Abstract
Human adenoviruses (HAdV) express either one or two virus-associated RNAs (VA RNAI or VA RNAII). The structure of VA RNA resembles human precursor microRNAs (pre-miRNA), and, like human pre-miRNA, VA RNA can be processed by DICER into small RNAs that resemble human miRNA. VA RNA-derived miRNA (mivaRNA) can mimic human miRNA post-transcriptional gene repression by binding to complementary sequences in the 3′ UTR of host mRNA. HAdV14 is a member of the B2 subspecies of species B adenovirus, and the emergent strain HAdV14p1 is associated with severe respiratory illness that can lead to acute respiratory distress syndrome. Utilizing small RNA sequencing, we identified four main mivaRNAs generated from the HAdV14/p1 VA RNA gene, two from each of the 5′ and 3′ regions of the terminal stem. There were temporal expression changes in the abundance of 5′ and 3′ mivaRNAs, with 3′ mivaRNAs more highly expressed early in infection and 5′ mivaRNAs more highly expressed later in infection. In addition, there are differences in expression between the emergent and reference strains, with HAdV14 expressing more mivaRNAs early during infection and HAdV14p1 having higher expression later during infection. HAdV14/p1 mivaRNAs were also shown to repress gene expression in a luciferase gene reporter system. Our results raise the question as to whether differential expression of mivaRNAs during HAdV14p1 infection could play a role in the increased pathogenesis associated with the emergent strain.
1. Introduction
Human adenoviruses (HAdVs) are non-enveloped viruses with a linear double-stranded DNA genome of ~36,000 bp. HAdVs are grouped into seven species (A–G) and consist of over 100 different types (consecutively numbered) based on genomic sequencing. HAdVs can infect the respiratory, gastrointestinal, and urinary tracts as well as the conjunctiva. Overall, HAdV usually results in mild, self-limited infections in immunocompetent individuals. HAdV14 is a member of the B2 subgroup of adenovirus and, as such, predominately causes kidney and urinary tract infections but can also be associated with respiratory infections. Outbreaks of emergent strains of Ad have resulted in severe and sometimes fatal infections in otherwise healthy people. In the past fifteen years, an emergent strain of HAdV14, HAdV14p1, emerged first in the U.S. and subsequently throughout the world [1,2,3,4,5,6]. HAdV14p1 was first identified as the causative agent of an outbreak of acute respiratory distress syndrome (ARDS) in U.S. military populations [7]. HAdV14p1 is 99.7% identical to HAdV14 yet displays increased lung pathogenesis in the Syrian hamster that models HAdV14p1 pathogenesis in humans [8,9]. Why HAdV14p1 has increased pathogenesis is still unclear.
The HAdV genome encodes roughly 40 proteins. In addition to protein encoding RNAs, non-coding RNAs (ncRNA) are also present in HAdV-infected cells [10,11,12]. Viral-associated RNA (VA RNA) are encoded by the HAdV genome and are the most abundant viral ncRNA found in infected cells [13,14]. All HAdV encode at least one VA RNA transcript ~160 nt in length, and approximately 80% of HAdV serotypes encode at least 2 VA RNAs (VA RNA-I and VA RNA-II), with VA RNA-I encoded by all HAdV serotypes [15,16]. VA RNAs are required for efficient viral replication, as the deletion of VA RNA-I reduces viral titer by 20-fold and the deletion of both VA RNA-I and VA RNA-II results in a 60-fold decrease in viral titer [17,18]. Unlike protein encoding HAdV RNAs, VA RNAs are transcribed by cellular RNA polymerase III and are expressed throughout the viral replication cycle [13].
VA RNA forms a complex secondary structure that is critical for its biological function [19,20,21]. Overall, despite their low sequence conservation among HAdV, VA RNAs share similar secondary structure and organization of the apical stem, central domain, and terminal stem [16,21]. The crystal structure of HAdV-2 VA RNA-I reveals a sharply bent, coaxially stacked, wobble-enriched, and pseudoknot-anchored molecule to allow VA RNA to interfere with nearly all host systems that interact with double-stranded RNA (dsRNA) [19]. The best characterized activity of VA RNA is its inhibition of protein kinase R (PKR) through the ability of VA RNA-I to bind and inhibit PKR dimerization [22,23,24]. PKR is part of the cellular innate immune response to viral infection, and, by sensing dsRNA produced during viral infection, PKR activation results in the global inhibition of cap-dependent protein synthesis. VA RNA also binds to 2′-5′ Oligoadenylate Synthetase 1 (OAS1), which results in the activation of OAS1 [25]. While the interaction of full-length VA RNA activates OAS1, the binding of cleaved VA RNAI (produced following DICER cleavage) to OAS1 inhibits OAS1 activity [11,26,27]. Interestingly, the cleaved VA RNAI has a higher affinity for OAS1 than full-length VA RNAI, suggesting that the inhibitory activity of cleaved VA RNAI may be the predominant activity of VA RNAI on OAS1 [28]. VA RNAI binds to the retinoic acid-inducible gene I (RIG-I). The triphosphorylyated 5′-end nucleotide of VA RNA triggers RIG-I signaling, which results in an increased type I interferon response [29,30,31,32]. These roles in modulating the cellular innate immune response to HAdV have been long understood.
Recently, new roles for VA RNAI in regulating cellular miRNA processes have been elucidated. Following transcription, VA RNAI is exported from the nucleus to the cytoplasm by Exportin 5 (Exp5). Exp5 exports RNA molecules (e.g., pre-miRNA and tRNAs) that contain a ‘mini-helix’ from the nucleus [33,34,35,36]. VA RNAI contains a ‘mini-helix’ at the end of the terminal stem, which results in VA RNA being a direct competitor with cellular small RNAs for interaction with Exp5 [37]. In addition to competing with pre-miRNA for nuclear export through Exp5, once in the cytoplasm, VA RNAs are cleaved by DICER, like pre-miRNA, and as VA RNA expression increases, more VA RNAs are associated with DICER than host pre-miRNA, resulting in decreased host miRNA expression [27,38,39]. DICER processes VA RNA into small RNAs that resemble host miRNA (called mivaRNA) that can be incorporated into the RNA-induced silencing complex (RISC) [37,38,40,41,42]. RISC association allows for mivaRNA to bind and repress target mRNA expression [38,40,41,42,43].
Group B2 HAdV members encode only the VA RNAI gene. Bioinformatic analysis of the HAdV14 genome showed that the VA RNAI gene of Ad14 is ~98% identical to the other group B2 members HAdV11, HAdV34, and HAdV35 [44]. In this report, we further characterize the VA RNA gene, identifying the intragenic promoter elements, transcriptional stop sites, potential OAS1 consensus sites, and GGGU/ACCC sites. Small RNA-seq was used to identify the mivaRNA produced during infection. Despite 100% sequence identity of the VA RNAs, differential expression of the mivaRNA exists between HAdV14- and HAdV14p1-infected A549 cells. We discuss the possibility that this differential mivaRNA expression could play a role in the pathogenesis of HAdV14p1.
2. Materials and Methods
2.1. Cells and Viruses
A549 cells (CCL-185, ATCC, Manassas, VA, USA) were grown at 37 °C and 5% CO2 in DMEM supplemented with 5% heat-inactivated bovine calf serum, 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM L-glutamine. A549 cells were monitored for mycoplasma contamination by PCR and validated by short tandem repeat markers (STR Authentication) (ATCC). HAdV14 deWit was obtained from ATCC, and HAdV14p1 (isolate 1986T) was obtained from the United States Navy [34,35,36]. Viruses were propagated and plaque titered (plaque-forming units (pfu)/mL) in A549 cells.
