A Renewed Appreciation of Helicoverpa armigera Nucleopolyhedrovirus BJ (Formerly Helicoverpa assulta Nucleopolyhedrovirus) with Whole Genome Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects Cell Lines and Virus Isolates
2.2. Identification of Virus by Restriction Endonuclease Analysis
2.3. Sequencing and ORF Finding
2.4. Sequence Analysis
3. Results and Discussion
3.1. Structure Characteristic of HearNPV-BJ Virion
3.2. Infective Properties of Various Insect Cell Lines of HearNPV-BJ
3.3. Restriction Enzyme Digest Mapping of HearNPV-BJ
3.4. Characterization of HearNPV-BJ Genome
3.5. Virus Species Demarcation Criteria
3.6. Phylogenetic Analysis of HearNPV-BJ
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibbs, A. Binomial nomenclature for virus species: A long view. Arch. Virol. 2020, 165, 3079–3083. [Google Scholar] [CrossRef] [PubMed]
- Murphy, F.A.; Fauquet, C.M.; Bishop, D.H.L.; Ghabrial, S.A.; Jarvis, A.W.; Martelli, G.P.; Mayo, M.A.; Summers, M.D. The International Committee on Taxonomy of Viruses. Arch. Virol. Suppl. 2000, 10, 1–586. [Google Scholar]
- Zhang, G.Y.; Sun, X.L.; Zhang, Z.X.; Zhang, Z.F.; Mo, F.F. Production and Effectiveness on the New Form ulation ofHeliothis Virus(NPV)Pesticide Emulsifiabie Suspension. Virol. Sin. 1995, 10, 242. [Google Scholar]
- Qin, Q. Production and Application of Helicoverpa armigera Nucleopolyhedrovirus Bio-pesticides Trademarked KEYUN on Large Scale. Biotechnol. Bull. 2008, 1, 467–470. [Google Scholar]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, W.J.; Wong, J.; Chun, G.; Lu, A.; McCutchen, B.F.; Presnail, J.K.; Herrmann, R.; Dolan, M.; Tingey, S.; et al. Comparative analysis of the complete genome sequences of Helicoverpa zea and Helicoverpa armigera nucleocapsid nucleopolyhedroviruses. J. Gen. Virol. 2002, 83, 673–684. [Google Scholar] [CrossRef]
- Chen, X.; IJkel, W.F.; Tarchini, R.; Sun, X.; Sandbrink, H.; Wang, H.; Peters, S.; Zuidema, D.; Lankhorst, R.K.; Vlak, J.M.; et al. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 2001, 82, 241–257. [Google Scholar] [CrossRef]
- Wang, H.L.; Ming, Q.L.; Zhao, C.H.; Wang, C.Z. Genetic basis of sex pheromone blend difference between Helicoverpa armigera (Hübner) and Helicoverpa assulta (Guenée) (Lepidoptera: Noctuidae). J. Insect Physiol. 2008, 54, 813–817. [Google Scholar] [CrossRef]
- Hu, Z.H.; Liu, M.F.; Jin, F.; Wang, Z.X.; Liu, X.Y.; Li, M.J.; Liang, B.F.; Xie, T.E. Nucleotide sequence of the Buzura suppressaria single nucleocapsid nuclear polyhedrosis virus polyhedrin gene. J. Gen. Virol 1993, 74 Pt 8, 1617–1620. [Google Scholar] [CrossRef]
- O’Reilly, D.R. Baculovirus Expression Vectors: A Laboratory Manual; Oxford University Press on Demand: Oxford, UK, 1992. [Google Scholar]
- Tang, P.; Zhang, H.; Li, Y.; Han, B.; Wang, G.; Qin, Q. Genomic sequencing and analyses of HearMNPV—A new Multinucleocapsid nucleopolyhedrovirus isolated from Helicoverpa armigera. Virol. J. 2012, 9, 1–18. [Google Scholar] [CrossRef]
