Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of Recombinant HIV-1 NCp7, NCp9, and NCp15
2.2. NMR Experiments
3. Results and Discussion
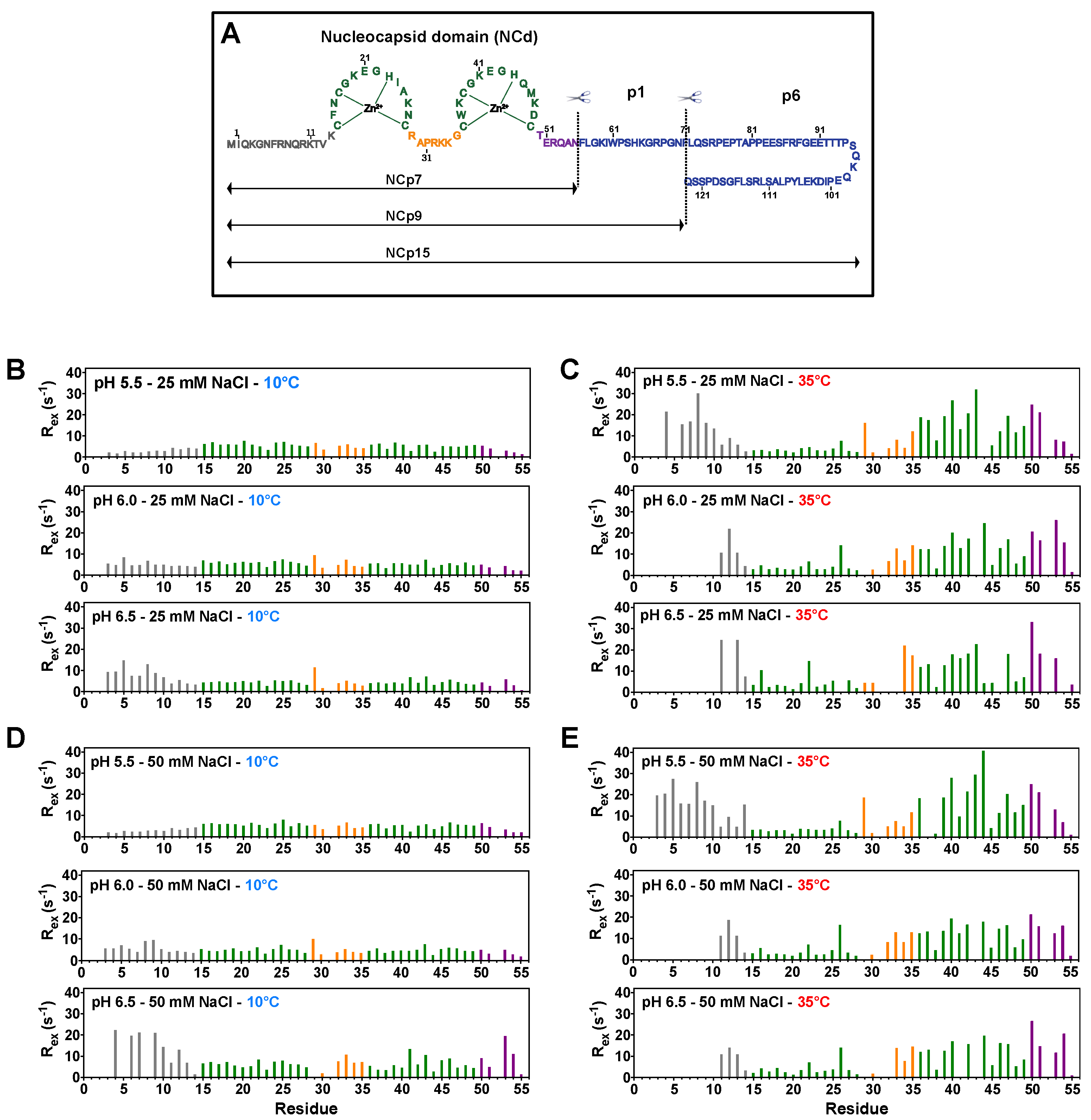
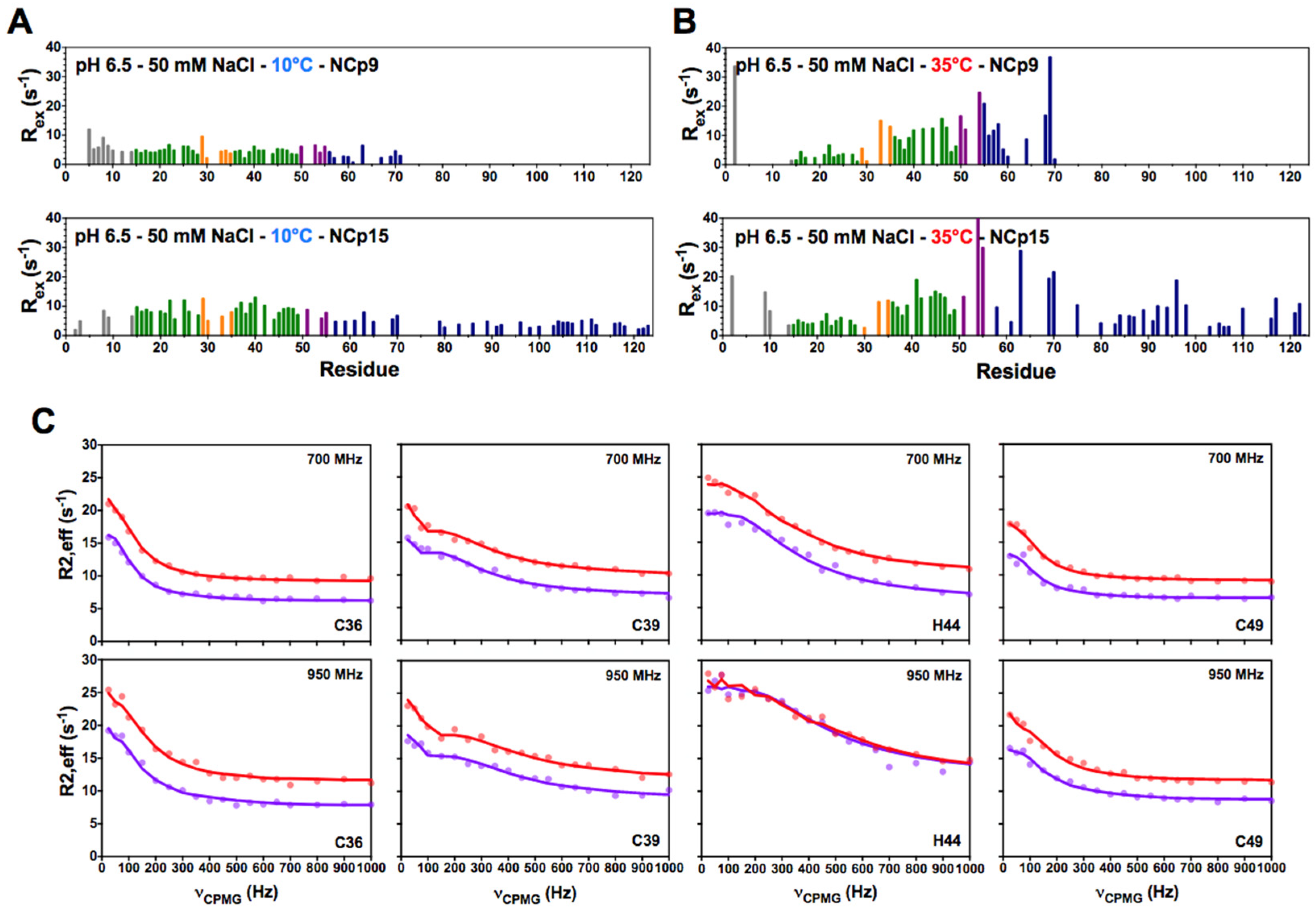
3.1. NCp7 and Its Precursors NCp9 and NCp15 Populate Two Minor States under Specific Temperature Conditions
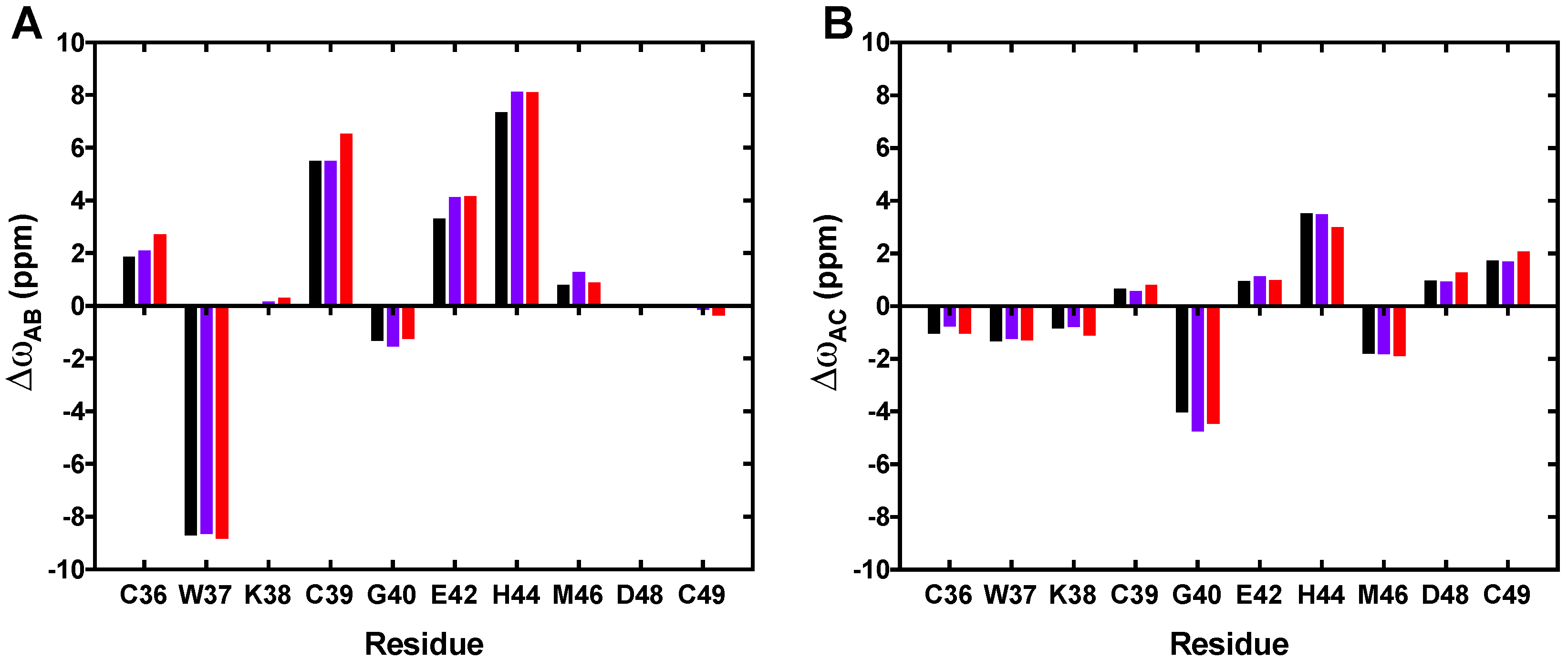
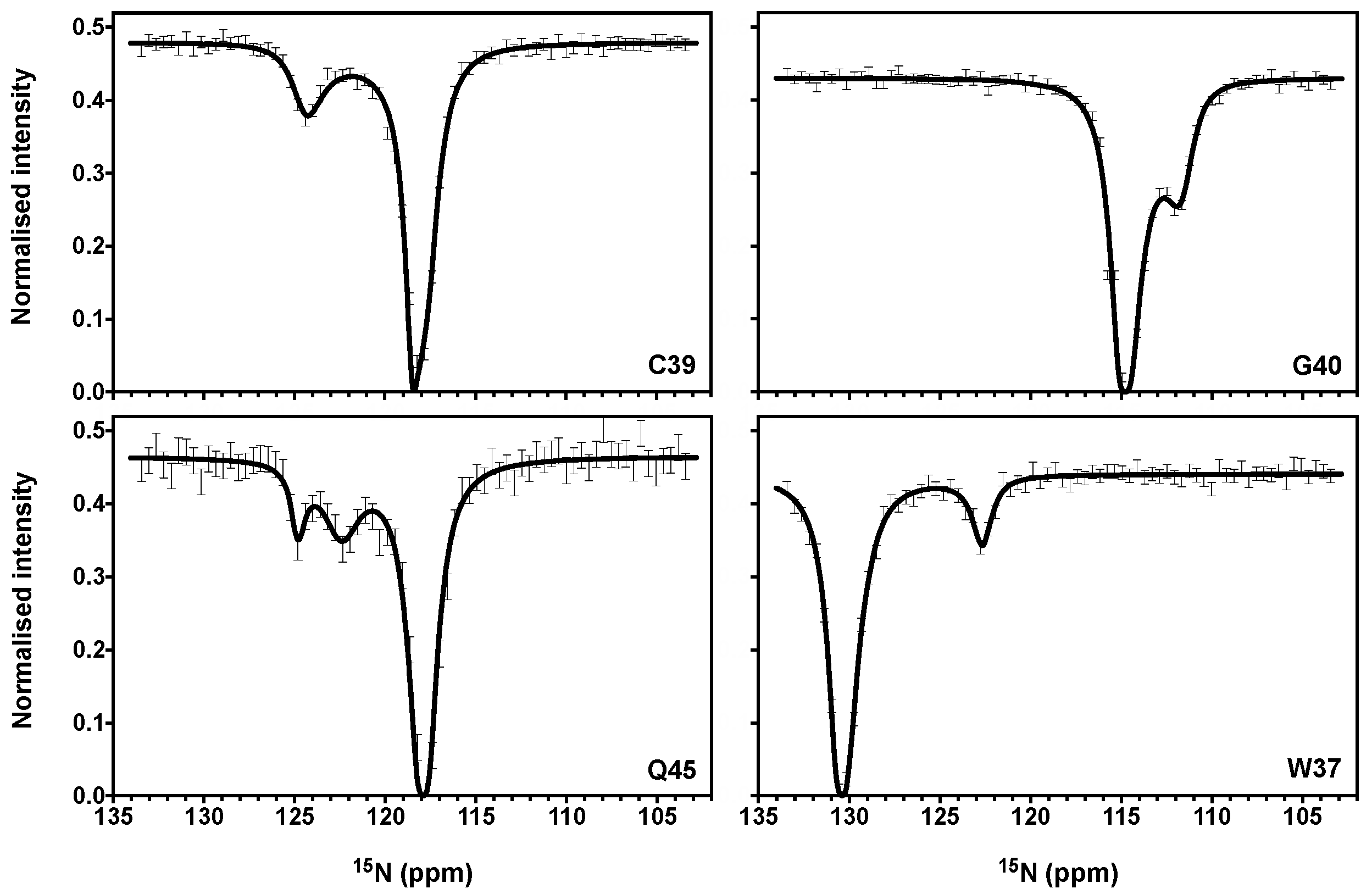
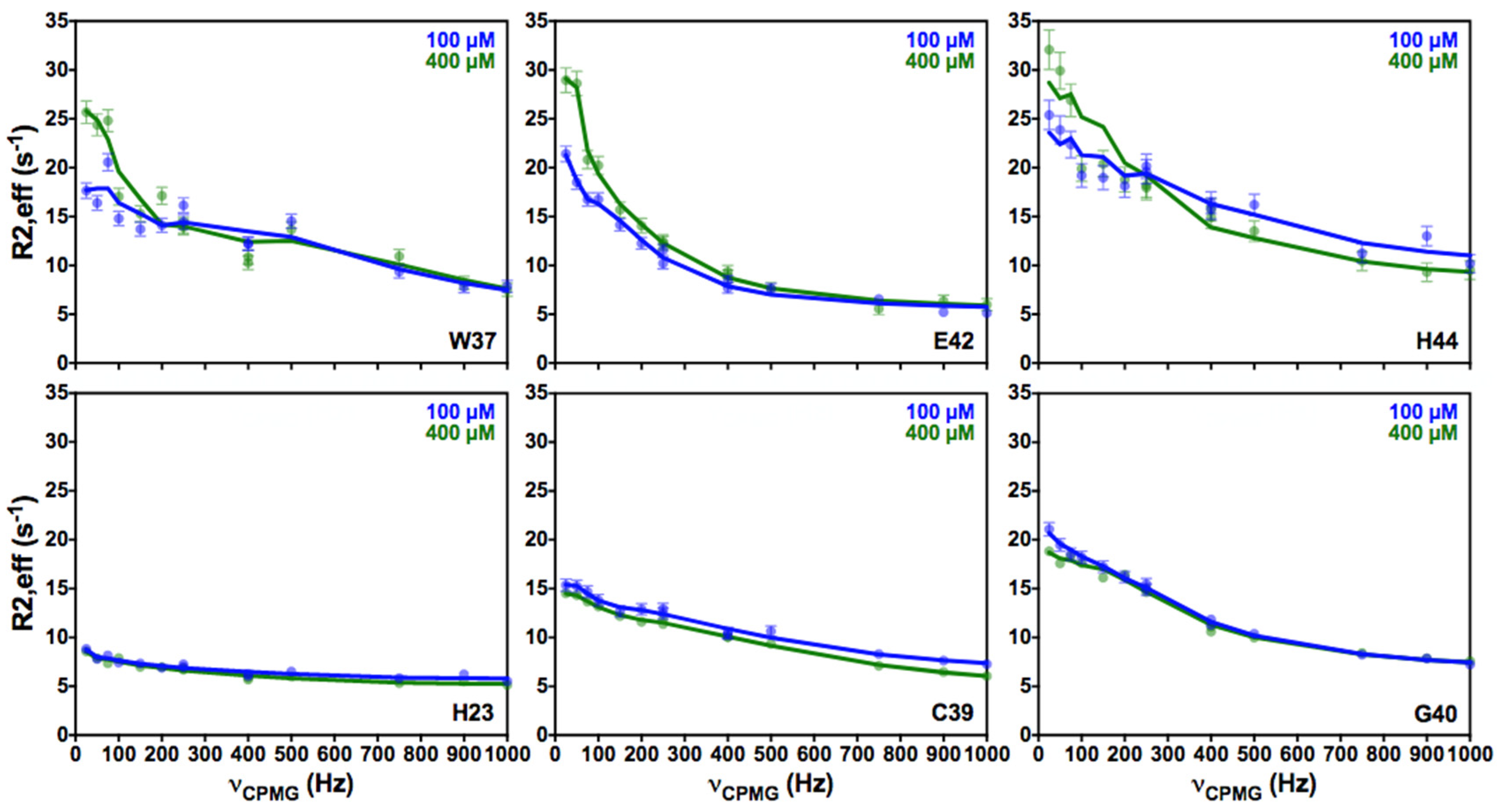
3.2. CEST Associated with CPMG Experiments on NCp7 and Analysis of the 15N Transverse Relaxation Rates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darlix, J.-L.; de Rocquigny, H.; Mauffret, O.; Mély, Y. Retrospective on the All-in-One Retroviral Nucleocapsid Protein. Virus Res. 2014, 193, 2–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, J.G.; Mitra, M.; Mascarenhas, A.; Musier-Forsyth, K. Role of HIV-1 Nucleocapsid Protein in HIV-1 Reverse Transcription. RNA Biol. 2010, 7, 754–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, J.A.; Gorelick, R.J. Nucleocapsid Protein Function in Early Infection Processes. Virus Res. 2008, 134, 39–63. [Google Scholar] [CrossRef] [Green Version]
- Darlix, J.-L.; Godet, J.; Ivanyi-Nagy, R.; Fossé, P.; Mauffret, O.; Mély, Y. Flexible Nature and Specific Functions of the HIV-1 Nucleocapsid Protein. J. Mol. Biol. 2011, 410, 565–581. [Google Scholar] [CrossRef]
- Godet, J.; Mély, Y. Biophysical Studies of the Nucleic Acid Chaperone Properties of the HIV-1 Nucleocapsid Protein. RNA Biol. 2010, 7, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, R.J.; Chabot, D.J.; Rein, A.; Henderson, L.E.; Arthur, L.O. The Two Zinc Fingers in the Human Immunodeficiency Virus Type 1 Nucleocapsid Protein Are Not Functionally Equivalent. J. Virol. 1993, 67, 4027–4036. [Google Scholar] [CrossRef] [Green Version]
- Mouhand, A.; Pasi, M.; Catala, M.; Zargarian, L.; Belfetmi, A.; Barraud, P.; Mauffret, O.; Tisné, C. Overview of the Nucleic-Acid Binding Properties of the HIV-1 Nucleocapsid Protein in Its Different Maturation States. Viruses 2020, 12, 1109. [Google Scholar] [CrossRef]