2.2. Infection of A549 Cells and Isolation of Total Cellular RNA
A549 cells were infected with either HAdV14 or HAdV14p1 at a multiplicity of infection (MOI) of 10 pfu/cell in suspension for 1 h at 37 °C, after which cells were plated and allowed to adhere until collected. Adherent and non-adherent cells were collected at 6, 12, 24, 36, and 48 h post-infection. Total RNA was isolated using miRNeasy kit (Qiagen, Germantown, MD, USA) with on column DNase treatment. The total RNA in each sample was quantified using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA), and quality was measured using the RNA6000 Nano chip on the Agilent 2100 Bioanalzyer (Agilent Technologies, Santa Clara, CA, USA). All samples had an RNA integrity number greater than 7.
2.3. Small RNA Library Preparation, Sequencing and Data Analysis
TruSeq Small RNA library prep kit (Illumina, San Diego, CA, USA) was used to create sequencing libraries. Specifically, adapters were ligated using the 5′ phosphate and 3′ hydroxyl groups common to most mature miRNAs. After adapter ligation, samples were reverse transcribed and amplified. Finally, the libraries were size selected using a 6% polyacrylamide gel and concentrated using ethanol precipitation. Purified libraries were normalized and pooled to create a double-stranded cDNA library and were sequenced on the Illumina MiSeq to render 50 base pair single end reads at the Loyola Stritch School of Medicine Genomics Facility. Adapter sequences were removed, and low-quality reads were trimmed from raw sequencing reads using Cutadapt (v. 1.11). The resulting reads were mapped to the HAdV14 deWit genome (GenBank accession number AY803294) in CLC Genomics Workbench (Qiagen).
2.4. Differential Expression Analysis
Differential expression was conducted with CLC Genomics Workbench. The VA RNA sequence was added to miRbase 22 and annotated to contain either total 5′ and 3′ reads or mivaRNA 5′A, 5′G, 3′A and 3′C seeds. Quantify miRNA 1.2 (CLC) was used with the following settings: allow for length-based isomiRs, no additional upstream bases, 5 additional downstream bases, no missing or mismatch bases, minimum sequence length = 18, maximum sequence length 25. Differential Expression for RNA-Seq 2.6 (CLC) was used for differential expression analysis with normalization set to Trimmed Mean of Means (TMM).
2.5. HAdV-14 mivaRNA 3′ UTR Luciferase Reporter Assays
The complement of the either the 5′ or 3′ seed sequences of Ad14 VA RNA were cloned into the multiple cloning site of pmirGLO Dual Luciferase Vector (Promega, Madison, WI, USA). Constructs were sequenced to confirm the correct orientation of the complimentary HAdV14 VA RNA seed sequences. Plasmids were purified using PureLink HiPure Plasmid Maxi Prep (Invitrogen) and transfected into A549 cells using Lipofectamine LTX (Invitrogen). Twenty-four hours after transfection, cells were infected with Ad14 at an MOI of 10. After an additional 24 h, luciferase activity was determined using a dual-luciferase assay (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. One-way ANOVA was followed by the Holm-Sidák test with p < 0.05 considered significant (Prism 9; GraphPad Software, San Diego, CA, USA).
2.6. Prediction of mivaRNA Host Gene Targets and Functional Enrichment Analysis
TargetScan and miRDB were used to predict cellular targets for mivaRNAs by seed sequences and filtered to remove duplicate targets. KEGG (https://www.genome.jp/kegg/, accessed on 2 May 2021) was used to predict the cellular pathways and processes potentially regulated by mivaRNA–target mRNA interaction.
2.7. RNA-Seq Library Preparation, Sequencing, Differential Expression Analysis, and Bioinformatic Analysis
Illumina stranded mRNA libraries were made from the total RNA preps from A549-infected cells. Sequencing was performed at University of Oregon Genomics & Cell Characterization Core (Eugene, OR, USA) on an Illumina HiSeq4000 set for pair ends 100 bp reads and demultiplexed. Reads were trimmed with Trim Reads 2.4 (CLC) before RNA-seq Analysis 2.21 (CLC) was performed, mapping to the human genome (Hg38) with a mismatch cost of 2, insertion cost of 3, deletion cost of 3, and length/similarity fractions set to 0.8. Paired reads were counted as a single read and expression value was set to reads per kilobase million (RPKM). Differential expression for RNA-Seq 2.4 (CLC) using gene expression values from RNA-seq analysis was performed and the results were filtered to the predicted mivaRNA targeted genes. Ingenuity Pathway Analysis (IPA) (Qiagen) was used for functional enrichment with the differential RNA-seq analysis from HAdV14-infected cells at 36 hpi vs. uninfected cells. FDR was set to 0.05 with absolute expression fold changes being >1.5 and expression intensity being >1.
3. Results
3.1. Analysis of HAdV14/HAdV14p1 VA RNA Gene Sequence and Secondary Structure
Like other HAdV group B2 members, HAdV14 and HAdV14p1 genomes encode only 1 VA RNA sequence that resembles VA RNA-I at nucleotides 10,452–10,613 [44]. Houng and colleagues have shown that the sequences of the VA RNA genes of HAdV14 and HAdV14p1 are 100% identical [9]. Analysis of the VA RNA-I sequence reveals two intragenic promoter elements (Box A (+13–23) and Box B (+55–65)) that are required for RNA polymerase III transcription (Figure 1A). VA RNA-I transcription in many serotypes has been shown to be initiated at one of two potential transcription start sites, either a G(+1) or three nucleotides upstream at A(−3) [16,27]. The HAdV14 VA RNA-I gene contains both possible start sites. Termination sequences (T1A and T1B) are found at +152–161. Additionally, there is a backup termination sequence that is found at +192–198. The VA RNA gene contains the conversed GGGU (+33–36) and ACCC (+120–123) that form the central domain (Figure 1A,B). Figure 1B shows a predicted secondary structure for the HAdV14 VA RNA molecule consisting of an apical stem, central domain, and terminal stem. HAdV VA RNA I contains two OAS1 consensus sites (WW(N9)WG) found in the central domain [45,46]. HAdV14 contains two OAS1 consensus sites at +36–48 and +93–105; both are found in the central domain.
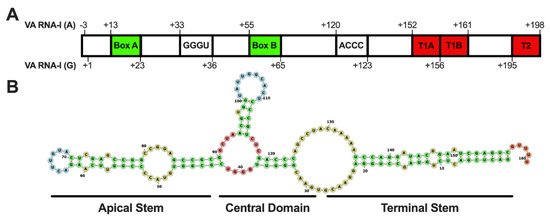
Figure 1.
Human adenovirus 14 VA RNA gene and transcript secondary structure. (A) The HAdV14 VA RNA gene contains two predicted start sites A (−3) and G (+1). RNA Pol III Box A and B promoter regions (green), two internal terminator sequences (red, T1A and T1B), and a backup terminator sequence (T2) are indicated. Two complementary sequences (GGGU and ACCC) are also present. (B) A predicted secondary structure shows the regions that form the terminal stem, central domain, and apical stems (RNAStructure [47]).
