The Characterization of a Novel Virus Discovered in the Yeast Pichia membranifaciens
Abstract
1. Introduction
2. Methods
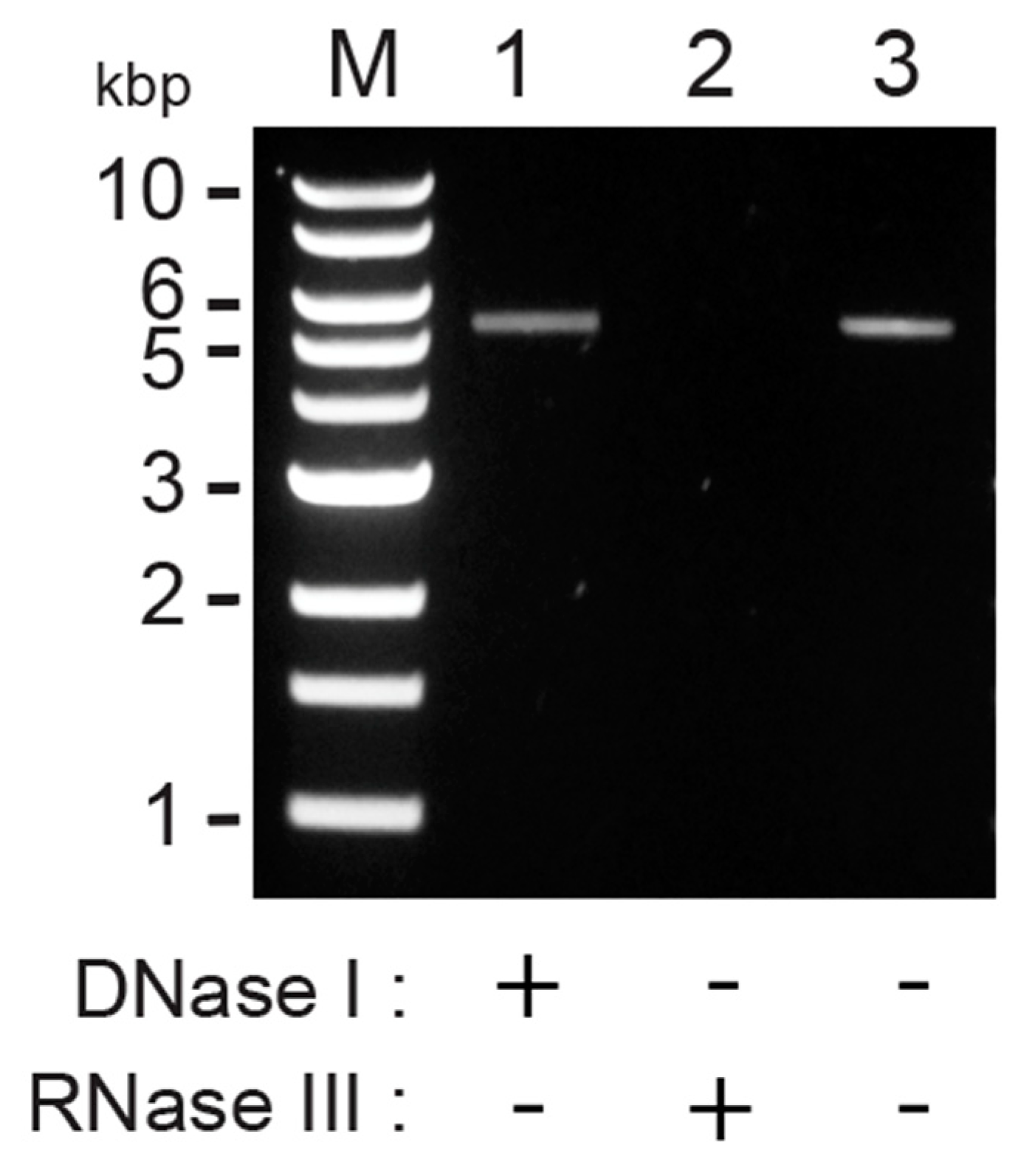
2.1. Purification and Digestion of dsRNAs with RNase III
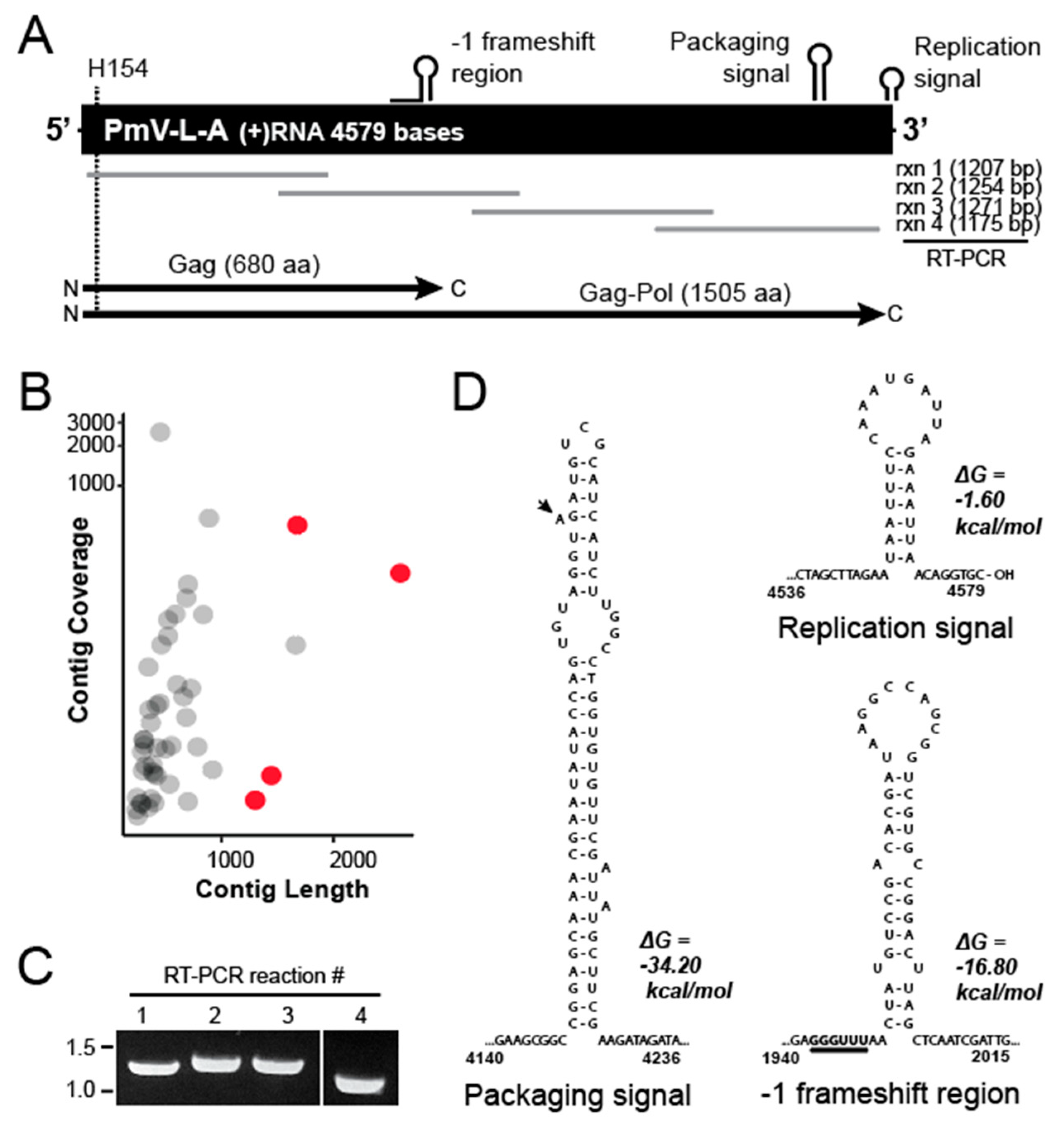
2.2. Determining the Genetic Sequence of dsRNAs
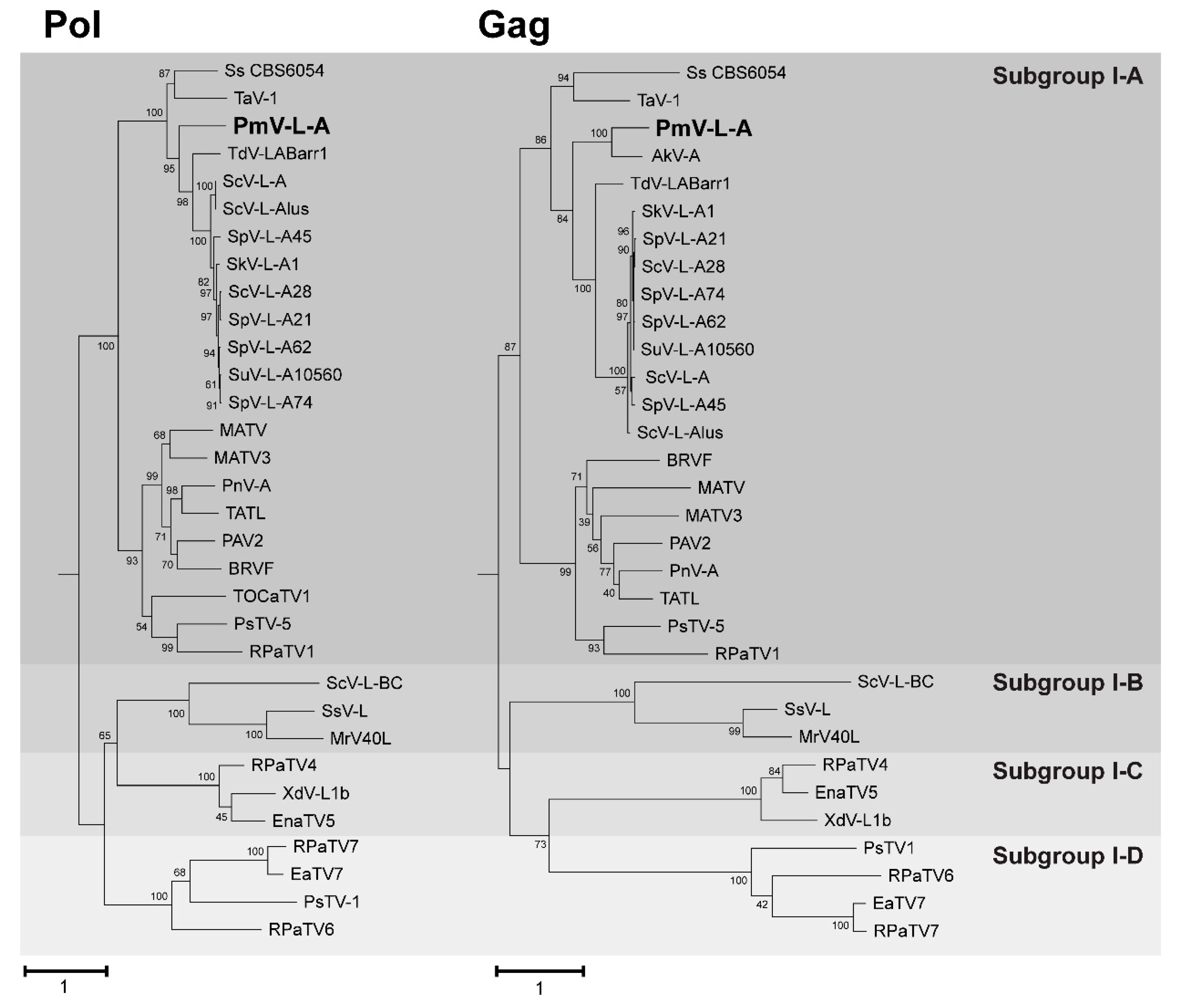
2.3. Phylogenetic Analysis of PmV-L-A
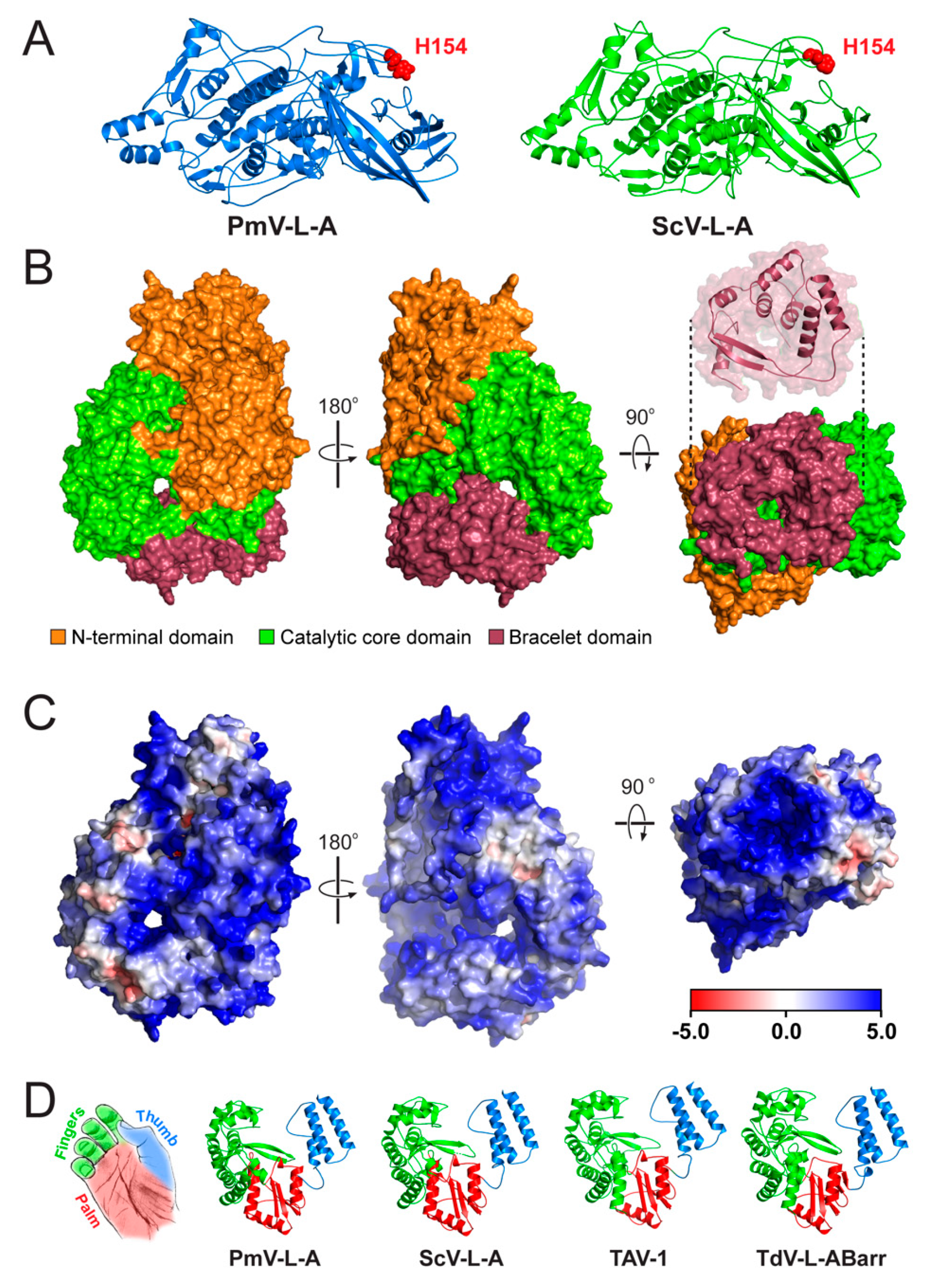
2.4. Molecular Modeling of the Gag and Pol Proteins of PmV-L-A
3. Results
3.1. Purification and Digestion of Double-Stranded RNAs from P. membranifaciens
3.2. Determining the Nucleic Acid Sequence of the dsRNA from P. membranifaciens
3.3. Phylogenetic Analysis of PmV-L-A
3.4. Structural Modeling of the Gag and Pol Proteins of PmV-L-A
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kurtzman, C.P. Pichia E.C. Hansen (1904). In The Yeast: A Taxonomic Study; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: Burlington, VT, USA, 2011; pp. 685–707. ISBN 9780080931272. [Google Scholar]
- Zhang, J.; Xie, J.; Zhou, Y.; Deng, L.; Yao, S.; Zeng, K. Inhibitory effect of Pichia membranaefaciens and Kloeckera apiculata against Monilinia fructicola and their biocontrol ability of brown rot in postharvest plum. Biol. Control 2017, 114, 51–58. [Google Scholar] [CrossRef]
- Santos, A.; Mauro, M.S.; Bravo, E.; Marquina, D. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 2009, 155, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Marquina, D. Killer toxin of Pichia membranifaciens and its possible use as a biocontrol agent against grey mould disease of grapevine. Microbiology 2004, 150, 2527–2534. [Google Scholar] [CrossRef] [PubMed]
