A Novel Phage Infecting the Marine Photoheterotrophic Bacterium Citromicrobium bathyomarinum
Abstract
1. Introduction
2. Materials and Methods
2.1. Phage Isolation and Purification
2.2. Transmission Electron Microscopy (TEM)
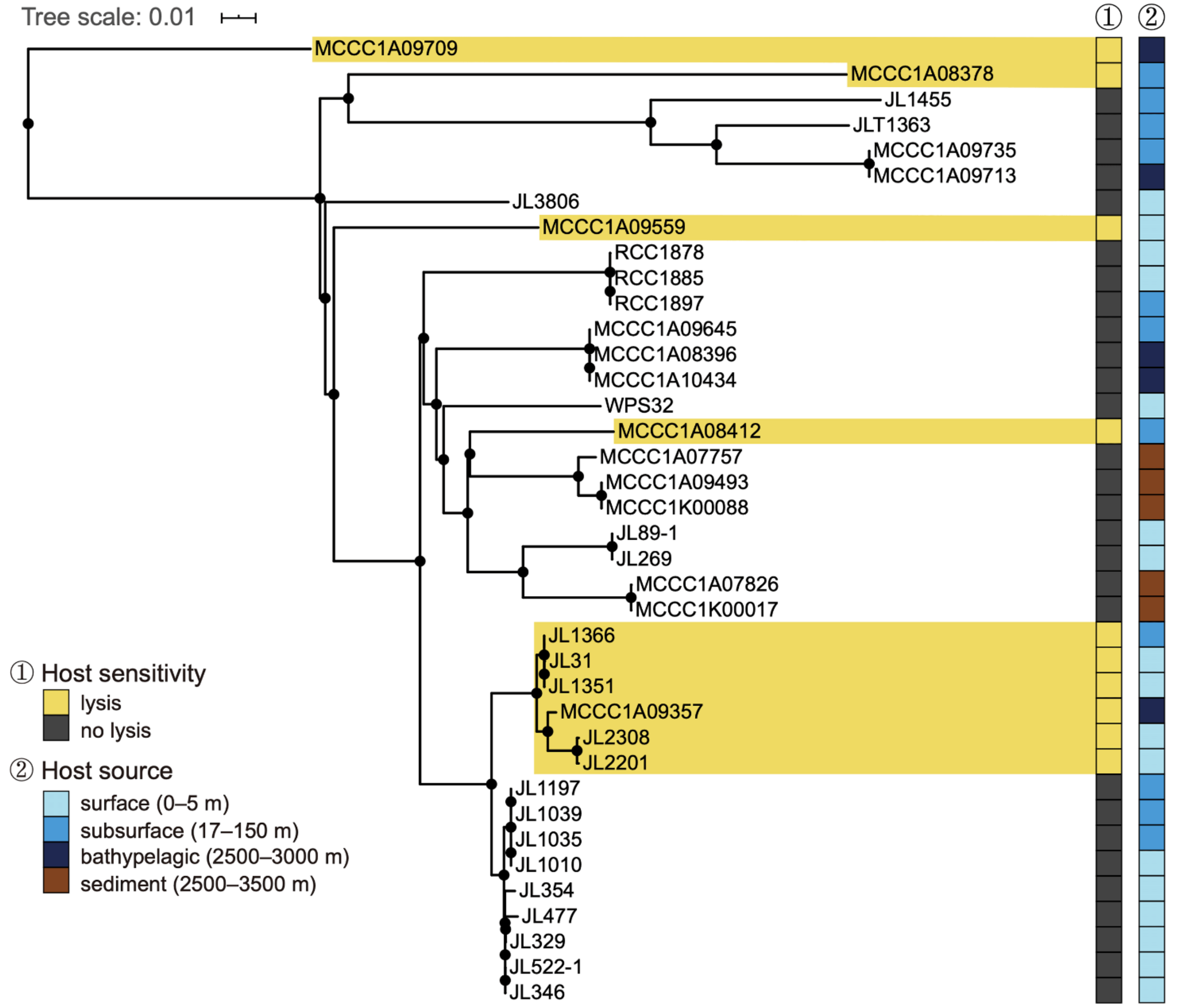
2.3. Determination of the Host Range
2.4. The One-Step Growth Curve
2.5. Phage DNA Extraction, Sequencing and In Silico Analyses
2.6. Structural Proteome and Data Processing
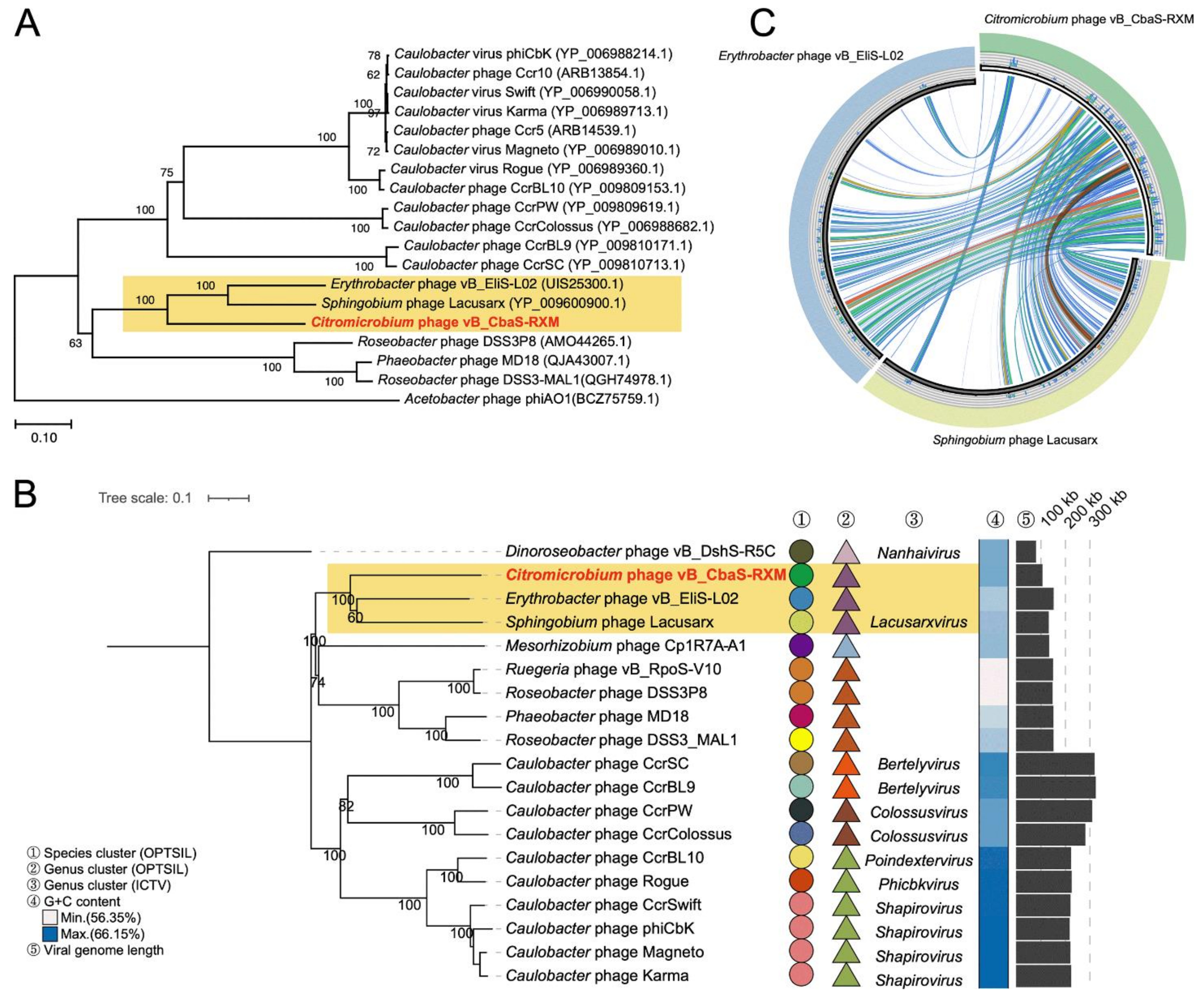
2.7. Phylogenetic Analysis
2.8. Calculation of Codon Usage (CU) and tRNA Relative Contribution Index (tRCI)
3. Results and Discussion
3.1. Biological Features of RXM
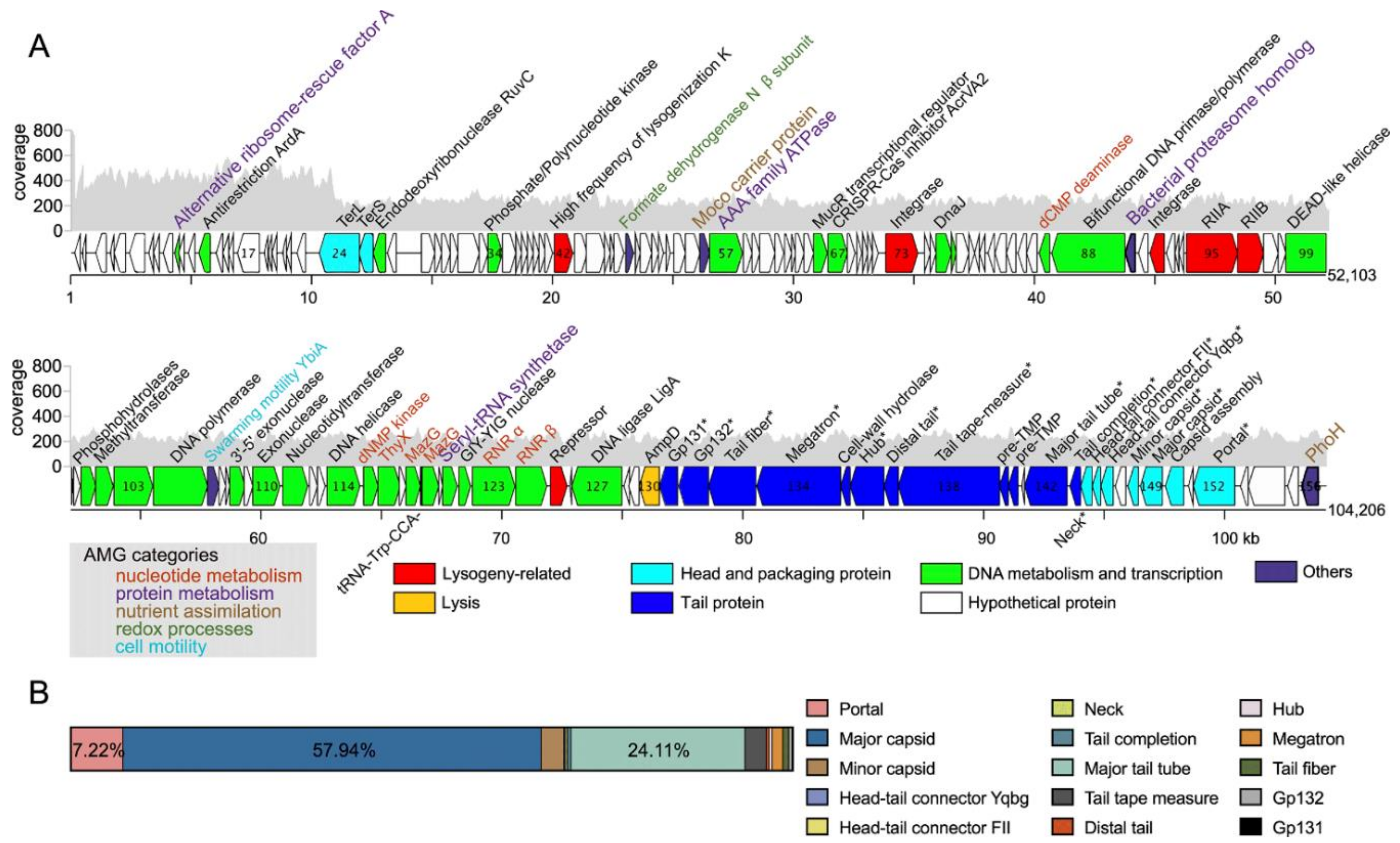
3.2. Genomic Features of RXM
3.3. RXM Represents a New Species of the Lacusarxvirus Genus
3.4. Phage–Host Interactions Inferred from Viral AMGs
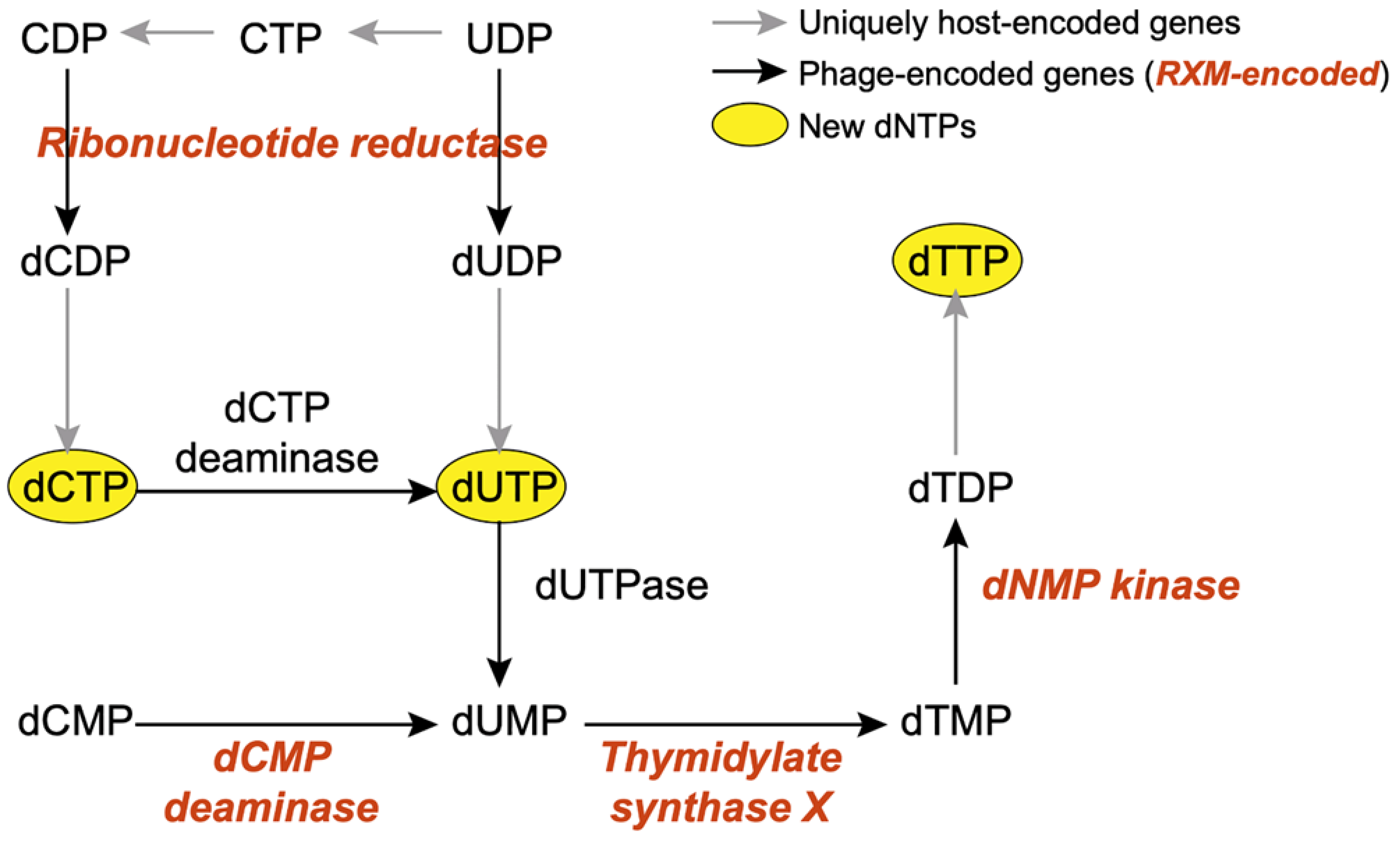
3.4.1. Nucleotide Metabolism
3.4.2. Protein Metabolism
3.4.3. Nutrient Assimilation
3.4.4. Swarming Motility
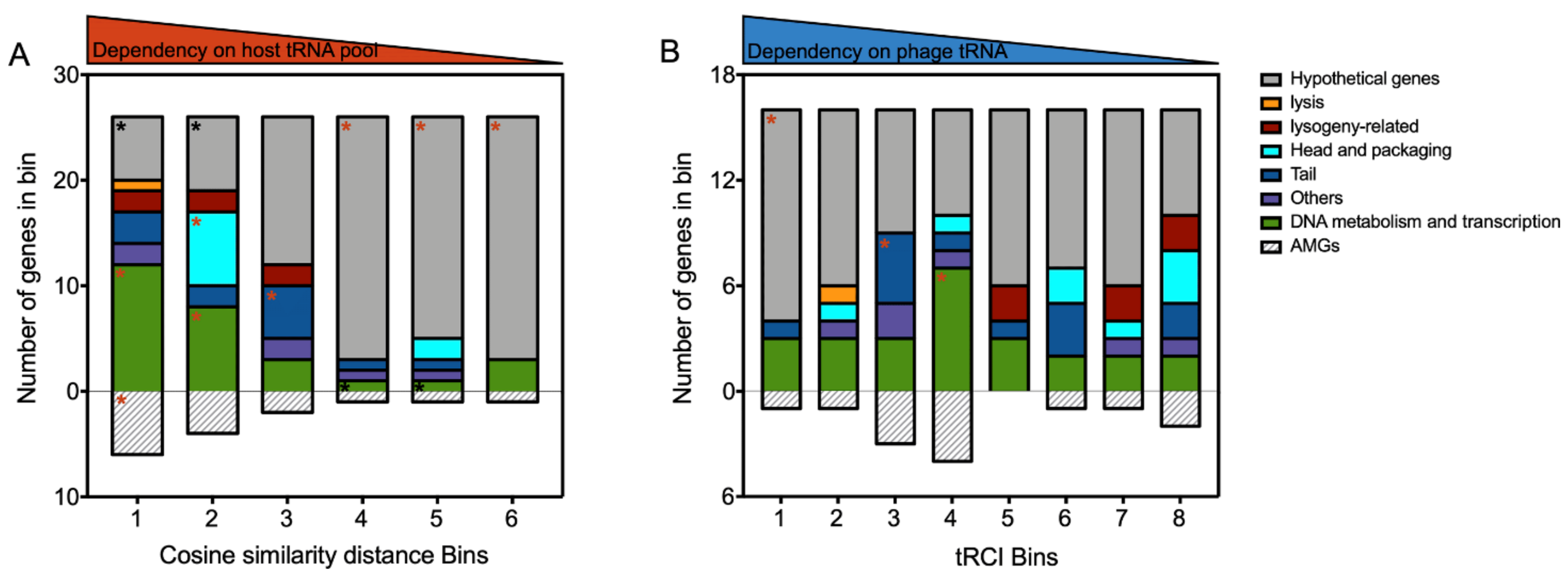
3.5. Viral tRNA Facilitates the Translation of Unique Genes That Differ in CU from the Host tRNA Inventory