3.2. Identification and Temporal Expression of HAdV14 Small RNAs
Previous studies have shown that the majority of small RNAs produced during HAdV infection map to the VA RNA gene(s). To examine the viral small RNAs produced during HAdV14 or HAdV14p1 infection, A549 cells were infected with either HAdV14 or HAdV14p1 (Figure 2). Four independent small RNA libraries from infection with either HAdV14 or HAdV14p1 per time point were sequenced. At 6 hpi, less than 1% of the reads mapped to the HAdV14 genome, then increased to ~20% at 24 hpi, then remained at ~15% at 36 and 48 hpi (Table 1). At all time points, >92% of all small RNAs that mapped to the HAdV14 genome mapped to VA RNA gene (Table 1), showing that the VA RNA gene is responsible for the vast majority of all HAdV14-encoded small RNAs. The diversity of the reads that aligned to the VA RNA gene increased during infection mainly by extensions or deletions at either the 5′ or 3′ ends of the small VA RNAs, with the total number of unique small VA RNAs similar between HAdV14 and HAdV14p1 (Supplementary Materials Table S1). The majority of the reads that aligned to the VA RNA gene mapped to the 5′ and 3′ ends of the terminal stem, with each small VA RNA comprising more than 1% of the total small RNA pool sequenced (Table 2). The number of unique small RNAs that mapped to the VA RNA gene increased from 6 to 24 hpi and then stabilized through 48 hpi (Figure 3A). Examination of the read counts showed temporal expression of small RNAs produced from the 5′ or 3′ ends of the VA RNA gene. At 6 and 12 hpi, the 3′ small RNAs are predominant, while the 5′ small RNAs show increased expression 36 and 48 hpi (Figure 3B–F). The same temporal expression patterns are seen in HAdV14p1-infected cells (Figure 3B–F). The differences in read counts between HAdV14- and HAdV14p1-infected cells suggest that there may be differential expression of 5′ and 3′ small VA RNAs produced during infection. Differential expression analysis showed that, for the most part, 5′ small VA RNAs during HAdV14p1 infection were not differentially expressed compared to HAdV14 infection, except at 12 hpi. In contrast, 3′ small VA RNAs were differentially expressed during HAdV14p1 infection compared to HAdV14 infection, except at 24 hpi.
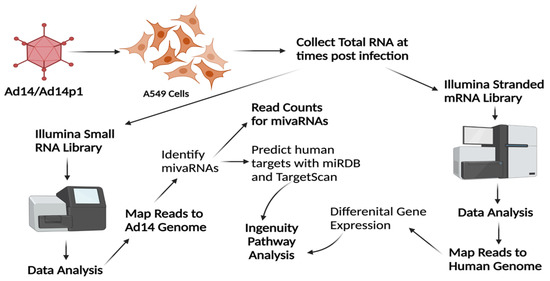
Figure 2.
RNA-seq and bioinformatics workflow. A549 cells were infected with HAdV14 or HAdV14p1 at an MOI of 10:1 and total RNA was collected at various times post-infection. To identify HAdV14 mivaRNAs, Illumina small RNA-seq libraries were constructed, sequenced on MiSeq, and reads were mapped to the HAdV14 genome to identify HAdV14 mivaRNAs. Quantitation of reads was performed with CLC Genomics Workbench. TargetScan and miRDB were used to predict human mRNA targets based on seed sequences of the identified mivaRNAs. To examine gene expression in A549 cells during infection, Illumina stranded mRNA-seq libraries were constructed, sequenced on a NextSeq 2000, reads mapped to the human genome, and differential gene expression determined. Ingenuity Pathway Analysis was used to explore differential expression of mivaRNA predicted target genes. Created with BioRender.com.

Table 1.
Summary of alignment of sequences reads to the adenovirus 14 genome.

Table 2.
Expression of HAdV14/p1-encoded small RNAs that are greater than 1% of total small RNA pool.
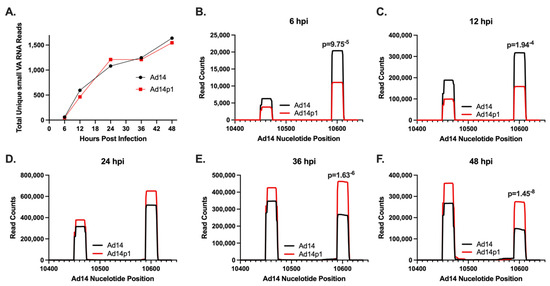
Figure 3.
Expression of HAdV14/p1 VA RNA-encoded small RNAs. (A) Total number of unique HAdV14 VA RNA-encoded small RNAs during HAdV14 (black) and HAdV14p1 (red) infection of A549 cells. (B–F) Number of small RNA reads to specific regions of the HAdV14 VA RNA gene during HAdV14 (black) and HAdV14p1 (red) infection. Nucleotides 10,450–10,473 and 10,590–10,613 correspond to the 5′ and 3′ terminal stem regions, respectively. Differential expression was determined with RNA-Seq 2.6 (CLC Genomics Workbench). FDR p-values 0.05 were considered significant and are shown.
3.3. Identification of Potential HAdV14/14p1 mivaRNAs
While there is diversity in the small RNAs that map to the VA RNA gene, repression of target gene expression involves the binding of miRNA to the target mRNA through the miRNA seed sequence, nucleotides 2–8 of the miRNA, and its complement on the target gene mRNA. Therefore, the seed sequences provide a better metric for both identifying and quantitating HAdV mivaRNAs. Unique miRNA seed sequences were identified for each small VA RNA molecule (Table 3). The total number of unique seed sequences increased during HAdV14 or HAdV14p1 infection and plateaued at 24 hpi (Figure 4A). The number of unique seed sequences was similar in both HAdV14- and HAdV14p1-infected cells (Figure 4A). To identify potential HAdV14/14p1 mivaRNAs, we identified the seed sequences that had total read counts >1000 and were found in small RNAs 20–33 nucleotides long. Eleven seed sequences were identified that met those criteria, and all of them mapped to either the 5′ or 3′ terminal ends of the VA RNA (Figure 4B). Based on the read counts, there are four main mivaRNAs produced during HAdV14/14p1 infection, two from both the 5′ and 3′ ends. The mivaRNAs from the 5′ end start at either the A (−3) or G (+1) and have been named mivaRNA 5′A or mivaRNA 5′G. At the 3′ end, the predominant mivaRNAs start at either C (138) or A (139) and have been designated mivaRNA 3′C and 3′A, respectively (Figure 4B). All four of the predominant mivaRNAs average 20 to 24 nucleotides in length, with the 5′ mivaRNAs showing the greatest length diversity, out to 50 nucleotides long (Supplementary Materials Table S1). Overall, >95% of the reads are between 20 and 24 nt. Using our read counts (Table 2) and unique seed sequence data (Table 3), we predict that there are five dominant DICER cleavage sites in HAdV-14/14p1 VA RNA. These predicted cleavage sites generate greater than 98% of the unique seed sequences that map to the VA RNA gene (Figure 4C). We predict at least four secondary cleavage sites that generate the unique seed sequences in Table 3 that have more than 1000 total counts (Figure 4C).

Table 3.
Unique miRNA seed sequences encoded in the HAdV14/p1 VA RNA.
