Avian Influenza a H9N2 Viruses in Morocco, 2018–2019
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. Samples Processing
2.2.1. RNA Extraction and Real Time RT-PCR
2.2.2. Virus Isolation
2.2.3. Full Genome Amplification and Sequencing of H9N2 Moroccan Isolates
2.3. Sequences and Phylogenetic Analyses
2.4. Statistical Analysis
3. Results
3.1. Case History and H9N2 Detection
3.2. Vaccination Status
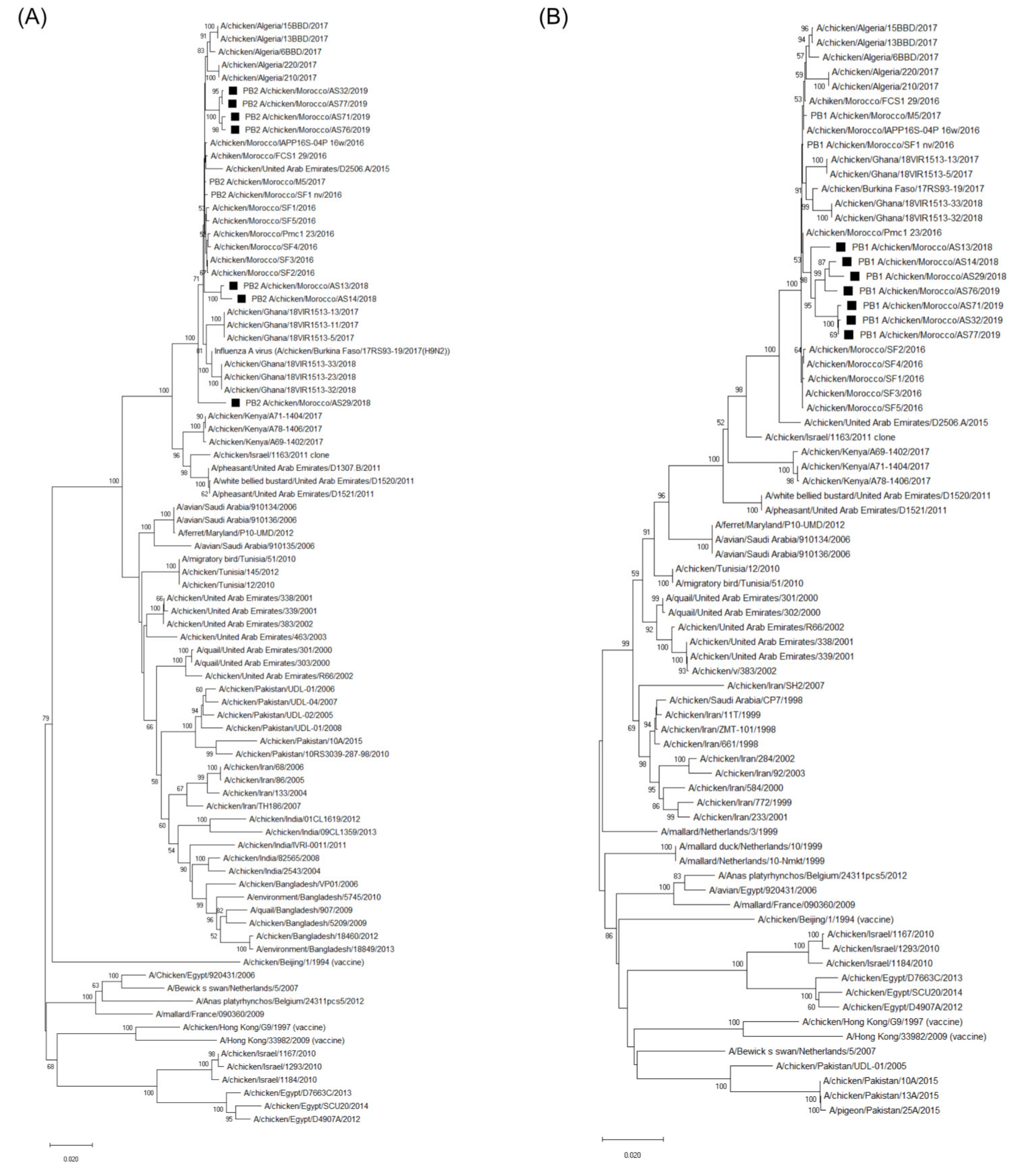
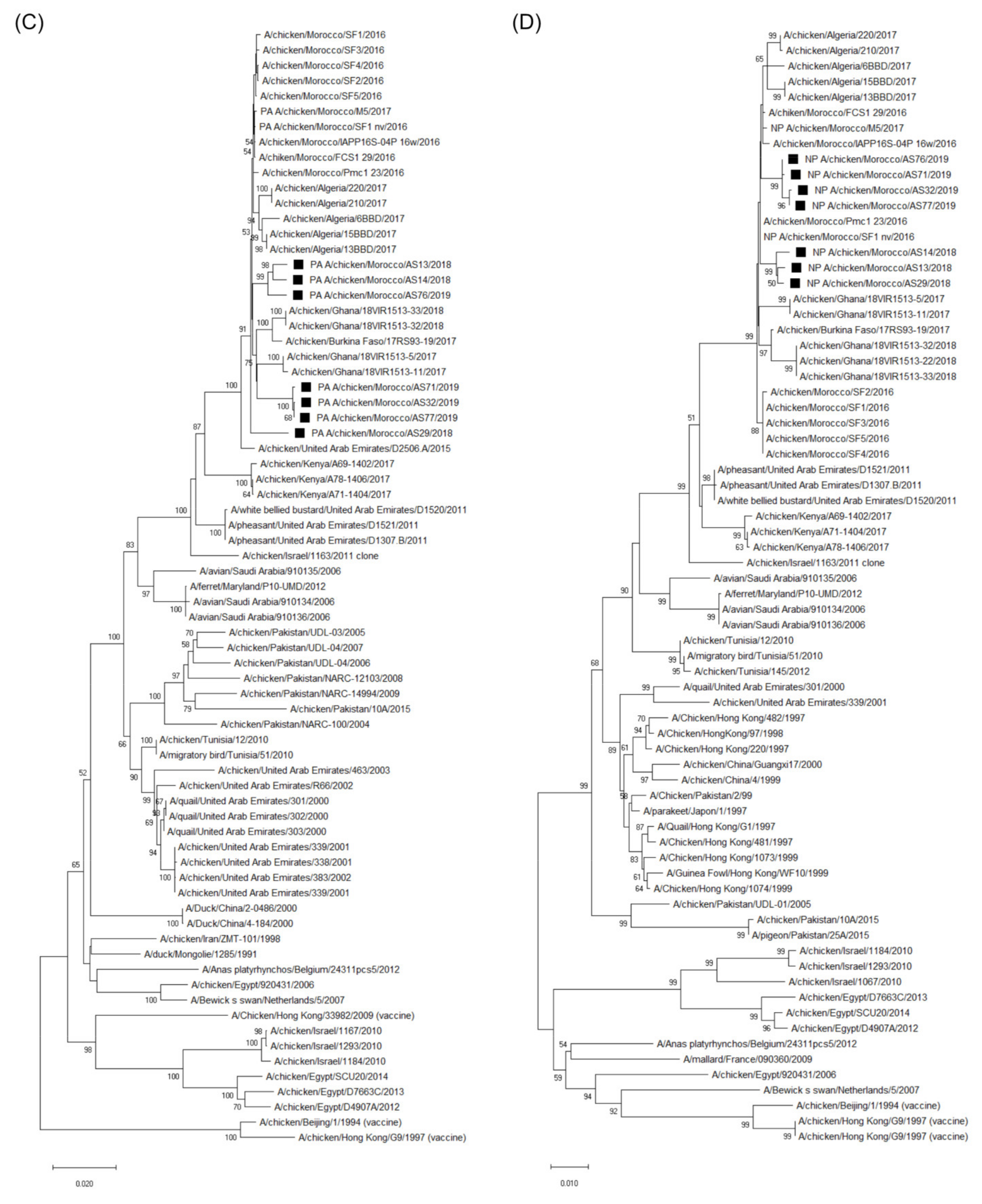
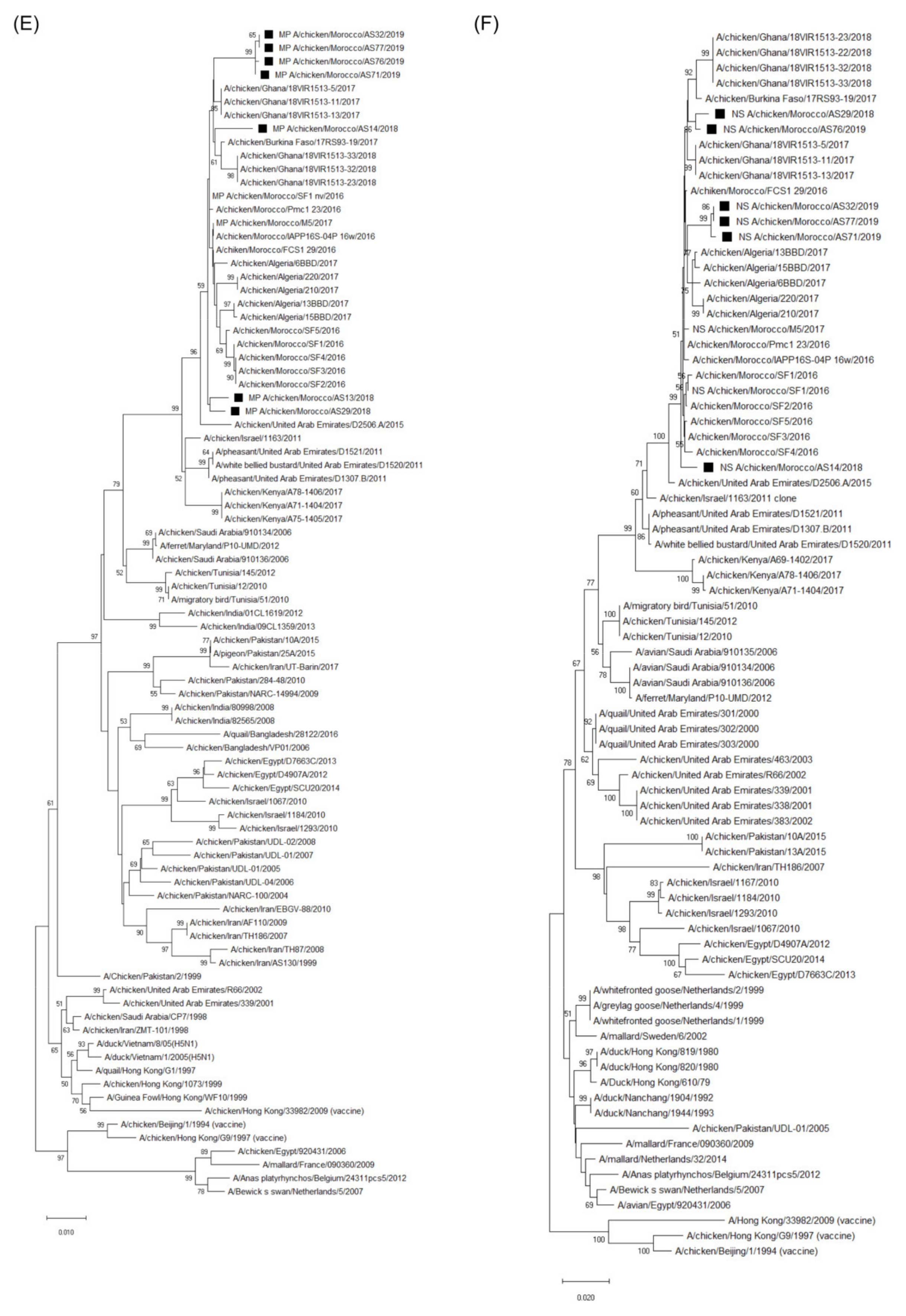
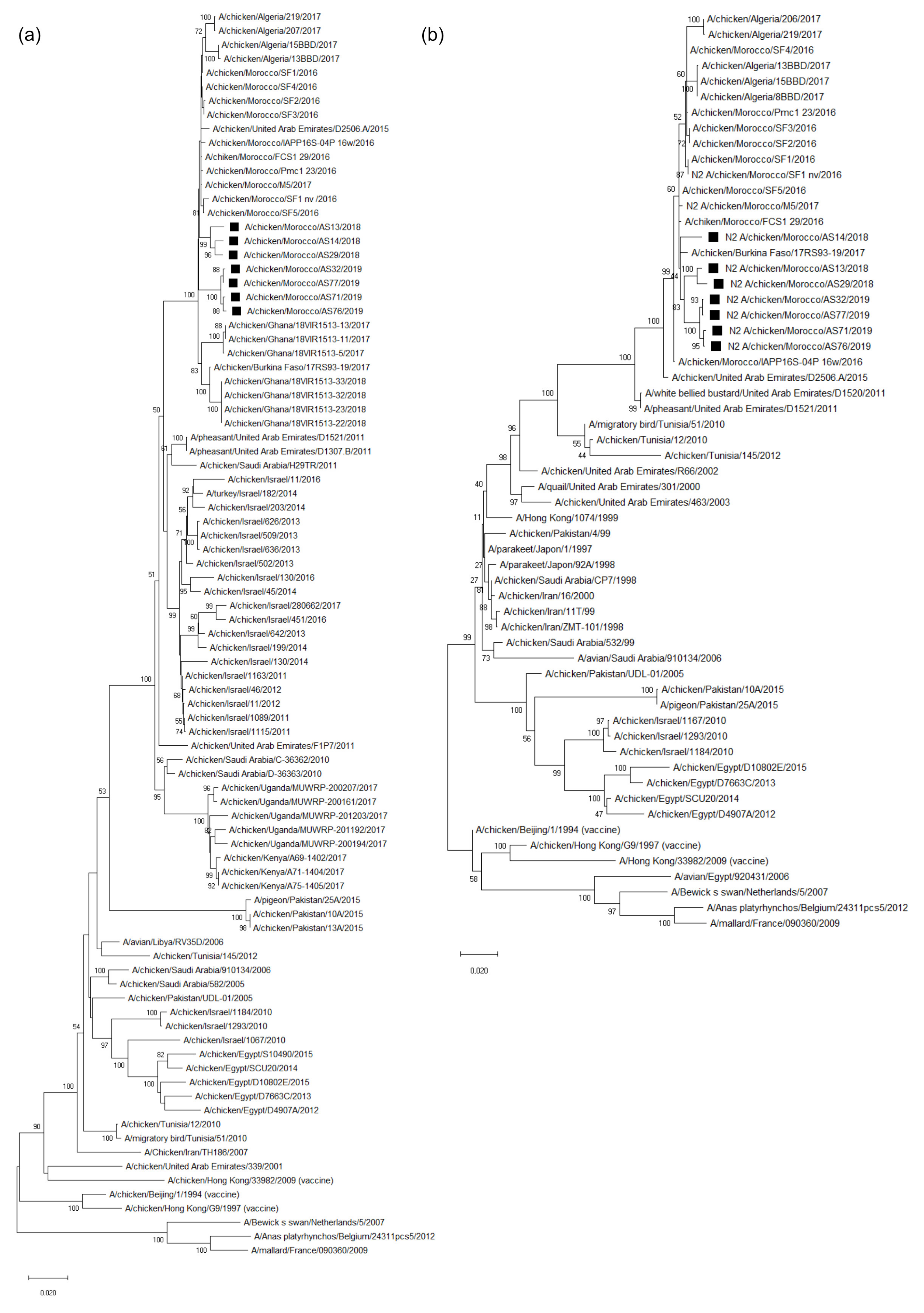
3.3. Molecular Characterization and Phylogenetic Analysis of the Eight Viral Segments
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Farm | Sampling Date | Location of Farm | Age of Birds | H9N2 Vaccination Status | H9N2 RT-qPCRCt Value | Farms Status |
|---|---|---|---|---|---|---|---|
| AS1 | F1 | 28/06/2018 | Rabat | 27D | V | 25 | P |
| AS2 | F2 | 16/07/2018 | Rabat | 46D | V | - | N |
| AS3 | 46D | - | |||||
| AS4 | 46D | - | |||||
| AS5 | F3 | 08/08/2018 | Oriental | 45D | NV | - | N |
| AS6 | 45D | - | |||||
| AS7 | F4 | 08/08/2018 | Oriental | 36D | NV | - | N |
| AS8 | F5 | 17/07/2018 | Casablanca | 28D | V | - | N |
| AS9 | F6 | 17/09/2018 | Temara | 43D | V | - | N |
| AS11 | F7 | 19/10/2018 | Meknes | 32D | NV | - | N |
| AS12B1 * | F8 | 23/10/2018 | Kenitra | 23D | NV | 20 | P |
| AS12B2 | 23/10/2018 | 23D | 33 | ||||
| AS13 *+ | F9 | 18/10/2018 | El hajeb | 32D | NV | 17 | P |
| AS14 *+ | F10 | 24/10/2018 | Meknes | 30D | NV | 18 | P |
| AS15 | F11 | 21/10/2018 | Meknes | 37D | NV | - | N |
| AS16 * | F12 | 17/10/2018 | Midelt | 40D | NV | 18 | P |
| AS17 | F13 | 11/12/2018 | Fes | 38D | NV | 26 | P |
| AS18 | F14 | 11/12/2018 | Fes | 35D | NV | 26 | P |
| AS19 | F15 | 11/12/2018 | Fes | 41D | NV | - | N |
| AS20 | F16 | 23/11/2018 | Fes | 32D | V | 25 | P |
| AS21 | F17 | 12/11/2018 | Salé | 36D | V | - | N |
| AS22 | F18 | 07/12/2019 | Meknes | 34D | NV | 22 | P |
| AS23 | 07/12/2019 | 34D | 26 | ||||
| AS24 | 07/12/2019 | 34D | 22 | ||||
| AS26 | F19 | 19/02/2019 | Benslimane | 30D | NV | - | N |
