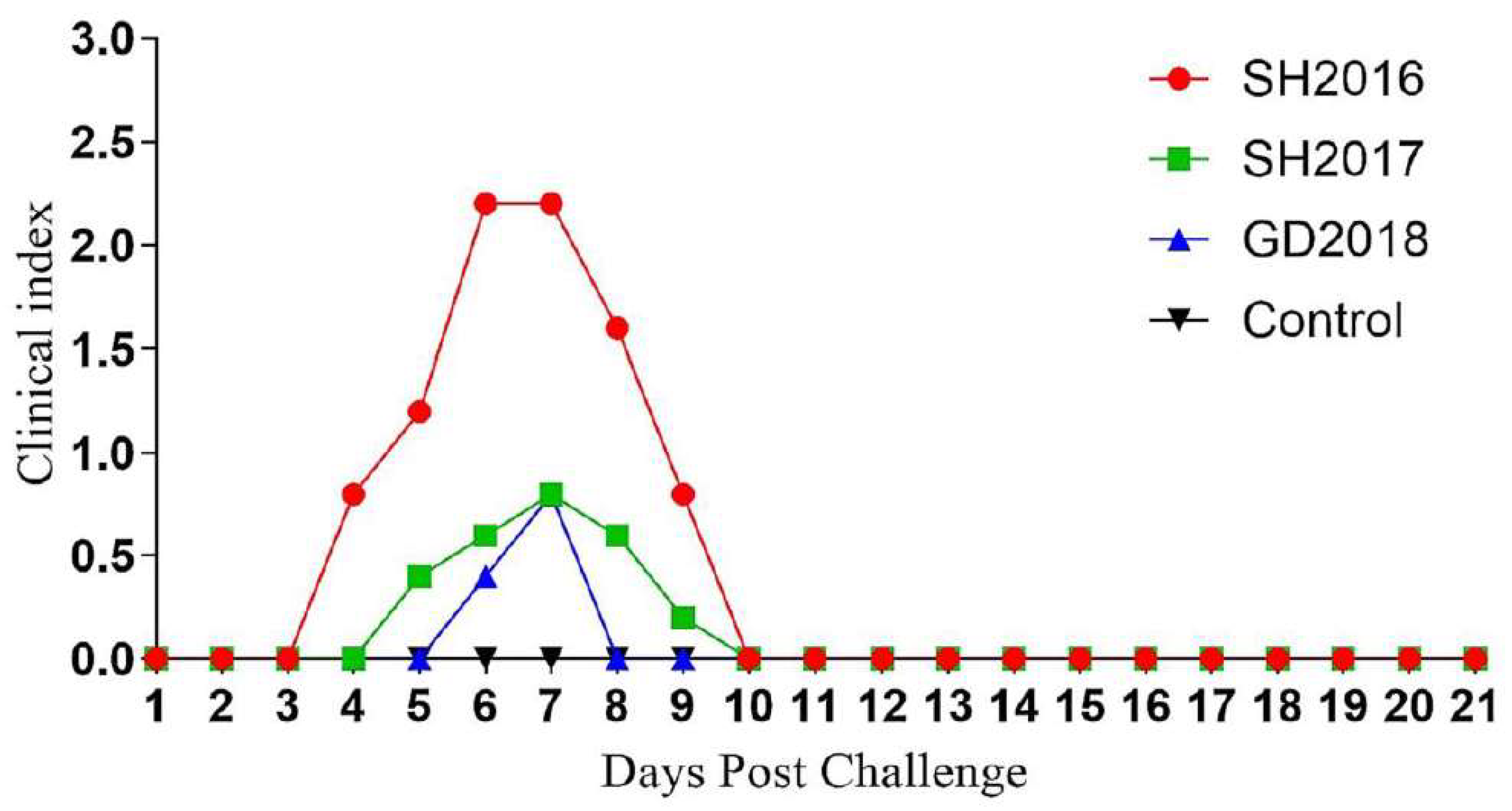
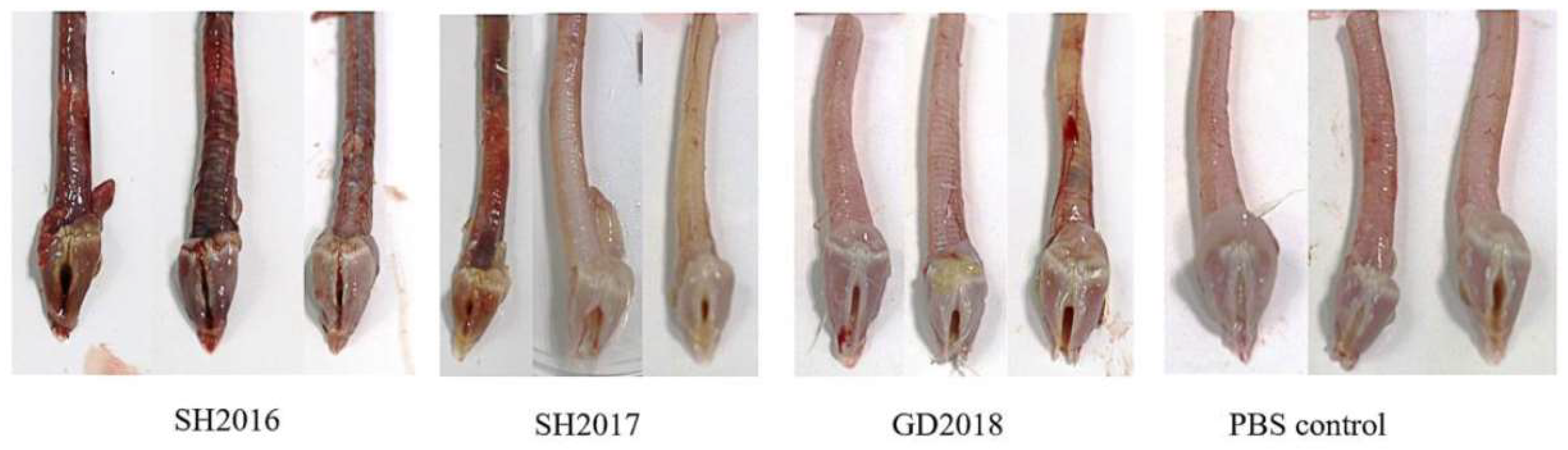
3.4. Pathogenicity of ILTVs in Chickens
3.5. Immune Response of Chickens Challenged with ILTVs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- King, A.; Carstens, E.B.; Lefkowitz, E.J. Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- May, H.G.; Tittsler, R.P. Tracheolaryngitis in poultry. J. Am. Vet. Med. Assoc. 1996, 40, 494–500. [Google Scholar]
- Bagust, T.; Jones, R.; Guy, J. Avian Infectious laryngotracheitis. Rev. Sci. Tech. 2000, 19, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Koski, D.M.; Predgen, A.S.; Trampel, D.W.; Conrad, S.K.; Narwold, D.R.; Hermann, J.R. Comparison of the pathogenicity of the USDA challenge virus strain to a field strain of infectious laryngotracheitis virus. Biologicals J. Int. Assoc. Biol. Standardizat. 2015, 43, 232–237. [Google Scholar] [CrossRef]
- Nazir, S.; Yegoraw, A.A.; Charlesworth, R.P.G.; Williamson, S.; Sharpe, S.; Walkden-Brown, S.W.; Gerber, P.F. Marked differences in virulence of three Australian field isolates of infectious laryngotracheitis virus in meat and layer chickens. Avian Pathol. 2020, 49, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Han, M.G.; Kim, S.J. Comparison of virulence and restriction endonuclease cleavage patterns of infectious laryngotracheitis viruses isolated in Korea. Avian Pathol. 2001, 30, 337–344. [Google Scholar] [CrossRef]
- Oldoni, I.; Rodríguez-Avila, A.; Riblet, S.M.; Zavala, G.; García, M. Pathogenicity and growth characteristics of selected infectious laryngotracheitis virus strains from the United States. Avian Pathol. 2009, 38, 47–53. [Google Scholar] [CrossRef]
- Han, M.G.; Kweon, C.H.; Mo, I.P.; Kim, S.J. Pathogenicity and vaccine efficacy of a thymidine kinase gene deleted infectious laryngotracheitis virus expressing the green fluorescent protein gene. Arch. Virol. 2002, 147, 1017–1031. [Google Scholar] [CrossRef]
- Fuchs, W.; Wiesner, D.; Veits, J.; Teifke, J.P.; Mettenleiter, T.C. In Vitro and In Vivo Relevance of Infectious Laryngotracheitis Virus gJ Proteins That Are Expressed from Spliced and Nonspliced mRNAs. J. Virol. 2005, 79, 705–716. [Google Scholar] [CrossRef]
- Devlin, J.M.; Browning, G.; Hartley, C.; Kirkpatrick, N.C.; Mahmoudian, A.; Noormohammadi, A.; Gilkerson, J. Glycoprotein G is a virulence factor in infectious laryngotracheitis virus. J. Gen. Virol. 2006, 87, 2839–2847. [Google Scholar] [CrossRef]