- Melchor, R.L.A.; Rosales, V.G.; Pérez, M.C.G.; Fernández, S.P.; Álvarez, G.O.; Mastache, J.M.N. Effectiveness of carboxylic acids from Pichia membranifaciens against coffee rust. Ciência E Agrotecnologia 2018, 42, 42–50. [Google Scholar] [CrossRef]
- Xu, X.; Chan, Z.; Xu, Y.; Tian, S. Effect of Pichia membranaefaciens combined with salicylic acid on controlling brown rot in peach fruit and the mechanisms involved. J. Sci. Food Agric. 2008, 88, 1786–1793. [Google Scholar] [CrossRef]
- Xu, X.; Tian, S. Reducing oxidative stress in sweet cherry fruit by Pichia membranaefaciens: A possible mode of action against Penicillium expansum. J. Appl. Microbiol. 2008, 105, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Qing, F.; Shiping, T. Postharvest Biological Control of Rhizopus Rot of Nectarine Fruits by Pichia membranefaciens. Plant Dis. 2000, 84, 1212–1216. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Ruiz, J.; Alonso, A.; Marquina, D.; Santos, A. The Biology of Pichia membranifaciens Killer Toxins. Toxins 2017, 9, 112. [Google Scholar] [CrossRef]
- Masih, E.I.; Paul, B. Secretion of β-1,3-Glucanases by the Yeast Pichia membranifaciens and Its Possible Role in the Biocontrol of Botrytis cinerea Causing Grey Mold Disease of the Grapevine. Curr. Microbiol. 2002, 44, 391–395. [Google Scholar] [CrossRef]
- Zhang, H.; Du, H.; Xu, Y. Volatile Organic Compound-Mediated Antifungal Activity of Pichia spp. and Its Effect on the Metabolic Profiles of Fermentation Communities. Appl. Environ. Microbiol. 2021, 87, e02992-20. [Google Scholar] [CrossRef] [PubMed]