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Yurkov, V.V.; Krieger, S.; Stackebrandt, E.; Beatty, J.T. Citromicrobium bathyomarinum, a novel aerobic bacterium isolated from deep-sea hydrothermal vent plume waters that contains photosynthetic pigment-protein complexes. J. Bacteriol. 1999, 181, 4517–4525. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Liu, Y.; Jeanthon, C.; Zhang, R.; Lin, W.; Yao, J.; Jiao, N. Geographic Impact on Genomic Divergence as Revealed by Comparison of Nine Citromicrobial Genomes. Appl. Environ. Microbiol. 2016, 82, 7205–7216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rathgeber, C.; Lince, M.T.; Alric, J.; Lang, A.S.; Humphrey, E.; Blankenship, R.E.; Verméglio, A.; Plumley, F.G.; Van Dover, C.L.; Beatty, J.T.; et al. Vertical distribution and characterization of aerobic phototrophic bacteria at the Juan de Fuca Ridge in the Pacific Ocean. Photosynth. Res. 2008, 97, 235–244. [Google Scholar] [CrossRef]
- Zeng, Y.; Wei, S.; Jiao, N. Genetic diversity of aerobic anoxygenic photosynthetic bacteria in open ocean surface waters and upper twilight zones. Mar. Biol. 2009, 156, 425–437. [Google Scholar] [CrossRef][Green Version]