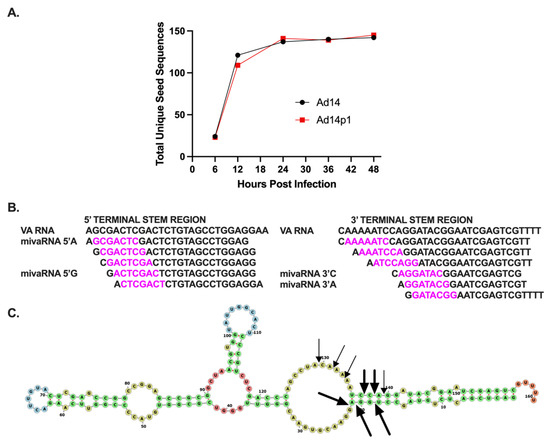
Figure 4.
Identification of HAdV14/p1 mivaRNAs. (A) Expression of total unique small VA RNA seed sequences during HAdV14 (black) and HAdV14p1 (red) infection of A549 cells. (B) Identification of unique seed sequences (purple) from small RNAs that are >1% of total small RNA pool. All aligned to either 5′ or 3′ terminal stem region of VA RNA. The four predominant small RNAs have been designated mivaRNA 5′A, mivaRNA 5′G, mivaRNA 3′C, and mivaRNA 3′A. (C) Predicted DICER cleavage sites indicated with bold arrows and regular arrows represent dominant and secondary sites, respectively, in the predicted secondary structure model (RNAStructure).
3.4. Differential Expression of mivaRNA during HAdV14 and HAdV14p1 Infection
Infection with HAdV14 or HAdV14p1 resulted in differential expression of 3′ small VA RNAs (Figure 3). With the dominant mivaRNAs identified, we performed differential expression analysis of each mivaRNA over time in HAdV14p1-infected vs. HAdV14-infected cells. There was no differential expression of mivaRNA 5′A during infection (Figure 5A). However, mivaRNA 5′G (Figure 5B) showed differential expression that was lower at 6 and 12 hpi in HAdV14p1-infected cells and then was higher in HAdV14p1-infected cells at 48 hpi. In HAdV14p1-infected cells, both of the 3′ mivaRNAs (3′C and 3′A) showed a lower expression at 6 and 12 hpi compared to HAdV-14-infected cells (Figure 5C,D). At later times (36 and 48 hpi), mivaRNA 3′C and 3′A were both expressed at higher levels during HAdV14p1 infection than during HAdV14 infection. Overall, the data show that there are temporal mivaRNA expression differences between HAdV14- and HAdV14p1-infected cells.
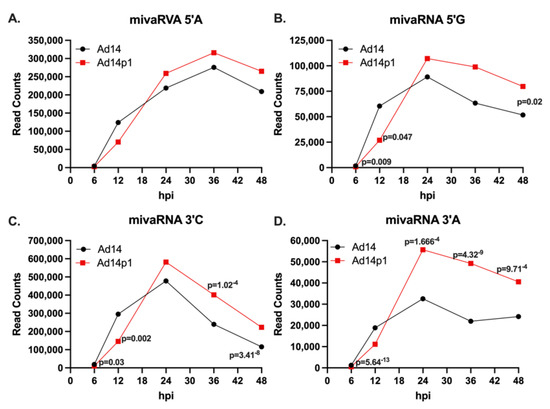
Figure 5.
Expression of mivaRNAs during infection. Total read counts of mivaRNA 5′A (A), mivaRNA 5′G (B), mivaRNA 3′C (C), or mivaRNA 3′A (D) from 4 replicative infections with either HAdV14 (black) or HAdV14p1 (red) in A549 cells. Differential expression was determined with RNA-Seq 2.6 (CLC Genomics Workbench). FDR p-values 0.05 were considered significant and are shown.
3.5. Expression of HAdV14 mivaRNA and A549 miRNA Expression during Infection
DICER processes pre-miRNAs and VA RNA into mature miRNA and mivaRNA, respectively [27,39]. During HAdV infection, it has been shown that VA RNAs can suppress miRNA biogenesis by competing with host pre-miRNAs for cleavage by DICER [38]. Infection with HAdV14p1 results in nearly a 50% reduction in cellular miRNA at 24 hpi that persists through full CPE (Table 4). To explore the relative abundance of HAdV14 mivaRNAs in relation to A549 miRNAs, the identified HAdV-14 mivaRNAs were added to the miRBase v22 list of human miRNAs to allow for counting of mivRNAs and host miRNAs in the CLC Genomics Workbench. At 6 hpi, only mivaRNA 3′C was one of the top 20 total miRNAs expressed (Table 5). Beginning at 12 hpi, mivaRNA 3′C, 5′A, and 5′G were found in the top six total miRNAs expressed and remained in the top six through full CPE at 48 hpi. After 6 hpi, all four predominant mivaRNAs were expressed at similar levels to the top cellular miRNAs. In general, increased expression of mivaRNAs resulted in decreased expression of all of the top 20 cellular miRNAs.

Table 4.
Summary of sequence reads that map to miRbase or HAdV14 VA RNA.

Table 5.
Cellular miRNA and mivaRNA expression during HAdV14 infection.
3.6. HAdV14 mivaRNA Are Functional miRNAs
Since HAdV VA RNAs are processed by DICER into mivaRNAs, we hypothesized that the mivaRNAs can repress gene expression like cellular miRNAs. To test the ability of HAdV14 mivaRNA to function as miRNA, the complement of each of the four main mivaRNA seed sequences were cloned into the 3′ UTR of luciferase in the pmirGLO Dual Luciferase Vector. Each reporter vector was transfected into A549 cells 24 h prior to infection with HAdV-14 at an MOI of 10, and luciferase activity was determined 24 hpi. As shown in Figure 6, HAdV-14 infection resulted in the repression of luciferase activity from all four mivaRNAs at 24 hpi. The strongest repression was seen from the 3′ mivaRNAs that are expressed at higher levels at 6 and 12 hpi (Figure 3B).
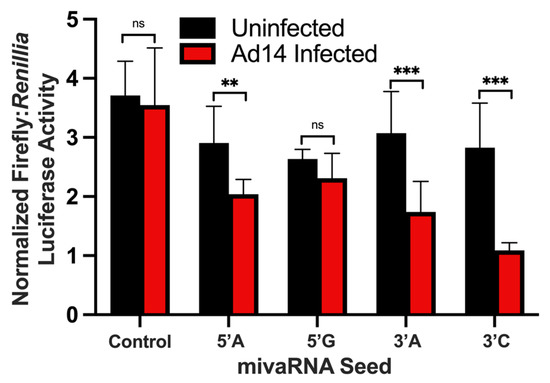
Figure 6.
Regulation of luciferase activity by mivaRNAs during viral infection. The complementary sequence of the mivaRNA seeds were cloned into pmirGLO Dual Luciferase Vector and transfected into A549 cells. Twenty-four hours after transfection, A549 cells were infected with Ad14 at an MOI of 10 and luciferase activity was determined 24 h after infection. Control seed is pmirGLO Vector with no mivaRNA complementary sequence. Luciferase activity is expressed as normalized Firefly:Renillia luciferase activity. Mean ± SD, n = 8, one-way ANOVA followed by Holm–Sidák test ** p < 0.01, *** p < 0.001.