- Jun, M.; Hajime, I. Obtainment of a Continuous Cell Line from the Larval Fat Bodies of the Mulberry Tiger Moth, Spilosoma imparilis (Lepidoptera:Arctiidae). Appl. Entomol. Zool. 1988, 23, 488–490. [Google Scholar]
- Li, X.; Liang, C.-Y.; Song, J.-H.; Chen, X.-W. The ORF 113 of Helicoverpa armigera single nucleopolyhedrovirus encodes a functional fibroblast growth factor. Virol. Sin. 2008, 23, 321–329. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.A.; Qin, Q.; Wang, Y.; Li, X.; Miao, L.; Yin, Z.; Zhang, A.; Qu, L.; Ding, C. A new cell line from larval fat bodies of the bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). In Vitro Cell Dev. Biol. Anim. 2006, 42, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, H.; Sun, X.; Chen, X.; Peng, C.; Pan, D.; Jehle, J.A.; Werf, W.V.D.; Vlak, J.M.; Hu, Z. Biological activity and field efficacy of a genetically modified Helicoverpa armigera single-nucleocapsid nucleopolyhedrovirus expressing an insect-selective toxin from a chimeric promoter. Biol. Control. 2004, 29, 124–137. [Google Scholar] [CrossRef]
- Hu, Z.H.; Arif, B.M.; Jin, F.; Martens, J.W.; Chen, X.W.; Sun, J.S.; Zuidema, D.; Goldbach, R.W.; Vlak, J.M. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1998, 79 Pt 11, 2841–2851. [Google Scholar] [CrossRef]
- Zhu, Z.; Yin, F.; Liu, X.; Hou, D.; Wang, J.; Zhang, L.; Arif, B.; Wang, H.; Deng, F.; Hu, Z. Genome sequence and analysis of Buzura suppressaria nucleopolyhedrovirus: A group II Alphabaculovirus. PLoS ONE 2014, 9, e86450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Blissard, G.W.; Rohrmann, G.F. Baculovirus Diversity and Molecular Biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef]
- Miele, S.A.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular insights on their diversity and conservation. Int. J. Evol. Biol. 2011, 2011, 379424. [Google Scholar] [CrossRef]
- Winstanley, D.; Rovesti, L. Insect Viruses as Biocontrol Agents; Springer: Berlin/Heidelberg, Germany, 1993. [Google Scholar]
- Boucias, D.G.; Pendland, J.C. Bacillus thuringiensis: Producer of Potent Insecticidal Toxins; Springer: Boston, MA, USA, 1998. [Google Scholar]
- Guang, Z.; Yuan, H. Original Study of Korean Isolate of HaSNPV. Virol. Sin. 2003, 18, 348–351. [Google Scholar]
- Lu, H.S. Insects Virus and Virus Disease of Insect; Science Press: Beijing, China, 1982. [Google Scholar]
- Deng, F.; Wang, R.; Fang, M.; Jiang, Y.; Xu, X.; Wang, H.; Chen, X.; Arif, B.M.; Guo, L.; Wang, H.; et al. Proteomics analysis of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus identified two new occlusion-derived virus-associated proteins, HA44 and HA100. J. Virol. 2007, 81, 9377–9385. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, G.R.; Zhang, J.M.; Hu, Y.Y. Sequence Analgsis of the BamHI-I Gene Fragment of Korean Isolate of Helicoverpa Armigera of the Single Nucleocapsid Nucleopolyhedrovirus. J.-Wuhan Univ. Nat. Sci. Ed. 2003, 49, 491–496. [Google Scholar]