- Williams, M.C.; Gorelick, R.J.; Musier-Forsyth, K. Specific Zinc-Finger Architecture Required for HIV-1 Nucleocapsid Protein’s Nucleic Acid Chaperone Function. Proc. Natl. Acad. Sci. USA 2002, 99, 8614–8619. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, W.; Naiyer, N.; Fichtenbaum, E.; Qualley, D.F.; McCauley, M.J.; Gorelick, R.J.; Rouzina, I.; Musier-Forsyth, K.; Williams, M.C. Single Aromatic Residue Location Alters Nucleic Acid Binding and Chaperone Function of FIV Nucleocapsid Protein. Virus Res. 2014, 193, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Boutant, E.; Bonzi, J.; Anton, H.; Nasim, M.B.; Cathagne, R.; Réal, E.; Dujardin, D.; Carl, P.; Didier, P.; Paillart, J.-C.; et al. Zinc Fingers in HIV-1 Gag Precursor Are Not Equivalent for GRNA Recruitment at the Plasma Membrane. Biophys. J. 2020, 119, 419–433. [Google Scholar] [CrossRef]
- Durand, S.; Seigneuret, F.; Burlaud-Gaillard, J.; Lemoine, R.; Tassi, M.-F.; Moreau, A.; Mougel, M.; Roingeard, P.; Tauber, C.; de Rocquigny, H. Quantitative Analysis of the Formation of Nucleoprotein Complexes between HIV-1 Gag Protein and Genomic RNA Using Transmission Electron Microscopy. J. Biol. Chem. 2021, 298, 101500. [Google Scholar] [CrossRef] [PubMed]
- Miller Jenkins, L.M.; Hara, T.; Durell, S.R.; Hayashi, R.; Inman, J.K.; Piquemal, J.-P.; Gresh, N.; Appella, E. Specificity of Acyl Transfer from 2-Mercaptobenzamide Thioesters to the HIV-1 Nucleocapsid Protein. J. Am. Chem. Soc. 2007, 129, 11067–11078. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Kovalenko, L.; Lyonnais, S.; Antaki, D.; Torbett, B.E.; Botta, M.; Mirambeau, G.; Mély, Y. Nucleocapsid Protein: A Desirable Target for Future Therapies Against HIV-1. Curr. Top. Microbiol. Immunol. 2015, 389, 53–92. [Google Scholar] [CrossRef] [PubMed]