3.7. Prediction of mivaRNA Host Targets and Functional Enrichment Analysis
Having established that mivaRNAs are expressed at levels similar to cellular miRNAs (Table 5) and are capable of repressing gene expression (Figure 6) of target genes that have the complementary seed sequence in the 3′ UTR of their mRNA, we sought to determine if there was any potential for HAdV14 mivaRNAs to repress cellular genes. TargetScan and mirDB were used to predict potential host target mRNAs using the seed sequences identified (Figure 4B). All but one of the mivaRNAs are unique seed sequences that are not found in human miRNAs. The 3′-mivaRNA-CAAAAATCCAGGATACGGAATCGAGTCGTT encodes the same seed sequence as human miR-584c-3p and was excluded from further analysis. The remaining 10 mivaRNA seeds were used to predict potential gene targets. Overall, there were 2184 predicted genes that could be targeted by the Ad14 mivaRNAs (Supplementary Materials Table S2). Of the 2184, 1919 were recognized by KEGG, and KEGG pathway analysis was performed on the predicted gene targets. The top pathways included many signal transduction pathways and cellular processes involved in viral infection, including metabolism, cytoskeleton/tight junctions/focal adhesion/cell adhesion molecules, mRNA transport/splicing/regulation, and cell death processes (Table 6 and Table S3).

Table 6.
Enrichment of mivaRNA targets by KEGG analysis.
3.8. Analysis of Predicted Target Gene Expression by RNA-Seq
Since our in vitro studies showed that the mivaRNAs can functionally repress luciferase gene expression (Figure 6), we sought to determine if mivaRNA might regulate expression of predicted target genes. RNA-seq analysis was performed on the total RNAs from cells at all time points. Differential expression analysis was performed against uninfected control RNA libraries. The genes were filtered to the 1919 genes recognized by KEGG analysis, and genes that were down regulated by at least 1.5-fold with an FDR of <0.05 were considered significantly down regulated. As seen in Figure 7A, from 6 hpi to 24 hpi, down regulated target genes increased from ~200 to ~370 for both HAdV14- and HAdV14p1-infected cells. Between 24 and 36 hpi, the number of down regulated genes plateaued (HAdV14) or slightly decreased (HAdV14p1) from the 24 hpi number. At 48 hpi (full CPE), the number of down regulated genes doubled for both HAdV14 and HAdV14p1 from the 36 hpi time point. At both 36 and 48 hpi, there were more down regulated target genes in HAdV14-infected cells than in HAdV14p1-infected cells. From 6 hpi to 24 hpi, 78% of down regulated targets were shared between HAdV14- and HAdV14p1-infected cells (Figure 7B). This similarity to target repression decreased at 36 and 48 hpi. To gain a better understanding of how the down regulated gene expression might alter cellular pathways, we performed IPA with the 36 hpi differential gene expression data sets. This time point was selected, as there should be sufficient repression observed at 36 hpi, and the time course of viral infection has not resulted in 4+ CPE. As seen in Table 7, the top ten pathways enriched from our RNA-seq data are very similar between HAdV14- and HAdV14p1-infected cells. The two pathways that are not the same in the top ten are not far out of the top ten (Supplementary Materials Table S4). Combined with the ability of mivaRNAs to functionally repress luciferase gene expression, these data suggest that mivaRNAs can regulate host gene expression during infection. However, further studies are needed to determine whether other viral genes or indirect effects of viral replication affect host gene expression.
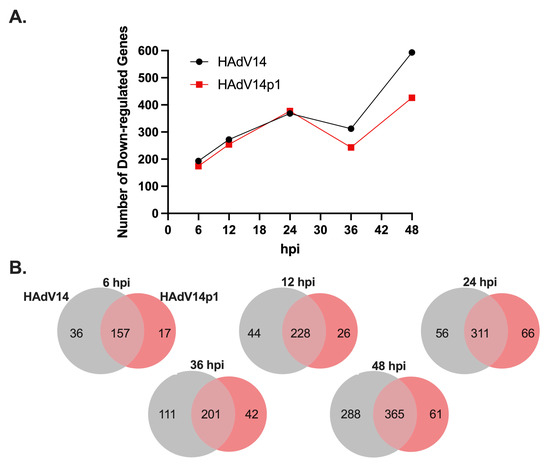
Figure 7.
Expression of mivaRNA predicted target genes. (A) Number of significantly down regulated (−1.5 fold, FDR p-value 0.05) mivaRNA target genes during HAdV14 (black) or HAdV14p1 (red) infection of A549 cells. (B) Venn diagram showing shared and unique down regulated target genes during HAdV14 (gray) and HAdV14p1 (red) infection.

Table 7.
IPA enrichment of top 10 canonical pathways at 36 hpi a.
4. Discussion
The adenoviral VA RNA gene is a multifunctional non-coding RNA molecule. VA RNAs are processed by DICER into mivaRNA that resemble and function like cellular miRNAs. While all human adenoviruses encode at least one VA RNA gene, the amount of sequence similarity between VA RNA genes from all human adenoviruses is limited between all human Ad, but is somewhat conserved within Ad groups. In this study, we sought to identify the mivaRNA produced during prototype HAdV14 deWit infection, assess predicted target gene expression, and compare mivaRNA expression with that of the emergent pathogenic strain HAdV14p1, using A549 cells that are permissive for HAdV14 and HAdV14p1 infection.
HAdV14, like other group B2 HAdV, only encode one VA RNA gene [44]. In HAdVs that only encode one VA RNA, that VA RNA resembles group C HAdV VA RNAI. Here, further bioinformatic analysis of the HAdV14 VA RNA gene has shown that the VA RNA gene contains both RNA Pol III Box A and B promoter sites as well as two internal terminator sequences and a backup terminator sequence. The HAdV14 VA RNA gene also encodes two potential OAS1 consensus activation sequences. Based on the predicted secondary structure of the HAdV14 VA RNA molecule, it includes a terminal stem, central domain, and apical stem. Overall, the bioinformatic results suggest that, like other HAdVs that only encode one single VA RNA gene, HAdV14 VA RNA resembles that of VA RNAI, rather than VA RNAII.
To characterize the mivaRNA produced from the HAdV14 VA RNA gene, we infected A549 cells and sampled mivaRNA expression through all phases of the Ad14 infectious cycle from very early (6 hpi) all the way through to full CPE (48 hpi). At all times, >90% of all small RNA reads that mapped to the HAdV14 genome mapped specifically to the VA RNA gene. These results are consistent with those seen during Ad5 infection of IMR-90 cells [48]. Zhao and colleagues first observed that there was temporal expression of mivaRNA molecules during the infection cycle [48]. We also observed temporal expression of mivaRNAs during HAdV14 infection. Our data showed that early during infection, the 3′ mivaRNAs are the most abundant HAdV14 mivaRNAs, while between 24 and 36 hpi, the 5′ mivaRNAs begin to be more abundant. This is opposite of what was observed following HAdV2-infected IMR-90 cells. The reason for this difference is unclear. The difference could be virus-specific (HAdV2 vs. HAdV14), cell type-specific (lung fibroblast, IMR-90, vs. lung epithelial, A549), or a combination. Supporting this theory are studies by Kamel et al., which showed that infection of 293 cells with HAdV5, HAdV11 (a group B2 HAdV), and HAdV37 resulted in more 3′ mivaRNA expression early during infection [27]. The same temporal expression patterns were seen during HAdV14p1 infection, as was observed by Zhao [48]. When we normalized mivaRNA expression, we observed that there was a significant difference in the expression of total mivaRNAs between HAdV14 and HAdV14p1 over time. Specifically, early in the infection cycle, HAdV14 mivaRNAs showed higher expression than HAdV14p1. After 24 hpi, this pattern is flipped, with HAdV14p1 mivaRNAs expressed at a higher level than HAdV14. Previous studies in our lab and others have shown that there is no difference in the replication rates of HAdV14 or HAdV14p1 in either tissue culture cells or in vivo in the lungs of Syrian hamsters, thus eliminating differences in viral replication as the reason for the differential expression of mivaRNAs [8,49]. The studies here were not designed to test whether this mivaRNA expression pattern has any role in the increased pathogenesis seen during HAdV14p1 infection. Studies are underway to test this possibility using a Syrian hamster model of differences in HAdV14p1 vs. HAdV14 lung pathogenesis [8,50].