| AS27 | F20 | 20/02/2019 | Rabat | 24D | NV | - | N |
| AAS28 | F21 | 04/11/2018 | Ait brahim (Fes) | 32D | V | 24 | P |
| AS29 *+ | F22 | 16/11/2018 | Fes | 36D | V | 16 | P |
| AS30 | F23 | 26/01/2019 | Hajeb | 20D | V | 24 | P |
| AS31 | F24 | 11/02/2019 | Sefrou | 30–36D | V | 17 | P |
| AS32 *+ | F25 | 12/02/2019 | Ain chegag | 37D | V | 19 | P |
| AS33 * | F26 | 13/02/2019 | Zerarda tahla | 40D | NV | 16 | P |
| AS34 | F27 | 25/01/2019 | Khemisset | 33D | NV | - | N |
| AS35 | F28 | 04/03/2019 | Meknes | 32D | - | N | |
| AS36 | F29 | 13/02/2019 | Fes | 34D | 14 | P | |
| AS37 | F30 | 13/02/2019 | Fes | 37D | V | - | N |
| AS38 | F31 | 01/02/2019 | Meknes | 32D | NV | - | N |
| AS39 | F32 | 12/02/2019 | Hadeb | 34D | NV | - | N |
| AS40 | F33 | 12/02/2019 | Salé | 42D | NV | 12 | P |
| AS41 | F34 | 25/02/2019 | Khemisset | 44D | V | - | N |
| AS42 | F35 | 25/02/2019 | Meknes | 35D | NV | - | N |
| AS43 * | F36 | 07/02/2019 | Casablanca | 29D | V | 16 | P |
| AS44 | F37 | 07/02/2019 | Casablanca | 32D | V | 16 | P |
| AS45 * | F38 | 10/02/2019 | Rabat | 45D | V | 17 | P |
| AS46 | 10/02/2019 | 45D | 14 | ||||
| AS47 | F39 | 11/02/2019 | Tiflet | 28D | NV | - | N |
| AS54 | F40 | 20/02/2019 | Rabat | 32D | V | 24 | P |
| AS55 | F41 | 20/02/2019 | Oriental | 37D | NV | - | N |
| AS56 * | F42 | 20/02/2019 | Oriental | 38D | NV | 26 | P |
| AS57 | F43 | 21/02/2019 | Taza | 29D | NV | 26 | P |
| AS58 | F44 | 21/02/2019 | Tahla | 36D | NV | - | P |
| AS59 | 21/02/2019 | 36D | 26 | ||||
| AS60 | F45 | 21/02/2019 | Tahla | 39D | NV | - | N |
| AS61 * | F46 | 24/02/2019 | Meknes | 44D | NV | 21 | P |
| AS62 | F47 | 24/02/2019 | Elhajeb | 37D | NV | 23 | P |
| AS63 | 24/02/2019 | 37D | 12 | ||||
| AS64 | F48 | 24/02/2019 | Elhajeb | 35D | NV | 26 | P |
| AS65 | F49 | 25/02/2019 | Meknes | 41D | NV | - | N |
| AS66 | F50 | 25/02/2019 | Meknes | 35D | NV | - | N |
| AS67 | F51 | 25/02/2019 | Meknes | 32D | NV | - | N |
| AS68 * | F52 | 26/02/2019 | Khenifra | 38D | NV | 14 | P |
| AS69 | F53 | 25/02/2019 | Khenifra | 28D | NV | 25 | P |