- Young, T.W.; Yagiu, M. A comparison of the killer character in different yeasts and its classification. Antonie van Leeuwenhoek 1978, 44, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, L.R.; Lee, M.D.; Crabtree, A.M.; Boyer, J.M.; Kizer, E.A.; Taggart, N.T.; Roslund, C.R.; Hunter, S.S.; Kennedy, C.B.; Willmore, C.G.; et al. The Species-Specific Acquisition and Diversification of a K1-like Family of Killer Toxins in Budding Yeasts of the Saccharomycotina. PLoS Genet. 2021, 17, e1009341. [Google Scholar] [CrossRef] [PubMed]
- Ghabrial, S.A.; Caston, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jiang, D. New Insights into Mycoviruses and Exploration for the Biological Control of Crop Fungal Diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Hypovirulence: Mycoviruses at the fungal–plant interface. Nat. Rev. Genet. 2005, 3, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Tarr, P.I.; Aline, R.F.; Smiley, B.L.; Scholler, J.; Keithly, J.; Stuart, K. LR1: A candidate RNA virus of Leishmania. Proc. Natl. Acad. Sci. USA 1988, 85, 9572–9575. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Ghabrial, S.A.; Fichorova, R.N.; Nibert, M.L. Trichomonasvirus: A new genus of protozoan viruses in the family Totiviridae. Arch. Virol. 2010, 156, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.L.; Yang, H.M.; Shen, K.A.; Wang, C.C. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc. Natl. Acad. Sci. USA 1993, 90, 8595–8599. [Google Scholar] [CrossRef]
- Herring, A.J.; Bevan, E.A. Virus-like Particles Associated with the Double-stranded RNA Species Found in Killer and Sensitive Strains of the Yeast Saccharomyces cerevisiae. J. Gen. Virol. 1974, 22, 387–394. [Google Scholar] [CrossRef]
- Huang, S.H.; Ghabrial, S.A. Organization and Expression of the Double-Stranded RNA Genome of Hel-minthosporium victoriae 190S Virus, a Totivirus Infecting a Plant Pathogenic Filamentous Fungus. Proc. Natl. Acad. Sci. USA 1996, 93, 12541–12546. [Google Scholar] [CrossRef]