Our results identified the core seed sequences produced during Ad14/Ad14p1 infection as coming from both the 5′ and 3′ terminal stems. Minor variants are also produced. How these arise is unclear. One possibility is that RNA pol III synthesis of VA RNAs might have some minor wobble in its exact initiation site, as these minor mivaRNA species show very slight variations in either the 5′ or 3′ ends. It is possible that this could have an effect on expression, as small differences in the 5′ start can result in a completely different mivaRNA seed sequence. We identified a group of long mivaRNAs produced from the 3′ terminal stem that start between 8 and 4 nucleotides upstream of the predominant 3′ mivaRNA. Interestingly, the longest mivaRNA produced from the 3′ terminal stem has the exact same seed sequencing as hsa-miR-584c-3p. It has been well documented that HAdV mivaRNAs can regulate gene expression, like gene-encoded miRNAs [26,40,42]. Using a luciferase reporter vector, we have shown that both the 5′ and 3′ HAdV-14 mivaRNAs can suppress gene expression during viral infection. Our results confirm that the 3′ mivaRNAs are more abundantly expressed than 5′ mivaRNAs early during infection, as the 3′ mivaRNAs showed stronger repression of luciferase activity than the 5′ mivaRNAs. mivaRNAs have been shown to associate with RISC complexes, but not all mivaRNAs associate with RISC complexes at the same rate [27,38,40,42]. This could account for the difference in repressive activity seen between the mivaRNAs in our reporter assay. The association of HAdV14 mivaRNAs with the RISC complex was not a focus of these studies, but is under investigation.
The ability of mivaRNAs to target and regulate host gene expression is an attractive theory for potential roles in HAdV replication. As a result, numerous studies have been conducted to determine potential target genes for mivaRNAs. Bioinformatic, microarray studies, and RNA-seq analysis have shown that many of the potential cellular gene targets for mivaRNAs are involved in DNA repair, cell growth, cell death, RNA metabolism, cell signaling pathways, and metabolism [26,42,48]. Our bioinformatic analysis (Table 6) on the predicted HAdV14 mivaRNA targets show similar cellular pathways to be potential targets, despite the lack of sequencing identity between HAdV14 seed sequences and HAdV2/5 seed sequences. Our IPA analysis (Table 7) also shows functional enrichment of the same cellular pathways from our RNA-seq data on HAdV14 mivaRNA target genes. This suggests that there is some conserved biological reason for mivaRNA expression during viral infection. However, studies have raised questions as to the role of mivaRNAs in viral replication. The mutation of the seed sequences of HAdV5 mivaRNAs had no impact on viral replication in HEK293 cells [51]. One underlying problem with studies (including these studies) on HAdV mivaRNAs is that they have been conducted in tissue culture cell lines rather than relevant primary cells or in a permissive animal model, which might clarify the role of mivaRNAs during infection in vivo. HAdV mivaRNAs have also been found in persistently infected lymphoid cells, suggesting a potential role in regulating lytic and persistent infection [52,53]. In addition to mivaRNAs from the VA RNA gene, another viral small RNA that maps to the Major Late Promoter region has been identified that regulates viral gene expression; we are in the process of exploring if there are other HAdV14 small viral RNAs [54].
In this study, we characterized the HAdV14 VA RNA gene and the mivaRNAs produced from it. We identified the predominant seed sequences and the cellular genes that the mivaRNAs can target. Our bioinformatic analysis using RNA-seq data of the expression of mivaRNA target genes showed that HAdV14 mivaRNA can regulate similar pathways seen from HAdV2/5 studies despite the lack of identity of the mivaRNA seed sequences. Despite 100% sequence identity later in infection, HAdV14p1-infected cells express significantly more mivaRNA than HAdV14-infected cells. Whether this difference has an impact on viral replication or pathogenesis is unknown. Due to a lack of studies in immunocompetent animal models, the role of HAdV mivaRNA in viral pathogenesis is largely unexplored. To answer questions about viral factors, such as mivaRNAs, we have developed an immunocompetent Syrian hamster model of HAdV14p1 lung pathogenesis that displays pathology similar to what is seen in the lungs of severely infected human patients [8,50]. We propose that the Syrian hamster model of HAdV14p1 lung pathogenesis could provide the model system to explore the roles of mivaRNA in viral replication and pathogenesis in vivo.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14050898/s1, Table S1: All small RNAs that map to HAdV14 VA RNA gene; Table S2: Predicted human genes targeted by mivaRNA; Table S3: Full KEGG analysis of all target genes; Table S4: Complete IPA pathways.
Author Contributions
All authors contributed significantly to the manuscript as follows: Conceptualization, E.R.M. and J.R.R.; methodology, E.R.M., H.M.B., H.-S.S. and J.R.R.; formal analysis, E.R.M. and J.R.R.; investigation, E.R.M., H.M.B., H.-S.S. and J.R.R.; data curation, E.R.M. and J.R.R.; writing—original draft preparation, J.R.R.; writing—review and editing, E.R.M. and J.R.R.; visualization, E.R.M. and J.R.R.; supervision, J.R.R.; project administration, J.R.R.; funding acquisition, J.R.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Idaho Center of Biological Research Excellence (COBRE) in Emerging and Re-emerging Infectious Diseases through the National Institute of General Medical Sciences, grant number P20GM109007 to J.R.R.; a Technology Access Award and Developmental Research Project from the Idaho INBRE through the National Institute of General Medical Sciences, grant number P20GM103408 to J.R.R.; Idaho INBRE Summer Research Fellowship, through the National Institute of General Medical Sciences, grant number P20GM103408 to H.S.S.; Department of Veterans Affairs Career Development Award, CDA-2-069-09S to J.R.R.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All RNA-seq data have been deposited to the Sequence Read Archive (SRA). Both the small RNA-seq and RNA-seq data are available from the bioproject PRJNA752359.
Acknowledgments
We would like to thank David Metzgar and Adriana Kajon for sharing emergent HAdV14p1 isolates, and Gina Kuffel and Michael Zilliox at the Loyola Stritch School of Medicine Genomics Facility for library preparation and small RNA sequencing. The authors are indebted to James L. Cook for his continued support of our research through thoughtful discussion and critical reading of this manuscript. We appreciate the efforts and hard work of Dennis Stevens, Amy Bryant, Carolyn Bohach, and Sam Minnich for providing research opportunities for undergraduate students in underfunded states such as Idaho and Nevada through their respective COBRE (D.S. and A.B) and INBRE (C.B. and S.M.) grants as mentioned above.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The views expressed here are those of the authors and do not reflect the opinion or policy of the Department of Veterans Affairs or the United States Government.