- Zheng, Q.; Chen, Q.; Xu, Y.; Suttle, C.A.; Jiao, N. A virus infecting marine photoheterotrophic Alphaproteobacteria (Citromicrobium spp.) defines a new lineage of ssDNA viruses. Front. Microbiol. 2018, 9, 1418. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ma, R.; Lai, J.; Chen, X.; Wang, L.; Yang, Y.; Wei, S.; Jiao, N.; Zhang, R. A novel phage infecting Alteromonas represents a distinct group of siphophages infecting diverse aquatic copiotrophs. mSphere 2021, 6, e0045421. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Taniguchi, T.; Itoh, T. MetaGeneAnnotator: Detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008, 15, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Tavares, P.; Petit, M.-A.; Guérois, R.; Zinn-Justin, S. Automated classification of tailed bacteriophages according to their neck organization. BMC Genom. 2014, 15, 1027. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Derbyshire, M.K.; DeWeese-Scott, C.; Gonzales, N.R.; Gwadz, M.; Hao, L.; He, S.; Hurwitz, D.I.; Jackson, J.D.; et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007, 35, D237–D240. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Enav, H.; Béjà, O.; Mandel-Gutfreund, Y. Cyanophage tRNAs may have a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts. ISME J. 2012, 6, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Ash, K.T.; Drake, K.M.; Gibbs, W.S.; Ely, B. Genomic diversity of type B3 bacteriophages of Caulobacter crescentus. Curr. Microbiol. 