- de Rocquigny, H.; Shvadchak, V.; Avilov, S.; Dong, C.Z.; Dietrich, U.; Darlix, J.-L.; Mély, Y. Targeting the Viral Nucleocapsid Protein in Anti-HIV-1 Therapy. Mini Rev. Med. Chem. 2008, 8, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, L.M.M.; Durell, S.R.; Maynard, A.T.; Stahl, S.J.; Inman, J.K.; Appella, E.; Legault, P.; Omichinski, J.G. Comparison of the Specificity of Interaction of Cellular and Viral Zinc-Binding Domains with 2-Mercaptobenzamide Thioesters. J. Am. Chem. Soc. 2006, 128, 11964–11976. [Google Scholar] [CrossRef]
- Miller Jenkins, L.M.; Byrd, J.C.; Hara, T.; Srivastava, P.; Mazur, S.J.; Stahl, S.J.; Inman, J.K.; Appella, E.; Omichinski, J.G.; Legault, P. Studies on the Mechanism of Inactivation of the HIV-1 Nucleocapsid Protein NCp7 with 2-Mercaptobenzamide Thioesters. J. Med. Chem. 2005, 48, 2847–2858. [Google Scholar] [CrossRef]
- Maynard, A.T.; Covell, D.G. Reactivity of Zinc Finger Cores: Analysis of Protein Packing and Electrostatic Screening. J. Am. Chem. Soc. 2001, 123, 1047–1058. [Google Scholar] [CrossRef]
- Maynard, A.T.; Huang, M.; Rice, W.G.; Covell, D.G. Reactivity of the HIV-1 Nucleocapsid Protein P7 Zinc Finger Domains from the Perspective of Density-Functional Theory. Proc. Natl. Acad. Sci. USA 1998, 95, 11578–11583. [Google Scholar] [CrossRef] [Green Version]
- Miller Jenkins, L.M.; Ott, D.E.; Hayashi, R.; Coren, L.V.; Wang, D.; Xu, Q.; Schito, M.L.; Inman, J.K.; Appella, D.H.; Appella, E. Small-Molecule Inactivation of HIV-1 NCp7 by Repetitive Intracellular Acyl Transfer. Nat. Chem. Biol. 2010, 6, 887–889. [Google Scholar] [CrossRef]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 Nucleocapsid Protein Bound to the SL3 Psi-RNA Recognition Element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef]
- Retureau, R.; Oguey, C.; Mauffret, O.; Hartmann, B. Structural Explorations of NCp7-Nucleic Acid Complexes Give Keys to Decipher the Binding Process. J. Mol. Biol. 2019, 431, 1966–1980. [Google Scholar] [CrossRef] [PubMed]