References
- Metzgar, D.; Osuna, M.; Kajon, A.E.; Hawksworth, A.W.; Irvine, M.; Russell, K.L. Abrupt Emergence of Diverse Species B Adenoviruses at US Military Recruit Training Centers. J. Infect. Dis. 2007, 196, 1465–1473. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Acute Respiratory Disease Associated with Adenovirus Serotype 14—Four States, 2006–2007. MMWR Morb. Mortal. Wkly. Rep. 2007, 56, 1181–1184. [Google Scholar]
- Huang, G.; Yu, D.; Zhu, Z.; Zhao, H.; Wang, P.; Gray, G.C.; Meng, L.; Xu, W. Outbreak of Febrile Respiratory Illness Associated with Human Adenovirus Type 14p1 in Gansu Province, China. Influ. Other Respir. 2013, 7, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Parcell, B.J.; McIntyre, P.G.; Yirrell, D.L.; Fraser, A.; Quinn, M.; Templeton, K.; Christie, S.; Romanes, F. Prison and Community Outbreak of Severe Respiratory Infection Due to Adenovirus Type 14p1 in Tayside, UK. J. Public Health 2015, 37, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Girouard, G.; Garceau, R.; Thibault, L.; Oussedik, Y.; Bastien, N.; Li, Y. Adenovirus Serotype 14 Infection, New Brunswick, Canada, 2011. Emerg. Infect. Dis. 2013, 19, 119–122. [Google Scholar] [CrossRef]
- Carr, M.J.; Kajon, A.E.; Lu, X.; Dunford, L.; O’Reilly, P.; Holder, P.; Gascun, C.F.D.; Coughlan, S.; Connell, J.; Erdman, D.D.; et al. Deaths Associated with Human Adenovirus-14p1 Infections, Europe, 2009–2010. Emerg. Infect. Dis. 2011, 17, 1402–1408. [Google Scholar] [CrossRef]
- Binn, L.N.; Sanchez, J.L.; Gaydos, J.C. Emergence of Adenovirus Type 14 in US Military Recruits—A New Challenge. J. Infect. Dis. 2007, 196, 1436–1437. [Google Scholar] [CrossRef]
- Radke, J.R.; Covert, H.J.; Bauer, F.; Ananthanarayanan, V.; Cook, J.L. Adenovirus 14p1 Immunopathogenesis during Lung Infection in the Syrian Hamster. Viruses 2020, 12, 595. [Google Scholar] [CrossRef]
- Houng, H.-S.H.; Gong, H.; Kajon, A.E.; Jones, M.S.; Kuschner, R.A.; Lyons, A.; Lott, L.; Lin, K.-H.; Metzgar, D. Genome Sequences of Human Adenovirus 14 Isolates from Mild Respiratory Cases and a Fatal Pneumonia, Isolated during 2006-2007 Epidemics in North America. Respir. Res. 2010, 11, 116. [Google Scholar] [CrossRef]
- Punga, T.; Darweesh, M.; Akusjärvi, G. Synthesis, Structure, and Function of Human Adenovirus Small Non-Coding RNAs. Viruses 2020, 12, 1182. [Google Scholar] [CrossRef]
- Vachon, V.K.; Conn, G.L. Adenovirus VA RNA: An Essential Pro-Viral Non-Coding RNA. Virus Res. 2015, 212, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Piedade, D.; Azevedo-Pereira, J.M. MicroRNAs as Important Players in Host–Adenovirus Interactions. Front. Microbiol. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Söderlund, H.; Pettersson, U.; Vennström, B.; Philipson, L.; Mathews, M.B. A New Species of Virus-Coded Low Molecular Weight RNA from Cells Infected with Adenovirus Type 2. Cell 1976, 7, 585–593. [Google Scholar] [CrossRef]
- Mathews, M.B. Genes for VA-RNA in Adenovirus 2. Cell 1975, 6, 223–230. [Google Scholar] [CrossRef]
- Mathews, M.B.; Shenk, T. Adenovirus Virus-Associated RNA and Translation Control. J. Virol. 1991, 65, 5657–5662. [Google Scholar] [CrossRef]
- Ma, Y.; Mathews, M.B. Structure, Function, and Evolution of Adenovirus-Associated RNA: A Phylogenetic Approach. J. Virol. 1996, 70, 5083–5099. [Google Scholar] [CrossRef]
- Thimmappaya, B.; Weinberger, C.; Schneider, R.J.; Shenk, T. Adenovirus VAI RNA Is Required for Efficient Translation of Viral MRNAs at Late Times after Infection. Cell 1982, 31, 543–551. [Google Scholar] [CrossRef]
- Bhat, R.A.; Thimmappaya, B. Adenovirus Mutants with DNA Sequence Perturbations in the Intragenic Promoter of VAI RNA Gene Allow the Enhanced Transcription of VAII RNA Gene in HeLa Cells. Nucleic Acids Res. 1984, 12, 7377–7388. [Google Scholar] [CrossRef]
- Hood, I.V.; Gordon, J.M.; Bou-Nader, C.; Henderson, F.E.; Bahmanjah, S.; Zhang, J. Crystal Structure of an Adenovirus Virus-Associated RNA. Nat. Commun. 2019, 10, 2871. [Google Scholar] [CrossRef]
- Coventry, V.K.; Conn, G.L. Analysis of Adenovirus VA RNA I Structure and Stability Using Compensatory Base Pair Modifications. Nucleic Acids Res. 2008, 36, 1645–1653. [Google Scholar] [CrossRef]
- Ma, Y.; Mathews, M.B. Comparative Analysis of the Structure and Function of Adenovirus Virus-Associated RNAs. J. Virol. 1993, 67, 6605–6617. [Google Scholar] [CrossRef] [PubMed]
- Kitajewski, J.; Schneider, R.J.; Safer, B.; Munemitsu, S.M.; Samuel, C.E.; Thimmappaya, B.; Shenk, T. Adenovirus VAI RNA Antagonizes the Antiviral Action of Interferon by Preventing Activation of the Interferon-Induced EIF-2α Kinase. Cell 1986, 45, 195–200. [Google Scholar] [CrossRef]
- Maran, A.; Mathews, M.B. Characterization of the Double-Stranded RNA Implicated in the Inhibition of Protein Synthesis in Cells Infected with a Mutant Adenovirus Defective for VA RNA. Virology 1988, 164, 106–113. [Google Scholar] [CrossRef]
- O’Malley, R.P.; Mariano, T.M.; Siekierka, J.; Mathews, M.B. A Mechanism for the Control of Protein Synthesis by Adenovirus VA RNAI. Cell 1986, 44, 391–400. [Google Scholar] [CrossRef]
- Desai, S.Y.; Patel, R.C.; Sen, G.C.; Malhotra, P.; Ghadge, G.D.; Thimmapaya, B. Activation of Interferon-Inducible 2′−5′ Oligoadenylate Synthetase by Adenoviral VAI RNA. J. Biol. Chem. 1995, 270, 3454–3461. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, O.; Carnero, E.; Abad, X.; Razquin, N.; Guruceaga, E.; Segura, V.; Fortes, P. Adenovirus VA RNA-Derived MiRNAs Target Cellular Genes Involved in Cell Growth, Gene Expression and DNA Repair. Nucleic Acids Res. 2010, 38, 750–763. [Google Scholar] [CrossRef]
- Kamel, W.; Segerman, B.; Punga, T.; Akusjärvi, G. Small RNA Sequence Analysis of Adenovirus VA RNA-Derived MiRNAs Reveals an Unexpected Serotype-Specific Difference in Structure and Abundance. PLoS ONE 2014, 9, e105746. [Google Scholar] [CrossRef]