| AS70 | F54 | 28/02/2019 | Salé | 23D | NV | 22 | P |
| AS71 *+ | F55 | 01/04/2019 | Sidi slimane | 14D | NV | 22 | P |
| AS72 | F56 | 11/12/2017 | Khenifra | 33D | NV | - | N |
| AS73 | F57 | 20/02/2019 | Khenifra | 36D | NV | 14 | P |
| AS74 | F58 | 11/04/2019 | Sidi slimane | 37D | NV | - | N |
| AS75 | F59 | 06/03/2019 | Salé | 29D | NV | - | N |
| AS76 *+ | F60 | 23/02/2019 | Meknes | 38D | NV | 20 | P |
| AS77 *+ | F61 | 17/01/2019 | Meknes | 29D | NV | 16 | P |
| AS78 | F62 | 01/04/2019 | Tiznit | 25D | NV | - | P |
| AS79 | 01/04/2019 | 25D | NV | 30 | |||
| AS80 | F63 | 01/04/2019 | Tiznit | 32D | NV | 30 | P |
| AS81 | 01/04/2019 | 32D | NV | 28 | |||
| AS82 | F64 | 01/04/2019 | Tiznit | 32D | NV | - | N |
| AS83 | 01/04/2019 | 37D | NV | - | |||
| AS84 | F65 | 01/04/2019 | Tiznit | 37D | NV | - | N |
| AS85 * | F66 | 01/04/2019 | Tiznit | 28D | NV | 16 | P |
| AS86 | F67 | 01/04/2019 | Tiznit | 37D | NV | - | N |
| AS87 | 01/04/2019 | 34D | NV | - | |||
| AS88 | F68 | 01/04/2019 | Tiznit | 34D | NV | - | N |
| AS89 | F69 | 01/04/2019 | Tiznit | 15D | NV | 26 | P |
| AS90 | F70 | 31/05/2019 | Rabat | 32D | NV | - | N |
| AS91 | 31/05/2019 | 32D | - | ||||
| AS92 | 31/05/2019 | 32D | - | ||||
| AS93 | 31/05/2019 | 32D | - | ||||
| AS94 | 31/05/2019 | 32D | - | ||||
| AS95 | 31/05/2019 | 32D | - | ||||
| AS96 | 31/05/2019 | 32D | - | ||||
| AS97 | 31/05/2019 | 32D | - | ||||
| AS98 | 31/05/2019 | 32D | - | ||||
| BS1 | F71 | 09/04/2019 | Fes | 28D | NV | - | N |
| BS2 | F72 | 09/04/2019 | Fes | 36D | NV | - | N |
| BS3 | F73 | 16/09/2019 | Ait moussa | 35D | NV | - | N |
| BS4 | 16/09/2019 | 35D | - | ||||
| BS5 | F74 | 08/10/2019 | Ait moussa | 36D | V | - | N |
| BS6 | 09/10/2019 | 36D | - | ||||
| BS7 | F75 | 23/10/2019 | Marrakech | 18D | NV | - | N |
| BS8 | F76 | 24/10/2019 | Haouz | 29D | NV | - | N |
| BS9 | F77 | 30/10/2019 | Marrakech | 34D | V | - | N |
| BS10 | F78 | 31/10/2019 | Rhamna | 13D | NV | - | N |
| BS11 | F79 | 05/11/2019 | Marrakech | 12D | NV | - | N |
| BS12 | F80 | 23/11/2019 | Casablanca | 30D | NV | 29 | P |