- Vuilleumier, C.; Bombarda, E.; Morellet, N.; Gérard, D.; Roques, B.P.; Mély, Y. Nucleic Acid Sequence Discrimination by the HIV-1 Nucleocapsid Protein NCp7: A Fluorescence Study. Biochemistry 1999, 38, 16816–16825. [Google Scholar] [CrossRef] [PubMed]
- Zargarian, L.; Tisné, C.; Barraud, P.; Xu, X.; Morellet, N.; René, B.; Mély, Y.; Fossé, P.; Mauffret, O. Dynamics of Linker Residues Modulate the Nucleic Acid Binding Properties of the HIV-1 Nucleocapsid Protein Zinc Fingers. PLoS ONE 2014, 9, e102150. [Google Scholar] [CrossRef]
- Avilov, S.V.; Piemont, E.; Shvadchak, V.; de Rocquigny, H.; Mély, Y. Probing Dynamics of HIV-1 Nucleocapsid Protein/Target Hexanucleotide Complexes by 2-Aminopurine. Nucleic Acids Res. 2008, 36, 885–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourbigot, S.; Ramalanjaona, N.; Boudier, C.; Salgado, G.F.J.; Roques, B.P.; Mély, Y.; Bouaziz, S.; Morellet, N. How the HIV-1 Nucleocapsid Protein Binds and Destabilises the (-)Primer Binding Site during Reverse Transcription. J. Mol. Biol. 2008, 383, 1112–1128. [Google Scholar] [CrossRef] [PubMed]
- Mitra, M.; Wang, W.; Vo, M.-N.; Rouzina, I.; Barany, G.; Musier-Forsyth, K. The N-Terminal Zinc Finger and Flanking Basic Domains Represent the Minimal Region of the Human Immunodeficiency Virus Type-1 Nucleocapsid Protein for Targeting Chaperone Function. Biochemistry 2013, 52, 8226–8236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouhand, A.; Belfetmi, A.; Catala, M.; Larue, V.; Zargarian, L.; Brachet, F.; Gorelick, R.J.; Van Heijenoort, C.; Mirambeau, G.; Barraud, P.; et al. Modulation of the HIV Nucleocapsid Dynamics Finely Tunes Its RNA-Binding Properties during Virion Genesis. Nucleic Acids Res. 2018, 46, 9699–9710. [Google Scholar] [CrossRef]
- Deshmukh, L.; Ghirlando, R.; Clore, G.M. Investigation of the Structure and Dynamics of the Capsid-Spacer Peptide 1-Nucleocapsid Fragment of the HIV-1 Gag Polyprotein by Solution NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 2014, 53, 1025–1028. [Google Scholar] [CrossRef] [Green Version]