- Chen, X.; Sun, X.; Hu, Z.; Li, M.; O’Reilly, D.R.; Zuidema, D.; Vlak, J.M. Genetic engineering of Helicoverpa armigera single-nucleocapsid nucleopolyhedrovirus as an improved pesticide. J. Invertebr. Pathol. 2000, 76, 140–146. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Tang, P.; Zhang, H.; Qin, Q.; Zhang, Z. Genome sequence and organization of the Mythimna (formerly Pseudaletia) unipuncta granulovirus Hawaiian strain. Sci. Rep. 2021, 11, 414. [Google Scholar] [CrossRef]
- Noune, C.; Hauxwell, C. Comparative Analysis of HaSNPV-AC53 and Derived Strains. Viruses 2016, 8, 280. [Google Scholar] [CrossRef]
- Guarino, L.A.; Gonzalez, M.A.; Summers, M.D. Complete Sequence and Enhancer Function of the Homologous DNA Regions of Autographa californica Nuclear Polyhedrosis Virus. J. Virol. 1986, 60, 224–229. [Google Scholar] [CrossRef]
- Pearson, M.; Bjornson, R.; Pearson, G.; Rohrmann, G. The Autographa californica baculovirus genome: Evidence for multiple replication origins. Science 1992, 257, 1382–1384. [Google Scholar] [CrossRef]
- Theilmann, D.A.; Stewart, S. Tandemly repeated sequence at the 3′ end of the IE-2 gene of the baculovirus Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus is an enhancer element. Virology 1992, 187, 97–106. [Google Scholar] [CrossRef]
- Cochran, M.A.; Faulkner, P. Location of Homologous DNA Sequences Interspersed at Five Regions in the Baculovirus AcMNPV Genome. J. Virol. 1983, 45, 961. [Google Scholar] [CrossRef]
- Guarino, L.A.; Summers, M.D. Interspersed Homologous DNA of Autographa californica Nuclear Polyhedrosis Virus Enhances Delayed-Early Gene Expression. J. Virol. 1986, 60, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, T.; Rohrmann, G.F.; Hashimoto, Y. Patterns of Genome Organization and Content in Lepidopteran Baculoviruses. Virology 2000, 278, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Hilton, S.; Winstanley, D. Identification and functional analysis of the origins of DNA replication in the Cydia pomonella granulovirus genome. J. Gen. Virol. 2007, 88, 1496–1504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Q.; Qin, Q.L.; Zhu, W.; Zhang, Z.F.; Li, Y.N.; Zhang, N.; Zhang, J.H. Genomic sequence analysis of Helicoverpa armigera nucleopolyhedrovirus isolated from Australia. Arch. Virol. 2014, 159, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, V.S. Replication of the baculovirus genome. Mol. Biol. 2003, 37, 288–299. [Google Scholar] [CrossRef]
- Miller, L.K.; Adang, M.J.; Browne, D. Protein Kinase Activity Associated with the Extracellular and Occluded Forms of the Baculovirus Autographa californica Nuclear Polyhedrosis Virus. J. Virol. 1983, 46, 275–278. [Google Scholar] [CrossRef]
- Wang, M.; Tuladhar, E.; Shen, S.; Wang, H.; van Oers, M.M.; Vlak, J.M.; Westenberg, M. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 2010, 84, 8821–8828. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 3rd ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2013. [Google Scholar]