2017, 74, 779–786. [Google Scholar] [CrossRef]
- Lohr, J.E.; Chen, F.; Hill, R.T. Genomic analysis of bacteriophage ϕJL001: Insights into its interaction with a sponge-associated alpha-proteobacterium. Appl. Environ. Microbiol. 2005, 71, 1598–1609. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Berry, J.D.; Russell, W.K.; Lessor, L.; Escobar-Garcia, D.A.; Hernandez, D.; Kane, A.; Keene, J.; Maddox, M.; Martin, R.; et al. The Caulobacter crescentus phage phiCbK: Genomics of a canonical phage. BMC Genom. 2012, 13, 542. [Google Scholar] [CrossRef]
- Nielsen, T.K.; Carstens, A.B.; Browne, P.; Lametsch, R.; Neve, H.; Kot, W.; Hansen, L.H. The first characterized phage against a member of the ecologically important sphingomonads reveals high dissimilarity against all other known phages. Sci. Rep. 2017, 7, 13566. [Google Scholar] [CrossRef]
- Torrents, E. Ribonucleotide reductases: Essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 2014, 4, 52. [Google Scholar] [CrossRef] [PubMed]
- Rihtman, B.; Bowman-Grahl, S.; Millard, A.; Corrigan, R.M.; Clokie, M.R.J.; Scanlan, D.J. Cyanophage MazG is a pyrophosphohydrolase but unable to hydrolyse magic spot nucleotides. Environ. Microbiol. Rep. 2019, 11, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Kurita, D.; Li, N.; Chen, Y.; Himeno, H.; Gao, N. Mechanistic insights into the alternative translation termination by ArfA and RF2. Nature 2017, 541, 550–553. [Google Scholar] [CrossRef] [PubMed]
- De Freitas Fernandes, A.; Serrão, V.H.B.; Scortecci, J.F.; Thiemann, O.H. Seryl-tRNA synthetase specificity for tRNA(Sec) in Bacterial Sec biosynthesis. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140438. [Google Scholar] [CrossRef]
- Izert, M.A.; Klimecka, M.M.; Górna, M.W. Applications of Bacterial Degrons and Degraders—Toward Targeted Protein Degradation in Bacteria. Front. Mol. Biosci. 2021, 8, 669762. [Google Scholar] [CrossRef]
- Fischer, K.; Llamas, A.; Tejada-Jimenez, M.; Schrader, N.; Kuper, J.; Ataya, F.S.; Galvan, A.; Mendel, R.R.; Fernandez, E.; Schwarz, G. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 2006, 281, 30186–30194. [Google Scholar] [CrossRef]
- Jormakka, M.; Törnroth, S.; Byrne, B.; Iwata, S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science 2002, 295, 1863–1868. [Google Scholar] [CrossRef]