| BS13 | F81 | 23/11/2019 | Casablanca | 21D | NV | 28 | P |
| BS14 | F82 | 23/11/2019 | Casablanca | 24D | NV | 33 | P |
| BS15 | F83 | 23/11/2019 | Casablanca | 24D | NV | 31 | P |
| BS18 | F84 | 25/11/2019 | Tiznit | 34D | V | - | N |
| BS19 | 25/11/2019 | 34D | - | ||||
| BS20 | 25/11/2019 | 34D | - | ||||
| BS21 | F85 | 05/12/2019 | Tiznit | 29D | V | - | N |
| BS22 | 05/12/2019 | 29D | - | ||||
| BS23 | 05/12/2019 | 29D | - | ||||
| BS47 | F86 | 14/11/2019 | Casablanca | 28D | V | 26 | P |
| BS48 | F87 | 14/11/2019 | Casablanca | 28D | V | 26 | P |
| BS49 | F88 | 14/11/2019 | Casablanca | 28D | V | 27 | P |
| BS50 | F89 | 24/10/2019 | Mohammedia | 28D | V | - | N |
| BS57 | F90 | 05/03/2020 | Casablanca | 34D | V | 29 | P |
| BS58 | 05/03/2020 | 34D | - | ||||
| BS59 | F91 | 05/03/2020 | Casablanca | 30D | V | - | P |
| BS60 | 05/03/2020 | 30D | 33 | ||||
| BS61 | F92 | 02/07/2020 | Tiznit | 42D | V | - | N |
| BS62 | F93 | 02/07/2020 | Tiznit | 34D | V | 30 | P |
| BS63 | F94 | 02/07/2020 | Tiznit | 30D | V | 30 | P |
| BS64 | F95 | 02/07/2020 | Tiznit | 22D | V | 30 | P |
| BS65 | F96 | 02/07/2020 | Tiznit | 30D | V | 29 | P |
| BS66 | F97 | 02/07/2020 | Tiznit | 35D | V | 32 | P |
| BS67 | F98 | 02/07/2020 | Tiznit | 29D | V | 27 | P |
| BS68 | F99 | 02/07/2020 | Tiznit | 21D | V | 29 | P |
| BS69 | F100 | 02/07/2020 | Tiznit | 13D | V | 27 | P |
| BS70 | F101 | 02/07/2020 | Tiznit | 31D | V | 28 | P |
| BS71 | F102 | 02/07/2020 | Tiznit | 44D | V | 30 | P |
| BS72 | F103 | 02/07/2020 | Tiznit | 36D | V | 28 | P |
| BS73 | F104 | 02/07/2020 | Tiznit | 24D | V | 29 | P |
| BS76 | F105 | 02/07/2020 | Tiznit | 42D | V | 30 | P |
| BS77 | 02/07/2020 | 42D | 28 | ||||
| BS78 | 02/07/2020 | 42D | 29 | ||||
| BS79 | 02/07/2020 | 42D | 27 | ||||
| BS80 | 02/07/2020 | 42D | 30 | ||||
| BS81 | 02/07/2020 | 42D | 32 | ||||
| BS82 | 02/07/2020 | 42D | 39 | ||||
| BS83 | 02/07/2020 | 42D | 39 | ||||
| BS84 | 02/07/2020 | 42D | 36 | ||||
| BS85 | F106 | 09/09/2020 | Casablanca | 33D | NV | 31 | P |
| BS86 | 09/09/2020 | 33D | 30 | ||||