- Shao, Q.; Jia, X.; Gao, Y.; Liu, Z.; Zhang, H.; Tan, Q.; Zhang, X.; Zhou, H.; Li, Y.; Wu, D.; et al. Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission. PLoS Pathog. 2021, 17, e1009396. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ochoa, W.F.; Sinkovits, R.S.; Poulos, B.T.; Ghabrial, S.A.; Lightner, D.V.; Baker, T.S.; Nibert, M.L. Infectious myonecrosis virus has a totivirus-like, 120-subunit capsid, but with fiber complexes at the fivefold axes. Proc. Natl. Acad. Sci. USA 2008, 105, 17526–17531. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.A.; Bevan, E.A. A New Species of Double-stranded RNA from Yeast. Nature 1972, 239, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; Martínez, A. Genome Organization of a New Double-Stranded RNA LA Helper Virus From Wine Torulaspora delbrueckii Killer Yeast as Compared with Its Saccharomyces Counterparts. Front. Microbiol. 2020, 11, 593846. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Gómez, P.; Esteban, R. Variation and Distribution of L-A Helper Totiviruses in Saccharomyces sensu stricto Yeasts Producing Different Killer Toxins. Toxins 2017, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Rowley, P.A.; Ho, B.; Bushong, S.; Johnson, A.; Sawyer, S.L. XRN1 Is a Species-Specific Virus Restriction Factor in Yeasts. PLoS Pathog. 2016, 12, e1005890. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, T.; Esteban, R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 2011, 108, 17667–17671. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.; Icho, T.; Wickner, R.B. A-1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. USA 1991, 88, 174–178. [Google Scholar] [CrossRef]