- Deshmukh, L.; Schwieters, C.D.; Grishaev, A.; Clore, G.M. Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering. Chemphyschem 2016, 17, 1548–1552. [Google Scholar] [CrossRef] [Green Version]
- Hansen, D.F.; Vallurupalli, P.; Kay, L.E. An Improved 15N Relaxation Dispersion Experiment for the Measurement of Millisecond Time-Scale Dynamics in Proteins. J. Phys. Chem. B 2008, 112, 5898–5904. [Google Scholar] [CrossRef]
- Vallurupalli, P.; Sekhar, A.; Yuwen, T.; Kay, L.E. Probing Conformational Dynamics in Biomolecules via Chemical Exchange Saturation Transfer: A Primer. J. Biomol. NMR 2017, 67, 243–271. [Google Scholar] [CrossRef] [PubMed]
- Vallurupalli, P.; Bouvignies, G.; Kay, L.E. Studying “Invisible” Excited Protein States in Slow Exchange with a Major State Conformation. J. Am. Chem. Soc. 2012, 134, 8148–8161. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, L.; Tugarinov, V.; Appella, D.H.; Clore, G.M. Targeting a Dark Excited State of HIV-1 Nucleocapsid by Antiretroviral Thioesters Revealed by NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 2687–2691. [Google Scholar] [CrossRef]
- Lee, B.M.; De Guzman, R.N.; Turner, B.G.; Tjandra, N.; Summers, M.F. Dynamical Behavior of the HIV-1 Nucleocapsid Protein. J. Mol. Biol. 1998, 279, 633–649. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G., 3rd; Kroenke, C.D.; Loria, J.P. Nuclear Magnetic Resonance Methods for Quantifying Microsecond-to-Millisecond Motions in Biological Macromolecules. Methods Enzymol. 2001, 339, 204–238. [Google Scholar]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. Nmrpipe—A Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Kneller, D.G. SPARKY 3; University of California: San Francisco, CA, USA, 2008. [Google Scholar]