| BS87 | F107 | 09/09/2020 | Casablanca | 27D | NV | 30 | P |
| BS88 | 09/09/2020 | 27D | 30 | ||||
| BS89 | 09/09/2020 | 27D | 29 | ||||
| BS90 | 09/09/2020 | 27D | 30 | ||||
| BS91 * | F108 | 10/09/2020 | Rabat | 29D | V | 24 | P |
| BS92 | 10/09/2020 | 29D | 29 | ||||
| BS93 | 10/09/2020 | 29D | 34 |
| 137 | 188 | 190 | 222 | 226 | 298 | 325 | 364 | 375 | 397 | 402 | 408 | 493 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SF1 | T | D | A | L | L | I | H | M | V | D | E | N | T |
| AS13 | T | D | T | L | L | I | H | I | V | D | E | N | T |
| AS14 | T | D | A | L | L | I | Q | M | V | D | E | N | I |
| AS29 | T | D | V | L | L | I | H | M | V | N | E | N | T |
| AS32 | T | D | A | L | L | V | H | M | V | D | D | N | T |
| AS71 | T | D | A | L | L | V | H | M | I | D | E | S | T |
| AS76 | T | N | A | L | Q | V | H | M | V | D | E | N | T |
| AS77 | T | D | A | L | Q | V | H | M | V | D | E | N | T |
| 8 | 16 | 31 | 42 | 46 | 50 | 56 | 57 | 58 | 60 | 65 | 80 | 88 | 101 | 116 | 127 | 254 | 261 | 290 | 329 | 332 | 368 | 385 | 390 | 400 | 416 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SF1 | I | T | T | Y | S | A | I | I | I | R | I | T | S | S | V | S | I | K | V | N | S | K | T | S | N | I |
| AS13 | I | T | T | Y | S | A | I | I | I | R | I | T | S | S | V | S | I | R | V | D | S | K | N | A | S | I |
| AS14 | I | T | T | Y | S | T | T | T | T | R | I | P | L | S | V | N | V | K | V | N | S | T | T | S | N | M |
| AS29 | I | T | T | H | P | A | I | I | I | K | I | T | S | S | V | S | I | R | V | N | S | K | T | A | S | I |
| AS32 | I | I | T | Y | S | A | I | I | I | R | T | T | S | S | I | S | I | K | V | N | S | K | T | S | S | I |
| AS71 | M | I | T | Y | S | A | I | I | I | R | T | T | S | A | V | S | I | K | A | N | F | K | T | S | S | I |
| AS76 | M | I | T | Y | S | A | I | I | I | R | T | T | S | A | V | S | I | K | V | N | F | K | T | S | S | I |
| AS77 | I | I | M | Y | S | A | I | I | I | R | T | T | S | S | I | S | I | K | V | N | S | K | T | S | S | I |
References
- Taxonomy. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 3 August 2021).