- Esteban, R.; Wickner, R.B. A deletion mutant of L-A double-stranded RNA replicates like M1 double-stranded RNA. J. Virol. 1988, 62, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, T.; Ribas, J.; Makhov, A.M.; Wickner, R.B. Pol of gag–pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature 1992, 359, 746–749. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Hisano, S.; Chiba, S.; Maruyama, K.; Andika, I.B.; Toyoda, K.; Fujimori, F.; Suzuki, N. Sequence and phylogenetic analyses of novel totivirus-like double-stranded RNAs from field-collected powdery mildew fungi. Virus Res. 2016, 213, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.S.; Brown, C.J.; Ytreberg, F.M.; Stenkamp, D.L. Predicting peak spectral sensitivities of vertebrate cone visual pigments using atomistic molecular simulations. PLoS Comput. Biol. 2018, 14, e1005974. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Blanc, A.; Ribas, J.C.; Wickner, R.B. His-154 Is Involved in the Linkage of the Saccharomyces cerevisiae LA Double-Stranded RNA Virus Gag Protein to the Cap Structure of mRNAs and Is Essential for M1 Satellite Virus Expression. Mol. Cell. Biol. 1994, 14, 2664–2674. [Google Scholar] [PubMed]
- Fujimura, T.; Esteban, R. Recognition of RNA Encapsidation Signal by the Yeast L-A Double-stranded RNA Virus. J. Biol. Chem. 2000, 275, 37118–37126. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Shen, X.-X.; Opulente, D.A.; Kominek, J.; Zhou, X.; Steenwyk, J.L.; Buh, K.V.; Haase, M.; Wisecaver, J.H.; Wang, M.; Doering, D.; et al. Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell 2018, 175, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Naitow, H.; Tang, J.; Canady, M.; Wickner, R.B.; Johnson, J.E. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat. Genet. 2002, 9, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; McDonald, S.M.; Tortorici, M.A.; Tao, Y.J.; Carpio, R.V.-D.; Nibert, M.L.; Patton, J.; Harrison, S.C. Mechanism for Coordinated RNA Packaging and Genome Replication by Rotavirus Polymerase VP1. Structure 2008, 16, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.M.; Tao, Y.J.; Patton, J.T. The ins and outs of four-tunneled Reoviridae RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2009, 19, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Routhier, E.; Bruenn, J. Functions of Conserved Motifs in the RNA-Dependent RNA Polymerase of a Yeast Double-Stranded RNA Virus. J. Virol. 1998, 72, 4427–4429. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Fink, G.R.; Bartel, D.P. Compatibility with Killer Explains the Rise of RNAi-Deficient Fungi. Science 2011, 333, 1592. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in Budding Yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Segers, G.C.; van Wezel, R.; Zhang, X.; Hong, Y.; Nuss, D.L. Hypovirus Papain-Like Protease p29 Suppresses RNA Silencing in the Natural Fungal Host and in a Heterologous Plant System. Eukaryot. Cell 2006, 5, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.M.; Andrewski, M.D.; Roossinck, M.J.; Keller, N.P. Aspergillus Mycoviruses Are Targets and Suppressors of RNA Silencing. Eukaryot. Cell 2008, 7, 350–357. [Google Scholar] [CrossRef]
- Kubota, K.; Ng, J.C.K. Lettuce chlorosis virus P23 Suppresses RNA Silencing and Induces Local Necrosis with Increased Severity at Raised Temperatures. Phytopathology 2016, 106, 653–662. [Google Scholar] [CrossRef][Green Version]