- Meng, H.; Deo, S.; Xiong, S.; Dzananovic, E.; Donald, L.J.; Dijk, C.W.; van McKenna, S.A. Regulation of the Interferon-Inducible 2′–5′-Oligoadenylate Synthetases by Adenovirus VAI RNA. J. Mol. Biol. 2012, 422, 635–649. [Google Scholar] [CrossRef]
- Vennström, B.; Pettersson, U.; Philipson, L. Initiation of Transcription in Nuclei Isolated from Adenovirus Infected Cells. Nucleic Acids Res. 1978, 5, 205–219. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawabata, K.; Kouyama, E.; Ishii, K.J.; Katayama, K.; Suzuki, T.; Kurachi, S.; Sakurai, F.; Akira, S.; Mizuguchi, H. Induction of Type I Interferon by Adenovirus-Encoded Small RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 17286–17291. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Minamitani, T.; Iwakiri, D.; Takada, K. Adenovirus Virus-Associated RNAs Induce Type I Interferon Expression through a RIG-I-Mediated Pathway. J. Virol. 2011, 85, 4035–4040. [Google Scholar] [CrossRef]
- Gwizdek, C.; Ossareh-Nazari, B.; Brownawell, A.M.; Doglio, A.; Bertrand, E.; Macara, I.G.; Dargemont, C. Exportin-5 Mediates Nuclear Export of Minihelix-Containing RNAs. J. Biol. Chem. 2003, 278, 5505–5508. [Google Scholar] [CrossRef]
- Gwizdek, C.; Bertrand, E.; Dargemont, C.; Lefebvre, J.-C.; Blanchard, J.-M.; Singer, R.H.; Doglio, A. Terminal Minihelix, a Novel RNA Motif That Directs Polymerase III Transcripts to the Cell Cytoplasm Terminal Minihelix and RNA Export. J. Biol. Chem. 2001, 276, 25910–25918. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Regener, K.; Schwappach, B.; Saffrich, R.; Paraskeva, E.; Hartmann, E.; Görlich, D. Exp5 Exports EEF1A via TRNA from Nuclei and Synergizes with Other Transport Pathways to Confine Translation to the Cytoplasm. EMBO J. 2002, 21, 6205–6215. [Google Scholar] [CrossRef]
- Brownawell, A.M.; Macara, I.G. Exportin-5, a Novel Karyopherin, Mediates Nuclear Export of Double-Stranded RNA Binding Proteins. J. Cell Biol. 2002, 156, 53–64. [Google Scholar] [CrossRef]
- Lu, S.; Cullen, B.R. Adenovirus VA1 Noncoding RNA Can Inhibit Small Interfering RNA and MicroRNA Biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar] [CrossRef]
- Andersson, M.G.; Haasnoot, P.C.J.; Xu, N.; Berenjian, S.; Berkhout, B.; Akusjärvi, G. Suppression of RNA Interference by Adenovirus Virus-Associated RNA. J. Virol. 2005, 79, 9556–9565. [Google Scholar] [CrossRef]
- Xu, N.; Segerman, B.; Zhou, X.; Akusjärvi, G. Adenovirus Virus-Associated RNAII-Derived Small RNAs Are Efficiently Incorporated into the RNA-Induced Silencing Complex and Associate with Polyribosomes. J. Virol. 2007, 81, 10540–10549. [Google Scholar] [CrossRef]
- Aparicio, O.; Razquin, N.; Zaratiegui, M.; Narvaiza, I.; Fortes, P. Adenovirus Virus-Associated RNA Is Processed to Functional Interfering RNAs Involved in Virus Production. J. Virol. 2006, 80, 1376–1384. [Google Scholar] [CrossRef]
- Sano, M.; Kato, Y.; Taira, K. Sequence-specific Interference by Small RNAs Derived from Adenovirus VAI RNA. FEBS Lett. 2006, 580, 1553–1564. [Google Scholar] [CrossRef]
- Bellutti, F.; Kauer, M.; Kneidinger, D.; Lion, T.; Klein, R. Identification of RISC-Associated Adenoviral MicroRNAs, a Subset of Their Direct Targets, and Global Changes in the Targetome upon Lytic Adenovirus 5 Infection. J. Virol. 2014, 89, 1608–1627. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Machitani, M.; Tachibana, M.; Sakurai, F.; Mizuguchi, H. A MicroRNA Derived from Adenovirus Virus-Associated RNAII Promotes Virus Infection via Post-Transcriptional Gene Silencing. J. Virol. 2019, 93, e01265-18. [Google Scholar] [CrossRef]
- Seto, J.; Walsh, M.P.; Mahadevan, P.; Purkayastha, A.; Clark, J.M.; Tibbetts, C.; Seto, D. Genomic and Bioinformatics Analyses of HAdV-14p, Reference Strain of a Re-Emerging Respiratory Pathogen and Analysis of B1/B2. Virus Res. 2009, 143, 94–105. [Google Scholar] [CrossRef]
- Kodym, R.; Kodym, E.; Story, M.D. 2′-5′-Oligoadenylate Synthetase Is Activated by a Specific RNA Sequence Motif. Biochem. Biophys. Res. Commun. 2009, 388, 317–322. [Google Scholar] [CrossRef]
- Vachon, V.K.; Calderon, B.M.; Conn, G.L. A Novel RNA Molecular Signature for Activation of 2′-5′ Oligoadenylate Synthetase-1. Nucleic Acids Res. 2015, 43, 544–552. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA Secondary Structure Prediction and Analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, M.; Pettersson, U. Identification of Adenovirus-Encoded Small RNAs by Deep RNA Sequencing. Virology 2013, 442, 148–155. [Google Scholar] [CrossRef]
- Anderson, B.D.; Barr, K.L.; Heil, G.L.; Friary, J.A.; Gray, G.C. A Comparison of Viral Fitness and Virulence between Emergent Adenovirus 14p1 and Prototype Adenovirus 14p Strains. J. Clin. Virol. 2012, 54, 265–268. [Google Scholar] [CrossRef][Green Version]
- Radke, J.R.; Yong, S.L.; Cook, J.L. Low-Level Expression of the E1B 20-Kilodalton Protein by Adenovirus 14p1 Enhances Viral Immunopathogenesis. J. Virol. 2016, 90, 497–505. [Google Scholar] [CrossRef]
- Kamel, W.; Segerman, B.; Öberg, D.; Punga, T.; Akusjärvi, G. The Adenovirus VA RNA-Derived MiRNAs Are Not Essential for Lytic Virus Growth in Tissue Culture Cells. Nucleic Acids Res. 2013, 41, 4802–4812. [Google Scholar] [CrossRef]
- Furuse, Y.; Ornelles, D.A.; Cullen, B.R. Persistently Adenovirus-Infected Lymphoid Cells Express MicroRNAs Derived from the Viral VAI and Especially VAII RNA. Virology 2013, 447, 140–145. [Google Scholar] [CrossRef]
- Assadian, F.; Kamel, W.; Laurell, G.; Svensson, C.; Punga, T.; Akusjärvi, G. Expression Profile of Epstein-Barr Virus and Human Adenovirus Small RNAs in Tonsillar B and T Lymphocytes. PLoS ONE 2017, 12, e0177275. [Google Scholar] [CrossRef]
- Kamel, W.; Akusjärvi, G. An Ago2-Associated Capped Transcriptional Start Site Small RNA Suppresses Adenovirus DNA Replication. RNA 2017, 23, 1700–1711. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).