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.; Mowery, J.D.; Bauchan, G.R.; et al. ICTV Virus Taxonomy Profile: Baculoviridae. J. Gen. Virol. 2019, 99, 1185–1186. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Michael, L.; Christina, K.; Koichiro, T. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar]
- Ivica, L.; Peer, B. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, W256–W259. [Google Scholar]
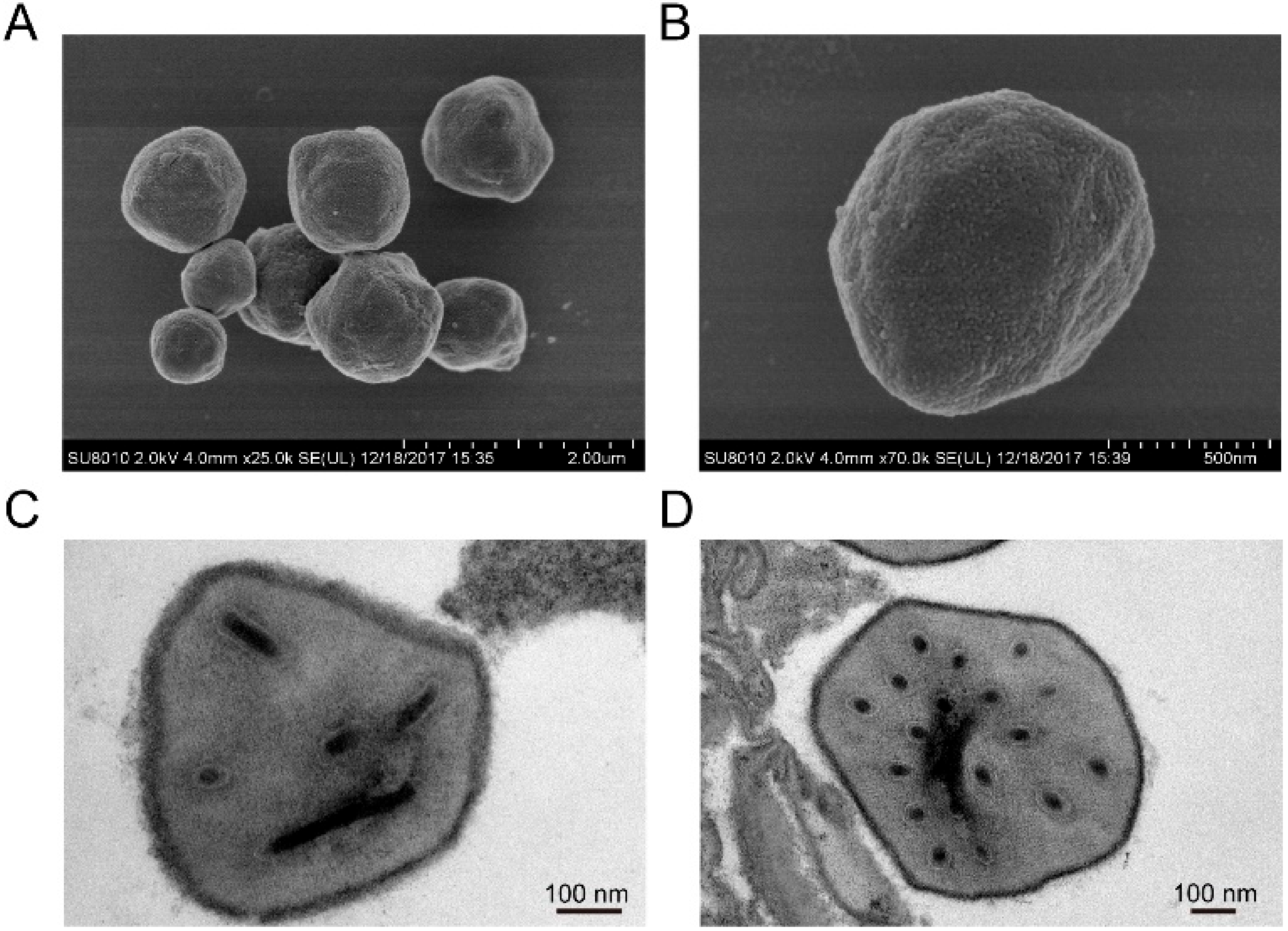
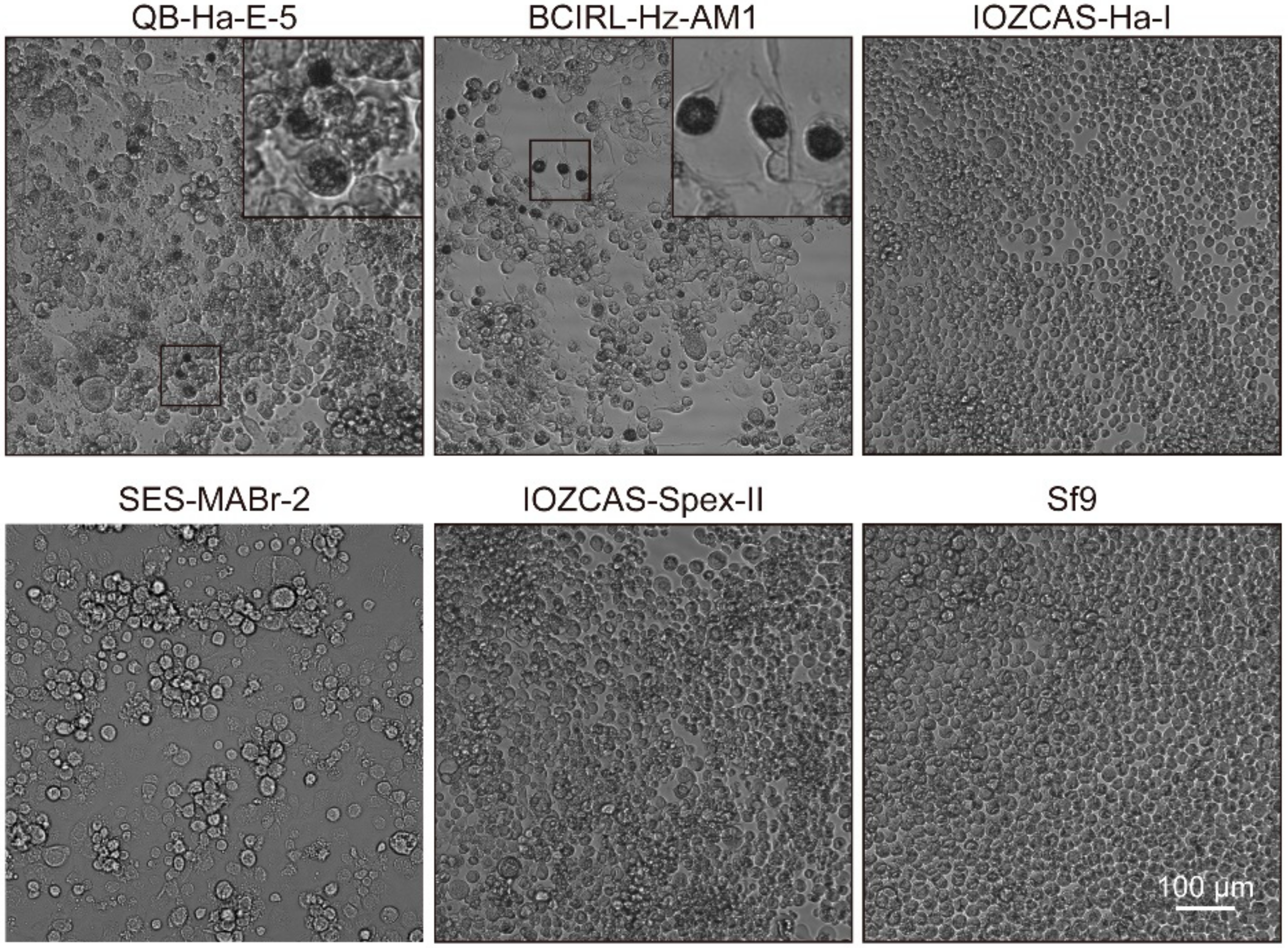

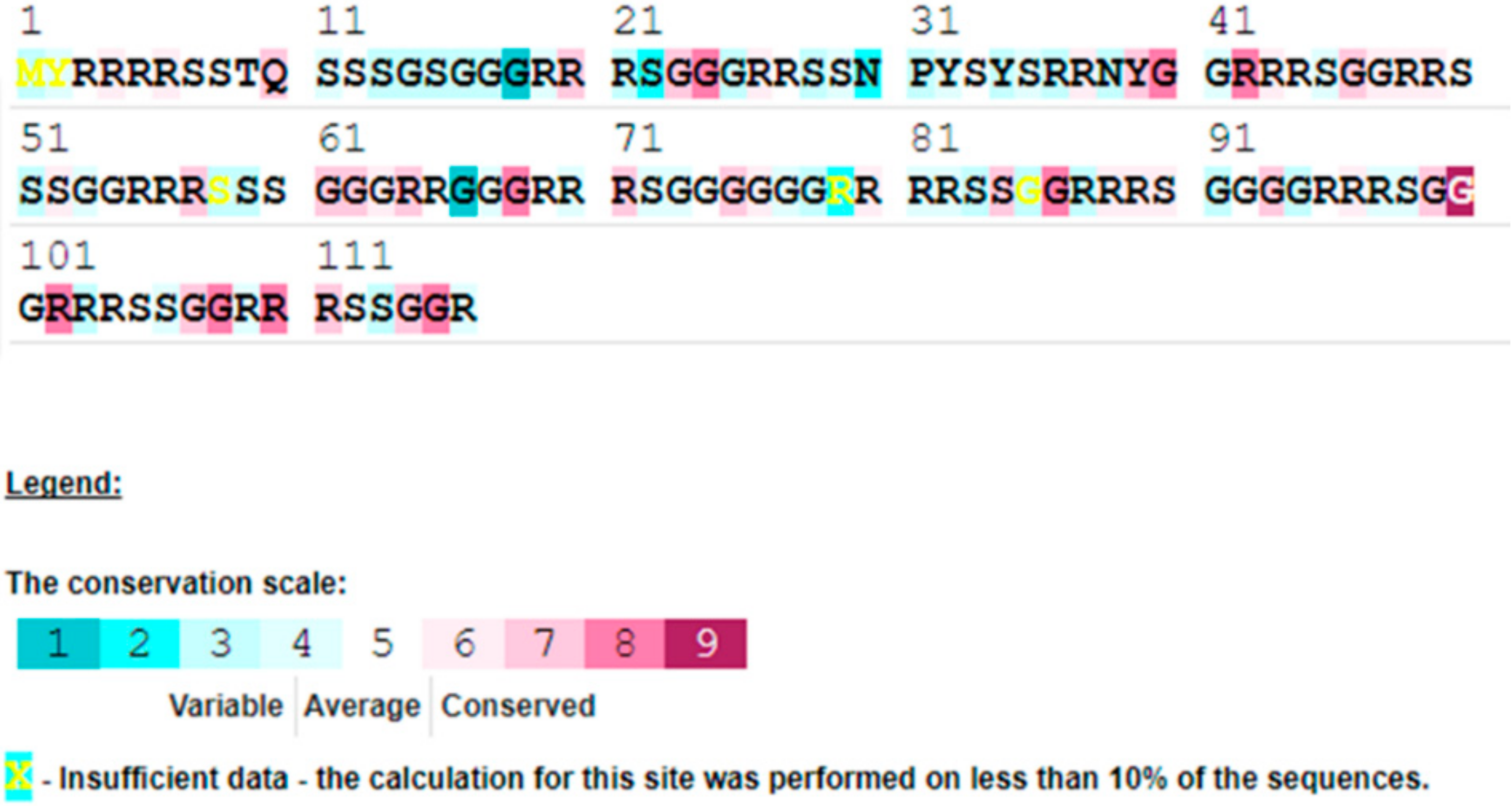
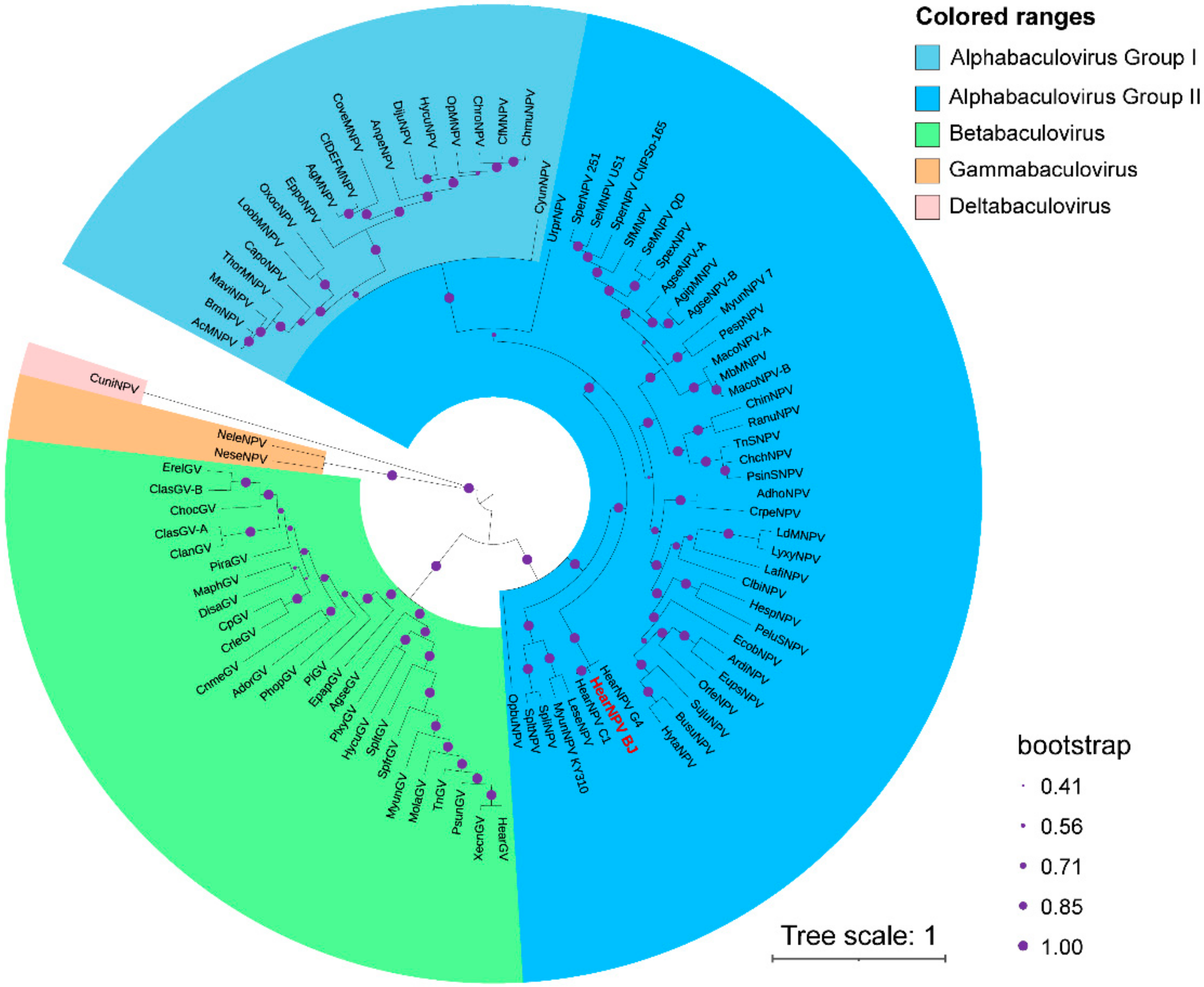
| ORF | From | To | Frame | Length (nt) | Length (aa) | Product | Function Domain Description and References |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 741 | + | 741 | 246 | polyhedrin | Polyhedrin; Provisional; PHA03389 |
| 2 | 1982 | 738 | − | 1245 | 414 | orf1236 | Wiskott Aldrich syndrome homology region 2 |
| 3 | 1997 | 2800 | + | 804 | 267 | pkinase | serine/threonine-protein kinase 1; Provisional; smart00246; Protein Kinases, catalytic domain; cl21453 |
| 4 | 5184 | 2923 | − | 2262 | 753 | hoar | --- |
| 5 | 5727 | 6584 | + | 858 | 285 | hypothetical protein | --- |
| 6 | 6949 | 7806 | + | 858 | 285 | ie-0 | Baculovirus immediate-early protein (IE-0); pfam05290 |
| 7 | 7823 | 9229 | + | 1407 | 468 | p49 * | Baculovirus Y142 protein; pfam04913 |
| 8 | 9240 | 9485 | + | 246 | 81 | odv-e18 * | Occlusion-derived virus envelope protein ODV-E18; pfam10717; Region: ODV-E18 |
| 9 | 9500 | 10,354 | + | 855 | 284 | odv-ec27 * | Aculovirus occlusion-derived virus envelope protein EC27; pfam05314 |
| 10 | 10,400 | 10,678 | + | 279 | 92 | hypothetical protein | Chitin-binding domain type 2; smart00494; Region: ChtBD2 |
| 11 | 11,310 | 10,705 | − | 606 | 201 | ep23 | Nucleopolyhedrovirus protein of unknown function (DUF884); pfam05959 |
| 12 | 11,352 | 13,355 | + | 2004 | 667 | ie-1 | Trans-activating transcriptional regulator; pfam03430 |
| 13 | 14,473 | 13,409 | − | 1065 | 354 | odv-e56 * | Baculoviral E56 protein, specific to ODV envelope; pfam04639 |
| 14 | 14,634 | 15,713 | + | 1080 | 359 | ORF16; me53 | Baculoviridae ME53; pfam06061 |
| 15 | 15,716 | 15,883 | + | 168 | 55 | hypothetical protein | --- |
| 16 | 15,936 | 16,217 | + | 282 | 93 | hypothetical protein | --- |
| 17 | 16,244 | 18,304 | + | 2061 | 686 | P74 * | Baculoviridae P74 N-terminal; pfam08404; Baculoviridae p74 conserved region; pfam04583 |
| 18 | 18,280 | 18,606 | + | 327 | 108 | unknown | --- |
| 19 | 19,507 | 18,704 | − | 804 | 267 | P26 | Nucleopolyhedrovirus p26 protein; pfam04766 |
| 20 | 20,462 | 19,899 | − | 564 | 187 | ORF23; lef-6 | --- |
| 21 | 21,447 | 20,476 | − | 972 | 323 | dbp | ssDNA binding protein; pfam04786 |
| 22 | 21,591 | 22,067 | + | 477 | 158 | hypothetical protein | Protein of unknown function (DUF424); pfam04242 |
| 23 | 25,062 | 24,295 | − | 768 | 255 | hypothetical protein | Protein of unknown function (DUF1247); pfam06851 |
| 24 | 24,902 | 25,153 | + | 252 | 83 | ubiquitin-like protein | --- |
| 25 | 25,217 | 25,723 | + | 507 | 168 | hypothetical protein | --- |
| 26 | 25,743 | 26,321 | + | 579 | 192 | el25 | --- |