- Capua, I.; Alexander, D.J. Avian Influenza: Recent Developments. Avian Pathol. 2004, 33, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizpour, A.; Goudarzi, H.; Charkhkar, S.; Momayez, R.; Hablolvarid, M.H. Experimental Study on Tissue Tropism and Dissemination of H9N2 Avian Influenza Virus and Ornithobacterium Rhinotracheale Co-Infection in SPF Chickens. J. Anim. Plant Sci. 2014, 24, 1655–1662. [Google Scholar]
- Seifi, S.; Asasi, K.; Mohammadi, A. Short Paper: An Experimental Study on Broiler Chicken Co-Infected with the Specimens Containing Avian Influenza (H9 Subtype) and Infectious Bronchitis (4/91 Strain) Viruses. Iran. J. Vet. Res. 2012, 13, 5. [Google Scholar]
- Homme, P.J.; Easterday, B.C. Avian Influenza Virus Infections. I. Characteristics of Influenza A/Turkey/Wisconsin/1966 Virus. Avian Dis. 1970, 14, 66–74. [Google Scholar] [CrossRef]
- Chen, B.L.; Zhang, Z.J.; Chen, W.B. Isolation and Preliminary Serological Characterization of Type A Influenza Viruses from Chickens. Chin. J. Vet. Med. 1994, 22, 3–5. [Google Scholar]
- Lee, D.-H.; Swayne, D.E.; Sharma, P.; Rehmani, S.F.; Wajid, A.; Suarez, D.L.; Afonso, C.L. H9N2 Low Pathogenic Avian Influenza in Pakistan (2012–2015). Vet. Rec. Open 2016, 3, e000171. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.; Liu, A.; Zhang, F.; Ling, Y.; Ou, C.; Hou, N.; He, C. Co-Infection of Broilers with Ornithobacterium Rhinotracheale and H9N2 Avian Influenza Virus. BMC Vet. Res. 2012, 8, 104. [Google Scholar] [CrossRef] [Green Version]
- Nili, H.; Asasi, K. Avian Influenza (H9N2) Outbreak in Iran. Avian Dis. 2003, 47, 828–831. [Google Scholar] [CrossRef]
- Monne, I.; Fusaro, A.; Al-Blowi, M.H.; Ismail, M.M.; Khan, O.A.; Dauphin, G.; Tripodi, A.; Salviato, A.; Marangon, S.; Capua, I.; et al. Co-Circulation of Two Sublineages of HPAI H5N1 Virus in the Kingdom of Saudi Arabia with Unique Molecular Signatures Suggesting Separate Introductions into the Commercial Poultry and Falconry Sectors. J. Gen. Virol. 2008, 89, 2691–2697. [Google Scholar] [CrossRef]
- Roussan, D.A.; Khawaldeh, G.Y.; Al Rifai, R.H.; Totanji, W.S.; Shaheen, I.A. Avian Influenza Virus H9 Subtype in Poultry Flocks in Jordan. Prev. Vet. Med. 2009, 88, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Wernery, U.; Shanmuganatham, K.K.; Krylov, P.S.; Joseph, S.; Friedman, K.; Krauss, S.; Webster, R.G. H9N2 Influenza Viruses from Birds Used in Falconry. Influenza Other Respir. Viruses 2013, 7, 1241–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tombari, W.; Paul, M.; Bettaieb, J.; Larbi, I.; Nsiri, J.; Elbehi, I.; Gribaa, L.; Ghram, A. Risk Factors and Characteristics of Low Pathogenic Avian Influenza Virus Isolated from Commercial Poultry in Tunisia. PLoS ONE 2013, 8, e53524. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; El-Shesheny, R.; Maatouq, A.M.; Moatasim, Y.; Shehata, M.M.; Bagato, O.; Rubrum, A.; Shanmuganatham, K.; Webby, R.J.; Ali, M.A.; et al. Genetic and Antigenic Evolution of H9N2 Avian Influenza Viruses Circulating in Egypt between 2011 and 2013. Arch. Virol. 2014, 159, 2861–2876. [Google Scholar] [CrossRef]
- Body, M.H.; Alrarawahi, A.H.; Alhubsy, S.S.; Saravanan, N.; Rajmony, S.; Mansoor, M.K. Characterization of Low Pathogenic Avian Influenza Virus Subtype H9N2 Isolated from Free-Living Mynah Birds (Acridotheres tristis) in the Sultanate of Oman. Avian Dis. 2015, 59, 329–334. [Google Scholar] [CrossRef]
- Kammon, A.; Heidari, A.; Dayhum, A.; Eldaghayes, I.; Sharif, M.; Monne, I.; Cattoli, G.; Asheg, A.; Farhat, M.; Kraim, E. Characterization of Avian Influenza and Newcastle Disease Viruses from Poultry in Libya. Avian Dis. 2015, 59, 422–430. [Google Scholar] [CrossRef]
- Group, T.S.H.W.; Schultz-Cherry, S.; Thomas, P. Assessing the Fitness of Distinct Clades of Influenza A (H9N2) Viruses. Emerg. Microbes Infect. 2013, 2, e75. [Google Scholar] [CrossRef]
- Jestin, V.; Manuguerra, J.C.; Eterradossi, N. Risque de Transmission à l’homme Des Virus Influenza Aviaires. Bull. Epidemiol. AFSSA 2003, 11, 1–3. [Google Scholar]