- Helferich, D.; Veits, J.; Teifke, J.P.; Mettenleiter, T.C.; Fuchs, W. The UL47 gene of avian infectious laryngotracheitis virus is not essential for in vitro replication but is relevant for virulence in chickens. J. Gen. Virol. 2007, 88, 732–742. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, C.; Wang, Y. Multiple Comparison Analysis of Two New Genomic Sequences of ILTV Strains from China with Other Strains from Different Geographic Regions. PLoS ONE 2015, 10, e0132747. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Zhao, Y.; Cui, X.; Zhang, X.; Cui, H.; Xue, M.; Wang, Y. Complete Genome Sequence of the First Chinese Virulent Infectious Laryngotracheitis Virus. PLoS ONE 2013, 8, e70154. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, O.; Hartley, C.A.; Devlin, J.M.; Browning, G.F.; Noormohammadi, A.H.; Lee, S.-W. Development and application of high-resolution melting analysis for the classification of infectious laryngotracheitis virus strains and detection of recombinant progeny. Arch Virol. 2019, 164, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12–R12. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Jones, R. Comparison of cultural methods for primary isolation of infectious laryngotracheitis virus from field material. Avian Pathol. 1988, 17, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.P.; Van Der Meer, F.; Checkley, S.; Joseph, T.; King, R.; Ravi, M.; Peters, D.; Fonseca, K.; Gagnon, C.A.; Provost, C.; et al. Analysis of Whole-Genome Sequences of Infectious laryngotracheitis Virus Isolates from Poultry Flocks in Canada: Evidence of Recombination. Viruses 2020, 12, 1302. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Vagnozzi, A.; Zavala, G.; Riblet, S.M.; Mundt, A.; García, M. Protection induced by commercially available live-attenuated and recombinant viral vector vaccines against infectious laryngotracheitis virus in broiler chickens. Avian Pathol. 2012, 41, 21–31. [Google Scholar] [CrossRef]
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef]
- Leib, D.A.; Bradbury, J.M.; Hart, C.A.; Mccarthy, K. Genome isomerism in two alphaherpesviruses:Herpesvirus saimiri-1(Herpesvirus tamarinus) and avian infectious laryngotracheitis virus. Arch. Virol. 1987, 93, 287–294. [Google Scholar] [CrossRef]
- Roizman, B. The Family Herpesviridae: A brief introduction. Fields Virol. 2001, 201, 2381–2397. [Google Scholar]
- Chandra, Y.G.; Lee, J.; Kong, B.-W. Genome sequence comparison of two United States live attenuated vaccines of infectious laryngotracheitis virus (ILTV). Virus Genes 2012, 44, 470–474. [Google Scholar] [CrossRef]