- Frank, A.C.; Wolfe, K. Evolutionary Capture of Viral and Plasmid DNA by Yeast Nuclear Chromosomes. Eukaryot. Cell 2009, 8, 1521–1531. [Google Scholar] [CrossRef]
- Khalifa, M.; MacDiarmid, R.M. A Novel Totivirus Naturally Occurring in Two Different Fungal Genera. Front. Microbiol. 2019, 10, 2318. [Google Scholar] [CrossRef] [PubMed]
- Ribas, J.; Wickner, R.B. RNA-dependent RNA polymerase consensus sequence of the L-A double-stranded RNA virus: Definition of essential domains. Proc. Natl. Acad. Sci. USA 1992, 89, 2185–2189. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, R.; Shu, B.; Jing, X.; Ye, H.-Q.; Gong, P. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat. Commun. 2020, 11, 2605. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.D.; Creagh, J.W.; Fredericks, L.R.; Crabtree, A.M.; Patel, J.S.; Rowley, P.A. The Characterization of a Novel Virus Discovered in the Yeast Pichia membranifaciens. Viruses 2022, 14, 594. https://doi.org/10.3390/v14030594
Lee MD, Creagh JW, Fredericks LR, Crabtree AM, Patel JS, Rowley PA. The Characterization of a Novel Virus Discovered in the Yeast Pichia membranifaciens. Viruses. 2022; 14(3):594. https://doi.org/10.3390/v14030594
Chicago/Turabian StyleLee, Mark D., Jack W. Creagh, Lance R. Fredericks, Angela M. Crabtree, Jagdish Suresh Patel, and Paul A. Rowley. 2022. "The Characterization of a Novel Virus Discovered in the Yeast Pichia membranifaciens" Viruses 14, no. 3: 594. https://doi.org/10.3390/v14030594
APA StyleLee, M. D., Creagh, J. W., Fredericks, L. R., Crabtree, A. M., Patel, J. S., & Rowley, P. A. (2022). The Characterization of a Novel Virus Discovered in the Yeast Pichia membranifaciens. Viruses, 14(3), 594. https://doi.org/10.3390/v14030594