- Iqbal, M.; Yaqub, T.; Mukhtar, N.; Shabbir, M.Z.; McCauley, J.W. Infectivity and Transmissibility of H9N2 Avian Influenza Virus in Chickens and Wild Terrestrial Birds. Vet. Res. 2013, 44, 100. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Zhang, Z.; Yu, Z.; Li, L.; Cheng, K.; Wang, T.; Huang, G.; Yang, S.; Zhao, Y.; Feng, N.; et al. Domestic Cats and Dogs Are Susceptible to H9N2 Avian Influenza Virus. Virus Res. 2013, 175, 52–57. [Google Scholar] [CrossRef]
- Peiris, M.; Yam, W.C.; Chan, K.H.; Ghose, P.; Shortridge, K.F. Influenza A H9N2: Aspects of Laboratory Diagnosis. J. Clin. Microbiol. 1999, 37, 3426–3427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiris, M.; Yuen, K.; Leung, C.; Chan, K.; Ip, P.; Lai, R.; Orr, W.; Shortridge, K. Human Infection with Influenza H9N2. Lancet 1999, 354, 916–917. [Google Scholar] [CrossRef]
- Guo, Y.; Li, J.; Cheng, X. Discovery of Men Infected by Avian Influenza A (H9N2) Virus. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 1999, 13, 105–108. [Google Scholar] [PubMed]
- Lin, Y.P.; Shaw, M.; Gregory, V.; Cameron, K.; Lim, W.; Klimov, A.; Subbarao, K.; Guan, Y.; Krauss, S.; Shortridge, K.; et al. Avian-to-Human Transmission of H9N2 Subtype Influenza A Viruses: Relationship between H9N2 and H5N1 Human Isolates. Proc. Natl. Acad. Sci. USA 2000, 97, 9654–9658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EL Houadfi, M.; Fellahi, S.; Nassik, S.; Guérin, J.-L.; Ducatez, M.F. First Outbreaks and Phylogenetic Analyses of Avian Influenza H9N2 Viruses Isolated from Poultry Flocks in Morocco. Virol. J. 2016, 13, 140. [Google Scholar] [CrossRef] [Green Version]
- Monne, I.; Ormelli, S.; Salviato, A.; Battisti, C.D.; Bettini, F.; Salomoni, A.; Drago, A.; Zecchin, B.; Capua, I.; Cattoli, G. Development and Validation of a One-Step Real-Time PCR Assay for Simultaneous Detection of Subtype H5, H7, and H9 Avian Influenza Viruses. J. Clin. Microbiol. 2008, 46, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barman, S.; Turner, J.C.M.; Hasan, M.K.; Akhtar, S.; El-Shesheny, R.; Franks, J.; Walker, D.; Seiler, P.; Friedman, K.; Kercher, L.; et al. Continuing Evolution of Highly Pathogenic H5N1 Viruses in Bangladeshi Live Poultry Markets. Emerg. Microbes Infect. 2019, 8, 650–661. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Peacock, T.P.; Benton, D.J.; Sadeyen, J.-R.; Chang, P.; Sealy, J.E.; Bryant, J.E.; Martin, S.R.; Shelton, H.; McCauley, J.W.; Barclay, W.S.; et al. Variability in H9N2 Haemagglutinin Receptor-Binding Preference and the PH of Fusion. Emerg. Microbes Infect. 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sealy, J.E.; Yaqub, T.; Peacock, T.P.; Chang, P.; Ermetal, B.; Clements, A.; Sadeyen, J.-R.; Mehboob, A.; Shelton, H.; Bryant, J.E.; et al. Association of Increased Receptor-Binding Avidity of Influenza A(H9N2) Viruses with Escape from Antibody-Based Immunity and Enhanced Zoonotic Potential. Emerg. Infect. Dis. 2018, 25, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, J.; Guo, J.; Deng, G.; Zhang, Q.; Wang, J.; He, X.; Wang, K.; Chen, J.; Li, Y.; et al. Genetics, Receptor Binding Property, and Transmissibility in Mammals of Naturally Isolated H9N2 Avian Influenza Viruses. PLoS Pathog. 2014, 10, e1004508. [Google Scholar] [CrossRef]
- Teng, Q.; Xu, D.; Shen, W.; Liu, Q.; Rong, G.; Li, X.; Yan, L.; Yang, J.; Chen, H.; Yu, H.; et al. A Single Mutation at Position 190 in Hemagglutinin Enhances Binding Affinity for Human Type Sialic Acid Receptor and Replication of H9N2 Avian Influenza Virus in Mice. J. Virol. 2016, 90, 9806–9825. [Google Scholar] [CrossRef] [Green Version]
- Hurt, A.C.; Holien, J.K.; Parker, M.W.; Barr, I.G. Oseltamivir Resistance and the H274Y Neuraminidase Mutation in Seasonal, Pandemic and Highly Pathogenic Influenza Viruses. Drugs 2009, 69, 2523–2531. [Google Scholar] [CrossRef]
- Suttie, A.; Deng, Y.-M.; Greenhill, A.R.; Dussart, P.; Horwood, P.F.; Karlsson, E.A. Inventory of Molecular Markers Affecting Biological Characteristics of Avian Influenza A Viruses. Virus Genes 2019, 55, 739–768. [Google Scholar] [CrossRef] [Green Version]
- Jonas, M.; Sahesti, A.; Murwijati, T.; Lestariningsih, C.L.; Irine, I.; Ayesda, C.S.; Prihartini, W.; Mahardika, G.N. Identification of Avian Influenza Virus Subtype H9N2 in Chicken Farms in Indonesia. Prev. Vet. Med. 2018, 159, 99–105. [Google Scholar] [CrossRef]