- Suzutani, T. The relationship between gene function and virulence in alpha-herpesviruses Nihon Rinsho. Jpn. J. Clin. Med. 2000, 58, 801–806. [Google Scholar]
- Atasoy, M.O.; Rohaim, M.A.; Munir, M. Simultaneous Deletion of Virulence Factors and Insertion of Antigens into the Infectious Laryngotracheitis Virus Using NHEJ-CRISPR/Cas9 and Cre–Lox System for Construction of a Stable Vaccine Vector. Vaccines 2019, 7, 207. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Veits, J.; Helferich, D.; Granzow, H.; Teifke, J.P.; Mettenleiter, T.C. Molecular biology of avian infectious laryngotracheitis virus. Veter- Res. 2007, 38, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Yadav, K.; Kumar, R.; Chaudhary, N.; Kumar, S. Glycoprotein D peptide-based diagnostic approach for the detection of avian infectious laryngotracheitis antibodies. Avian Pathol. 2019, 48, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, R.J.; Atanasiu, D.; Cairns, T.M.; Gallagher, J.R.; Krummenacher, C.; Cohen, G.H. Herpes Virus Fusion and Entry: A Story with Many Characters. Viruses 2012, 4, 800–832. [Google Scholar] [CrossRef]
- Coppo, M.J.; Hartley, C.A.; Devlin, J.M. Immune responses to infectious laryngotracheitis virus. Dev. Comp. Immunol. 2013, 41, 454–462. [Google Scholar] [CrossRef]
- Devlin, J.M.; Browning, G.F.; Gilkerson, J.R. A glycoprotein I- and glycoprotein E-deficient mutant of infectious laryngotracheitis virus exhibits impaired cell-to-cell spread in cultured cells. Arch. Virol. 2006, 151, 1281–1289. [Google Scholar] [CrossRef]
- Mundt, A.; Mundt, E.; Hogan, R.J.; García, M. Glycoprotein J of infectious laryngotracheitis virus is required for efficient egress of infectious virions from cells. J. Gen. Virol. 2011, 92, 2586–2589. [Google Scholar] [CrossRef]
- Huang, Q.; Mat-Arip, Y.; Guo, P. Sequencing of a 5.5-kb DNA Fragment and Identification of a Gene Coding for a Subunit of the Helicase/Primase Complex of Avian Laryngotracheitis virus (ILTV). Virus Genes 1997, 15, 119–121. [Google Scholar] [CrossRef]
- Wang, Z.; Qiao, Y.; Chen, Z.; Liang, Y.; Cui, L.; Zhang, Y.; Li, X.; Xu, L.; Wei, P.; Liu, S.; et al. Fos Facilitates Gallid Alpha-Herpesvirus 1 Infection by Transcriptional Control of Host Metabolic Genes and Viral Immediate Early Gene. Viruses 2021, 13, 1110. [Google Scholar] [CrossRef]
- Liao, Y.; Lupiani, B.; Ai-Mahmood, M.; Reddy, S.M. Marek’s disease virus US3 protein kinase phosphorylates chicken HDAC 1 and 2 and regulates viral replication and pathogenesis. PLOS Pathog. 2021, 17, e1009307. [Google Scholar] [CrossRef]