| 27 | 27,318 | 26,380 | − | 939 | 312 | 39K Protein | Baculovirus 33KDa late protein (PP31); pfam05311 |
| 28 | 27,667 | 27,284 | − | 384 | 127 | lef11 | Baculovirus LEF-11 protein; pfam06385 |
| 29 | 28,352 | 27,636 | − | 717 | 238 | hypothetical protein | Nudix hydrolase |
| 30 | 28,583 | 29,662 | + | 1080 | 359 | unknown | --- |
| 31 | 30,975 | 29,737 | − | 1239 | 412 | p47 * | Viral transcription regulator p47; Provisional; PHA03391 |
| 32 | 31,048 | 31,719 | + | 672 | 223 | lef12 | Nucleopolyhedrovirus LEF-12 protein; pfam06256 |
| 33 | 31,805 | 32,047 | + | 243 | 80 | hypothetical protein | --- |
| 34 | 34,749 | 32,044 | − | 2706 | 901 | lef8 * | DNA-directed RNA polymerase subunit beta-like protein; Provisional; PHA03394 |
| 35 | 34,802 | 35,386 | + | 585 | 194 | hypothetical protein | RNA recognition motif (RRM) superfamily; cl17169 |
| 36 | 35,527 | 35,679 | + | 153 | 50 | hypothetical protein | --- |
| 37 | 37,456 | 35,687 | − | 1770 | 589 | chitinase | Early set domain associated with the catalytic domain of sugar utilizing enzymes at either the N or C terminus; cl09101; Glyco_18 domain; smart00636 |
| 38 | 38,042 | 37,500 | − | 543 | 180 | hypothetical protein | Protein of unknown function (DUF2616); pfam11077 |
| 39 | 38,161 | 38,571 | + | 411 | 136 | Ac53 * | Baculovirus U-box/Ring-like domain; pfam05883 |
| 40 | 39,714 | 38,578 | − | 1137 | 378 | hypothetical protein | Bacterial protein of unknown function (DUF853) |
| 41 | 39,722 | 39,949 | + | 228 | 75 | hypothetical protein | --- |
| 42 | 39,909 | 40,124 | + | 216 | 71 | lef10 | Late expression factor |
| 43 | 39,997 | 41,052 | + | 1056 | 351 | vp1054 * | Baculovirus VP1054 protein; pfam05789 |
| 44 | 41,172 | 41,378 | + | 207 | 68 | hypothetical protein | --- |
| 45 | 41,379 | 41,519 | + | 141 | 46 | hypothetical protein | --- |
| 46 | 41,857 | 42,222 | + | 366 | 121 | unknown | Nucleopolyhedrovirus protein of unknown function (DUF918); pfam06033 |
| 47 | 42,909 | 42,427 | − | 483 | 160 | hypothetical protein | ChaB; cl01887; Region: ChaB |
| 48 | 42,921 | 43,190 | + | 270 | 89 | hypothetical protein | ChaB; pfam06150; Region: ChaB |
| 49 | 44,056 | 43,402 | − | 654 | 217 | 25K FP protein frameshift | Baculovirus FP protein; pfam03258 |
| 50 | 44,522 | 46,081 | + | 1560 | 519 | lef9 * | Late expression factor 9; Provisional; PHA03396 |
| 51 | 47,244 | 46,141 | − | 1104 | 367 | cathepsin-like cysteine proteinase | Cathepsin propeptide inhibitor domain (I29); smart00848; Papain family cysteine protease; pfam00112 |
| 52 | 47,896 | 47,309 | − | 588 | 195 | hypothetical protein | Virulence factor Mce family protein; TIGR00996 |
| 53 | 48,806 | 47,967 | − | 840 | 279 | glycoprotein GP37 | pherodin-like protein; Provisional; PHA03387 |
| 54 | 49,957 | 51,030 | + | 1074 | 357 | bro-a | BRO family, N-terminal domain; pfam02498 |
| 55 | 51,785 | 52,495 | + | 711 | 236 | he65 | --- |
| 56 | 53,323 | 52,570 | − | 753 | 250 | iap-2 | Baculoviral inhibition of apoptosis protein repeat domain; cd00022; RING finger; cd16713 |