- Mulder, F.A.; Skrynnikov, N.R.; Hon, B.; Dahlquist, F.W.; Kay, L.E. Measurement of Slow (Micros-Ms) Time Scale Dynamics in Protein Side Chains by (15)N Relaxation Dispersion NMR Spectroscopy: Application to Asn and Gln Residues in a Cavity Mutant of T4 Lysozyme. J. Am. Chem. Soc. 2001, 123, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Millet, O.; Loria, J.P.; Kroenke, C.D.; Pons, M.; Palmer, A.G. The Static Magnetic Field Dependence of Chemical Exchange Linebroadening Defines the NMR Chemical Shift Time Scale. J. Am. Chem. Soc. 2000, 122, 2867–2877. [Google Scholar] [CrossRef]
- Morin, S.; Linnet, T.E.; Lescanne, M.; Schanda, P.; Thompson, G.S.; Tollinger, M.; Teilum, K.; Gagné, S.; Marion, D.; Griesinger, C.; et al. Relax: The Analysis of Biomolecular Kinetics and Thermodynamics Using NMR Relaxation Dispersion Data. Bioinformatics 2019, 35, 4205. [Google Scholar] [CrossRef]
- Kumari, P.; Frey, L.; Sobol, A.; Lakomek, N.-A.; Riek, R. 15N Transverse Relaxation Measurements for the Characterization of Μs-Ms Dynamics Are Deteriorated by the Deuterium Isotope Effect on 15N Resulting from Solvent Exchange. J. Biomol. NMR 2018, 72, 125–137. [Google Scholar] [CrossRef] [Green Version]
- McConnell, H.M. Reaction Rates by Nuclear Magnetic Resonance. J. Chem. Phys. 1958, 28, 430–431. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Wu, K.-P.; Baum, J. Fast Hydrogen Exchange Affects 15N Relaxation Measurements in Intrinsically Disordered Proteins. J. Biomol. NMR 2013, 55, 249–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvignies, G.; Hansen, D.F.; Vallurupalli, P.; Kay, L.E. Divided-Evolution-Based Pulse Scheme for Quantifying Exchange Processes in Proteins: Powerful Complement to Relaxation Dispersion Experiments. J. Am. Chem. Soc. 2011, 133, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Fawzi, N.L.; Ying, J.; Ghirlando, R.; Torchia, D.A.; Clore, G.M. Atomic-Resolution Dynamics on the Surface of Amyloid-β Protofibrils Probed by Solution NMR. Nature 2011, 480, 268–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamanos, T.K.; Tugarinov, V.; Clore, G.M. Unraveling the Structure and Dynamics of the Human DNAJB6b Chaperone by NMR Reveals Insights into Hsp40-Mediated Proteostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 21529–21538. [Google Scholar] [CrossRef] [Green Version]