- Blakey, J.; Stoute, S.; Crossley, B.; Mete, A. Retrospective analysis of infectious laryngotracheitis in backyard chicken flocks in California, 2007–2017, and determination of strain origin by partial ICP4 sequencing. J. Vet. Diagn. Invest. 2019, 31, 350–358. [Google Scholar] [CrossRef] [PubMed]
- McMenamy, M.; McKillen, J.; Hjertner, B.; Kiss, I.; Yacoub, A.; Leijon, M.; Duffy, C.; Belák, S.; Welsh, M.; Allan, G. Development and comparison of a Primer-Probe Energy Transfer based assay and a 5′ conjugated Minor Groove Binder assay for sensitive real-time PCR detection of infectious laryngotracheitis virus. J. Virol. Methods 2011, 175, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-G.; Ma, J.; Xue, C.-Y.; Wang, W.; Guo, C.; Chen, F.; Qin, J.-P.; Huang, N.-H.; Bi, Y.-Z.; Cao, Y.-C. Dynamic distribution and tissue tropism of infectious laryngotracheitis virus in experimentally infected chickens. Arch. Virol. 2012, 158, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Hui-Xin, L.I.; Han, Z.X.; Kong, X.G.; Liu, S.W. Pathogenicity study of infectious laryngotracheitis virus LJS09. Chin. J. Prev. Vet. Med. 2015, 3, 181–184. [Google Scholar]
- Pekosz, A.; Griot, C.; Nathanson, N.; Gonzalez-Scarano, F. Tropism of Bunyaviruses: Evidence for a G1 Glycoprotein-Mediated Entry Pathway Common to the California Serogroup. Virology 1995, 214, 339–348. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Zhang, Z.; Su, X.; Lu, H.; Li, X.; Yuan, C.; Liu, Q.; Teng, Q.; Geri, L.; Li, Z. Biological Characteristics of Infectious Laryngotracheitis Viruses Isolated in China. Viruses 2022, 14, 1200. https://doi.org/10.3390/v14061200
Wu M, Zhang Z, Su X, Lu H, Li X, Yuan C, Liu Q, Teng Q, Geri L, Li Z. Biological Characteristics of Infectious Laryngotracheitis Viruses Isolated in China. Viruses. 2022; 14(6):1200. https://doi.org/10.3390/v14061200
Chicago/Turabian StyleWu, Mi, Zhifei Zhang, Xin Su, Haipeng Lu, Xuesong Li, Chunxiu Yuan, Qinfang Liu, Qiaoyang Teng, Letu Geri, and Zejun Li. 2022. "Biological Characteristics of Infectious Laryngotracheitis Viruses Isolated in China" Viruses 14, no. 6: 1200. https://doi.org/10.3390/v14061200
APA StyleWu, M., Zhang, Z., Su, X., Lu, H., Li, X., Yuan, C., Liu, Q., Teng, Q., Geri, L., & Li, Z. (2022). Biological Characteristics of Infectious Laryngotracheitis Viruses Isolated in China. Viruses, 14(6), 1200. https://doi.org/10.3390/v14061200