| 57 | 54,195 | 53,371 | − | 825 | 274 | hypothetical protein | FtsJ-like methyltransferase; pfam01728 |
| 58 | 54,565 | 54,164 | − | 402 | 133 | Ac68 * | Protein of unknown function (DUF708); pfam05341 |
| 59 | 54,585 | 55,724 | + | 1140 | 379 | lef3 | Nucleopolyhedrovirus late expression factor 3(LEF-3); pfam05847 |
| 60 | 58,190 | 55,833 | − | 2358 | 785 | Desmop * | Viral Desmoplakin N-terminus; pfam06771; Domain of unknown function (DUF4200); pfam13863 |
| 61 | 58,221 | 61,283 | + | 3063 | 1020 | DNA polymerase * | DNA polymerase type-B family; smart00486; DnaQ-like (or DEDD) 3’-5’ exonuclease domain superfamily; cl10012 |
| 62 | 61,818 | 61,360 | − | 459 | 152 | hypothetical protein | --- |
| 63 | 62,267 | 61,884 | − | 384 | 127 | hypothetical protein | Protein of unknown function (DUF1160); pfam06648 |
| 64 | 62,273 | 62,530 | 258 | 85 | hypothetical protein | --- | |
| 65 | 63,809 | 62,571 | − | 1239 | 412 | vlf-1 * | Very late expression factor 1; Provisional; PHA03397; DNA breaking-rejoining enzymes, C-terminal catalytic domain; cl00213 |
| 66 | 64,154 | 63,822 | − | 333 | 110 | Ac78 * | Nucleopolyhedrovirus protein of unknown function (DUF912); pfam06024 |
| 67 | 65,191 | 64,223 | − | 969 | 322 | P40/Gp41 * | Tructural glycoprotein p40/gp41 conserved region |
| 68 | 65,121 | 65,846 | + | 726 | 241 | Ac81 * | --- |
| 69 | 66,396 | 65,719 | − | 678 | 225 | hypothetical protein | Telokin-like protein-20 (TLP-20) domain; cd00235 |
| 70 | 66,326 | 68,776 | + | 2451 | 816 | VP91 capsid protein * | Viral capsid protein 91 N-terminal; pfam08475; Chitin-binding domain type 2; smart00494 |
| 71 | 69,755 | 68,904 | − | 852 | 283 | CG30 | Chromosome segregation ATPase; COG1196 |
| 72 | 70,725 | 69,844 | − | 882 | 293 | vp39 capsid * | Baculovirus major capsid protein VP39; pfam04501 |
| 73 | 70,724 | 72,109 | + | 1386 | 461 | lef-4 * | Late expression factor 4 (LEF-4); pfam05098 |
| 74 | 72,926 | 72,162 | − | 765 | 254 | P33 * | Baculovirus P33; pfam05214 |
| 75 | 72,928 | 73,416 | + | 489 | 162 | P18 * | Protein of unknown function (DUF682); pfam05081 |
| 76 | 73,462 | 74,154 | + | 693 | 230 | odv-e25 * | Occlusion-derived virus envelope protein E25; pfam05274 |
| 77 | 74,683 | 74,186 | − | 498 | 165 | hypothetical protein | Chitin-binding domain type 2; smart00494 |
| 78 | 78,463 | 74,702 | − | 3762 | 1253 | helicase * | Baculovirus DNA helicase; pfam04735 |
| 79 | 78,420 | 78,941 | + | 522 | 173 | Ac96 * | Baculovirus 19 kDa protein conserved region; pfam04798 |
| 80 | 79,965 | 79,000 | − | 966 | 321 | 38k * | Viral phosphatase superfamily protein; Provisional |
| 81 | 79,861 | 80,808 | + | 948 | 315 | lef-5 * | Baculoviridae late expression factor 5; pfam04838; Baculoviridae late expression factor 5 C-terminal domain; pfam11792 |
| 82 | 81,152 | 80,802 | − | 351 | 116 | p6.9 protein * | --- |
| 83 | 82,326 | 81,217 | − | 1110 | 369 | P40 * | Baculovirus protein of unknown function (DUF844); pfam05815 |