- Miller Jenkins, L.M.; Paine, E.L.; Deshmukh, L.; Nikolayevskiy, H.; Lyons, G.C.; Scerba, M.T.; George Rosenker, K.; Luecke, H.F.; Louis, J.M.; Chertova, E.; et al. Inhibition of HIV Maturation via Selective Unfolding and Cross-Linking of Gag Polyprotein by a Mercaptobenzamide Acetylator. J. Am. Chem. Soc. 2019, 141, 8327–8338. [Google Scholar] [CrossRef]
- Ramboarina, S.; Srividya, N.; Atkinson, R.A.; Morellet, N.; Roques, B.P.; Lefèvre, J.-F.; Mély, Y.; Kieffer, B. Effects of Temperature on the Dynamic Behaviour of the HIV-1 Nucleocapsid NCp7 and Its DNA Complex. J. Mol. Biol. 2002, 316, 611–627. [Google Scholar] [CrossRef]
- Lapadat-Tapolsky, M.; Pernelle, C.; Borie, C.; Darlix, J.L. Analysis of the Nucleic Acid Annealing Activities of Nucleocapsid Protein from HIV-1. Nucleic Acids Res. 1995, 23, 2434–2441. [Google Scholar] [CrossRef]
- Tsuchihashi, Z.; Brown, P.O. DNA Strand Exchange and Selective DNA Annealing Promoted by the Human Immunodeficiency Virus Type 1 Nucleocapsid Protein. J. Virol. 1994, 68, 5863–5870. [Google Scholar] [CrossRef] [Green Version]
- Tisné, C.; Roques, B.P.; Dardel, F. The Annealing Mechanism of HIV-1 Reverse Transcription Primer onto the Viral Genome. J. Biol. Chem. 2004, 279, 3588–3595. [Google Scholar] [CrossRef] [Green Version]
- Ren, W.; Ji, D.; Xu, X. Metal Cofactor Modulated Folding and Target Recognition of HIV-1 NCp7. PLoS ONE 2018, 13, e0196662. [Google Scholar] [CrossRef] [Green Version]
- Mely, Y.; De Rocquigny, H.; Morellet, N.; Roques, B.P.; Gerad, D. Zinc Binding to the HIV-1 Nucleocapsid Protein: A Thermodynamic Investigation by Fluorescence Spectroscopy. Biochemistry 1996, 35, 5175–5182. [Google Scholar] [CrossRef] [PubMed]
| NCp7 | NCp9 | NCp15 | |
|---|---|---|---|
| pA | 0.945 ± 0.002 | 0.883 ± 0.009 | 0.870 ± 0.007 |