- El Khantour, A.; El Houadfi, M.; Saâdia, N.; Tligui, N.; el Mellouli, F.; Sikht, F.-Z.; Ducatez, M.; Soulaymani, A.; Fellahi, S. Protective Efficacy Evaluation of Four Inactivated Commercial Vaccines Against Low Pathogenic Avian Influenza H9N2 Virus Under Experimental Conditions in Broiler Chickens. Avian Dis. 2021, 65, 351–357. [Google Scholar] [CrossRef]
- Park, K.J.; Kwon, H.; Song, M.-S.; Pascua, P.N.Q.; Baek, Y.H.; Lee, J.H.; Jang, H.-L.; Lim, J.-Y.; Mo, I.-P.; Moon, H.-J.; et al. Rapid Evolution of Low-Pathogenic H9N2 Avian Influenza Viruses Following Poultry Vaccination Programmes. J. Gen. Virol. 2011, 92, 36–50. [Google Scholar] [CrossRef]
- Alexander, D.J. An Overview of the Epidemiology of Avian Influenza. Vaccine 2007, 25, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, T.; Lambrecht, B.; Marché, S.; Steensels, M.; Van Borm, S.; Bublot, M. Influenza Vaccines and Vaccination Strategies in Birds. Comp. Immunol. Microbiol. Infect. Dis. 2008, 31, 121–165. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.G.; Lee, Y.J.; Kim, Y.J.; Lee, E.K.; Jeong, O.M.; Sung, H.W.; Kim, J.H.; Kwon, J.H. An Inactivated Vaccine to Control the Current H9N2 Low Pathogenic Avian Influenza in Korea. J. Vet. Sci. 2008, 9, 67. [Google Scholar] [CrossRef] [Green Version]
- Wan, H.; Perez, D.R. Amino Acid 226 in the Hemagglutinin of H9N2 Influenza Viruses Determines Cell Tropism and Replication in Human Airway Epithelial Cells. JVI 2007, 81, 5181–5191. [Google Scholar] [CrossRef] [Green Version]
- Kawaoka, Y.; Webster, R.G. Sequence Requirements for Cleavage Activation of Influenza Virus Hemagglutinin Expressed in Mammalian Cells. Proc. Natl. Acad. Sci. USA 1988, 85, 324–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Segment | Strain | ||||||
|---|---|---|---|---|---|---|---|
| AS13 | AS14 | AS29 | AS32 | AS71 | AS76 | AS77 | |
| PB2 | MW165151 | MW165079 | MW165121 | MW165089 | MW165136 | MW165110 | MW165106 |
| PB1 | MW165154 | MW165125 | MW165122 | MW165088 | MW165142 | MW165113 | MW165101 |
| PA | MW165158 | MW165082 | MW165117 | MW165085 | MW165139 | MW165116 | MW165103 |
| HA | MW165152 | MW165084 | MW165120 | MW165090 | MW165137 | MW165111 | MW165105 |
| NP | MW165157 | MW165083 | MW165124 | MW165086 | MW165140 | MW165109 | MW165108 |
| NA | MW165155 | MW165078 | MW165119 | MW165092 | MW165135 | MW165115 | MW165104 |
| NS | MW165156 | MW165080 | MW165123 | MW165091 | MW165141 | MW165114 | MW165102 |
| M | MW165153 | MW165081 | MW165118 | MW165087 | MW165138 | MW165112 | MW165107 |
| Number of Farms | Positive Farms | Positivity Rate | |
|---|---|---|---|
| Fes-Meknes | 34 | 20 | 59% |
| Rabat-Sale-Kenitra | 18 | 8 | 44% |
| Casablanca-Settat | 16 | 13 | 81% |
| Draa-Tafilalet | 1 | 1 | 100% |
| BeniMellal-Khenifra | 4 | 3 | 75% |
| Souss-Massa | 26 | 17 | 65% |
| Oriental | 4 | 1 | 25% |
| Marrakech-Safi | 5 | 0 | 0% |
| Morocco (Total) | 108 | 63 | 58% |
| Vaccinated | 44 (41%) | 31 | 70% |
| Unvaccinated | 64 (59%) | 32 | 50% |
| HA * RBS | NA | M2 | |||
|---|---|---|---|---|---|
| 190 | Q226L | 227 | 274 | S31N | |
| SF1 ** | V | L | I | Q | N |
| AS13 | T | L | I | Q | N |
| AS14 | A | L | I | Q | N |
| AS29 | V | L | I | Q | N |
| AS32 | A | L | I | Q | N |
| AS71 | A | L | I | Q | N |
| AS76 | A | Q | I | Q | N |
| AS77 | A | Q | I | Q | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikht, F.-Z.; Ducatez, M.; Touzani, C.D.; Rubrum, A.; Webby, R.; El Houadfi, M.; Tligui, N.-S.; Camus, C.; Fellahi, S. Avian Influenza a H9N2 Viruses in Morocco, 2018–2019. Viruses 2022, 14, 529. https://doi.org/10.3390/v14030529
Sikht F-Z, Ducatez M, Touzani CD, Rubrum A, Webby R, El Houadfi M, Tligui N-S, Camus C, Fellahi S. Avian Influenza a H9N2 Viruses in Morocco, 2018–2019. Viruses. 2022; 14(3):529. https://doi.org/10.3390/v14030529
Chicago/Turabian StyleSikht, Fatima-Zohra, Mariette Ducatez, Charifa Drissi Touzani, Adam Rubrum, Richard Webby, Mohammed El Houadfi, Nour-Said Tligui, Christelle Camus, and Siham Fellahi. 2022. "Avian Influenza a H9N2 Viruses in Morocco, 2018–2019" Viruses 14, no. 3: 529. https://doi.org/10.3390/v14030529
APA StyleSikht, F.-Z., Ducatez, M., Touzani, C. D., Rubrum, A., Webby, R., El Houadfi, M., Tligui, N.-S., Camus, C., & Fellahi, S. (2022). Avian Influenza a H9N2 Viruses in Morocco, 2018–2019. Viruses, 14(3), 529. https://doi.org/10.3390/v14030529