| 84 | 82,740 | 82,372 | − | 369 | 122 | hypothetical protein | Protein of unknown function (DUF1098); pfam06497 |
| 85 | 83,873 | 82,740 | − | 1134 | 377 | P48 * | Baculovirus P48 protein; pfam04878 |
| 86 | 83,969 | 85,786 | + | 1818 | 605 | capsid-associated protein VP80 | Nucleopolyhedrovirus capsid protein P87; pfam07267 |
| 87 | 85,974 | 87,059 | + | 1086 | 361 | odv-ec43 * | Protein of unknown function (DUF673); pfam05054 |
| 88 | 87,105 | 87,389 | + | 285 | 94 | hypothetical protein | --- |
| 89 | 89,474 | 87,456 | − | 2019 | 672 | ORF99; odv-e66; HaORF96 | Occlusion-derived virus envelope protein E66 |
| 90 | 90,325 | 89,495 | − | 831 | 276 | hypothetical protein | Glycosyltransferase family A (GT-A) includes diverse families of glycosyl transferases with a common GT-A type structural fold; cl11394 |
| 91 | 92,636 | 93,235 | + | 600 | 199 | Pif-3 * | per os infectivity factor 3; Provisional; PHA03399 |
| 92 | 93,239 | 93,595 | + | 357 | 118 | ORF101 | --- |
| 93 | 93,691 | 95,223 | + | 1533 | 510 | hypothetical protein | Poly (ADP-ribose) glycohydrolase (PARG); pfam05028 |
| 94 | 95,302 | 96,063 | + | 762 | 253 | hypothetical protein | Baculovirus protein of unknown function (DUF816) |
| 95 | 96,078 | 96,410 | + | 333 | 110 | hypothetical protein | --- |
| 96 | 97,274 | 96,468 | − | 807 | 268 | iap-3 | Baculoviral inhibition of apoptosis protein repeat domain; cd00022 |
| 97 | 99,042 | 97,537 | − | 1506 | 501 | bro-c | BRO family, N-terminal domain; pfam02498 |
| 98 | 99,210 | 99,689 | + | 480 | 159 | sod | Copper/zinc superoxide dismutase (SODC); pfam00080 |
| 99 | 99,696 | 101,069 | + | 1374 | 457 | hypothetical protein | --- |
| 100 | 101,700 | 101,122 | − | 579 | 192 | hypothetical protein | --- |
| 101 | 101,870 | 102,226 | + | 357 | 118 | hypothetical protein | --- |
| 102 | 102,237 | 102,503 | + | 267 | 88 | hypothetical protein | --- |
| 103 | 102,571 | 104,157 | + | 1587 | 528 | Pif-1 * | Per os infectivity; pfam05092 |
| 104 | 105,318 | 104,413 | − | 906 | 301 | hypothetical protein | Acidic and basic fibroblast growth factor family; FGFs are mitogens; cd00058 |
| 105 | 106,725 | 105,445 | − | 1281 | 426 | alkaline exonuclease * | Inhibitor of Apoptosis domain; pfam00653 |
| 106 | 107,134 | 106,745 | − | 390 | 129 | hypothetical protein | Protein of unknown function (DUF1477); pfam07346 |
| 107 | 109,422 | 108,496 | − | 927 | 308 | unknown | --- |
| 108 | 109,621 | 109,836 | + | 216 | 71 | hypothetical protein | --- |
| 109 | 110,679 | 109,954 | − | 726 | 241 | lef2 * | lef-2; pfam03041 |
| 110 | 110,950 | 110,546 | − | 405 | 134 | unknown | --- |
| 111 | 111,041 | 111,787 | + | 747 | 248 | p24 capsid | Baculovirus P24 capsid protein; pfam05073 |
| 112 | 111,849 | 112,139 | + | 291 | 96 | gp16 | --- |
| 113 | 112,191 | 113,213 | + | 1023 | 340 | polyhedrin envelope protein | Baculovirus polyhedron envelope protein, PEP, N terminus; pfam04512; Baculovirus polyhedron envelope protein, PEP, C terminus; pfam04513 |
| 114 | 113,292 | 113,756 | + | 465 | 154 | hypothetical protein | --- |