| pB | 0.027 ± 0.002 | 0.086 ± 0.008 | 0.087 ± 0.007 |
| pC | 0.027 ± 0.001 | 0.030 ± 0.001 | 0.041 ± 0.001 |
| kex_AB (s−1) | 331 ± 31 | 66 ± 7 | 64 ± 5 |
| kex_AC (s−1) | 418 ± 13 | 227 ± 14 | 169 ± 9 |
| kex_BC (s−1) | 93 ± 15 | ~0 | ~0 |
| CESTH44,W37,C39 + CPMGH44,W37,C39 | CESTH44,W37,C39 + CPMG10residues | CPMG10residues | CESTH44,W37,C39 | CESTH44,W37,C39,G40,E42 + CPMGH44,W37,C39 | |
|---|---|---|---|---|---|
| pA | 0.959 ± 0.001 | 0.957 ± 0.001 | 0.945 ± 0.002 | 0.967 ± 0.002 | 0.961 ± 0.001 |
| pB | 0.015 ± 0.001 | 0.016± 0.001 | 0.026 ± 0.002 | 0.013 ± 0.001 | 0.016 ± 0.001 |
| pC | 0.025 ± 0.001 | 0.026± 0.001 | 0.028 ± 0.001 | 0.019 ± 0.003 | 0.022 ± 0.001 |
| kex_AB (s−1) | 462 ± 40 | 507 ± 42 | 349 ± 38 | 297 ± 72 | 504 ± 25 |
| kex_AC (s−1) | 593 ± 45 | 524 ± 36 | 415 ± 24 | 149 ± 48 | 630 ± 24 |
| kex_BC (s1)) | 122 ± 56 | 10 ± 30 | 79 ± 24 | 347 ± 68 | 73 ± 32 |
| Χ2reduced | 1.77 | 1.51 | 1.12 | 1.20 | 2.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mouhand, A.; Zargarian, L.; Belfetmi, A.; Catala, M.; Pasi, M.; Lescop, E.; Tisné, C.; Mauffret, O. Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain. Viruses 2022, 14, 632. https://doi.org/10.3390/v14030632
Mouhand A, Zargarian L, Belfetmi A, Catala M, Pasi M, Lescop E, Tisné C, Mauffret O. Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain. Viruses. 2022; 14(3):632. https://doi.org/10.3390/v14030632
Chicago/Turabian StyleMouhand, Assia, Loussiné Zargarian, Anissa Belfetmi, Marjorie Catala, Marco Pasi, Ewen Lescop, Carine Tisné, and Olivier Mauffret. 2022. "Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain" Viruses 14, no. 3: 632. https://doi.org/10.3390/v14030632
APA StyleMouhand, A., Zargarian, L., Belfetmi, A., Catala, M., Pasi, M., Lescop, E., Tisné, C., & Mauffret, O. (2022). Investigation of the Low-Populated Excited States of the HIV-1 Nucleocapsid Domain. Viruses, 14(3), 632. https://doi.org/10.3390/v14030632