| 115 | 113,886 | 114,476 | + | 591 | 196 | odv-c21 | --- |
| 116 | 115,689 | 114,520 | − | 1170 | 389 | 38.7kD protein | BRO family, N-terminal domain; pfam02498; Protein of unknown function (DUF3627); pfam12299 |
| 117 | 116,428 | 115,691 | − | 738 | 245 | lef1 * | Eukaryotic and archaeal DNA primase small subunit; pfam01896 |
| 118 | 116,831 | 116,403 | − | 429 | 142 | hypothetical protein | --- |
| 119 | 116,976 | 118,523 | + | 1548 | 515 | egt | Ecdysteroid UDP-glucosyltransferase; Provisional; PHA03392; UDP-glucoronosyl and UDP-glucosyl transferase; pfam00201 |
| 120 | 118,723 | 119,301 | + | 579 | 192 | hypothetical protein | --- |
| 121 | 119,252 | 120,052 | + | 801 | 266 | bv-ec31 | Protein of unknown function (DUF1251); pfam06856 |
| 122 | 122,976 | 120,133 | − | 2844 | 947 | hypothetical protein | --- |
| 123 | 123,382 | 123,891 | + | 510 | 169 | pkip-1 | Pkip-1 protein; pfam06878 |
| 124 | 124,755 | 123,958 | − | 798 | 265 | arif1 | Actin-rearrangement-inducing factor (Arif-1); pfam06770 |
| 125 | 125,016 | 126,164 | + | 1149 | 382 | Pif-2 * | Baculovirus hypothetical protein; pfam04631 |
| 126 | 128,238 | 126,205 | − | 2034 | 677 | fusion prot‘ein | Protein of unknown function (DUF3609); pfam12259 |
| 127 | 128,925 | 128,380 | − | 546 | 181 | hypothetical protein | --- |
| 128 | 129,107 | 129,694 | − | 588 | 195 | hypothetical protein | --- |
| lef8-lef9-polh | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| HearNPV-BJ | - | 0.00542 | 0.00140 | 0.00340 | 0.00946 | 0.44794 |
| HearNPV-Au | 0.00542 | - | 0.00441 | 0.00240 | 0.00521 | 0.44516 |
| HearNPV-C1 | 0.00140 | 0.00441 | - | 0.00200 | 0.00966 | 0.44835 |
| HearNPV-G4 | 0.00340 | 0.00240 | 0.00200 | - | 0.00763 | 0.44676 |
| HzSNPV | 0.00946 | 0.00521 | 0.00966 | 0.00763 | - | 0.44509 |
| AcMNPV | 0.44794 | 0.44516 | 0.44835 | 0.44676 | 0.44509 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Liu, X.; Tang, K.; Zhang, Z.; Zhang, H.; Li, Y. A Renewed Appreciation of Helicoverpa armigera Nucleopolyhedrovirus BJ (Formerly Helicoverpa assulta Nucleopolyhedrovirus) with Whole Genome Sequencing. Viruses 2022, 14, 618. https://doi.org/10.3390/v14030618
Zhao L, Liu X, Tang K, Zhang Z, Zhang H, Li Y. A Renewed Appreciation of Helicoverpa armigera Nucleopolyhedrovirus BJ (Formerly Helicoverpa assulta Nucleopolyhedrovirus) with Whole Genome Sequencing. Viruses. 2022; 14(3):618. https://doi.org/10.3390/v14030618
Chicago/Turabian StyleZhao, Lulu, Xingjian Liu, Kai Tang, Zhifang Zhang, Huan Zhang, and Yinü Li. 2022. "A Renewed Appreciation of Helicoverpa armigera Nucleopolyhedrovirus BJ (Formerly Helicoverpa assulta Nucleopolyhedrovirus) with Whole Genome Sequencing" Viruses 14, no. 3: 618. https://doi.org/10.3390/v14030618
APA StyleZhao, L., Liu, X., Tang, K., Zhang, Z., Zhang, H., & Li, Y. (2022). A Renewed Appreciation of Helicoverpa armigera Nucleopolyhedrovirus BJ (Formerly Helicoverpa assulta Nucleopolyhedrovirus) with Whole Genome Sequencing. Viruses, 14(3), 618. https://doi.org/10.3390/v14030618