- Peters, D.L.; McCutcheon, J.G.; Stothard, P.; Dennis, J.J. Novel Stenotrophomonas maltophilia temperate phage DLP4 is capable of lysogenic conversion. BMC Genom. 2019, 20, 300. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, R.; Wang, N.; Cai, L.; Tong, Y.; Sun, Q.; Chen, F.; Jiao, N. Novel phage–host interactions and evolution as revealed by a cyanomyovirus isolated from an estuarine environment. Environ. Microbiol. 2018, 20, 2974–2989. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, R.; Shao, S.; Wei, S.; Ye, J.; Yang, Y.; Jiao, N.; Zhang, R. A Novel Phage Infecting the Marine Photoheterotrophic Bacterium Citromicrobium bathyomarinum. Viruses 2022, 14, 512. https://doi.org/10.3390/v14030512
Ma R, Shao S, Wei S, Ye J, Yang Y, Jiao N, Zhang R. A Novel Phage Infecting the Marine Photoheterotrophic Bacterium Citromicrobium bathyomarinum. Viruses. 2022; 14(3):512. https://doi.org/10.3390/v14030512
Chicago/Turabian StyleMa, Ruijie, Shuai Shao, Shuzhen Wei, Junlei Ye, Yahui Yang, Nianzhi Jiao, and Rui Zhang. 2022. "A Novel Phage Infecting the Marine Photoheterotrophic Bacterium Citromicrobium bathyomarinum" Viruses 14, no. 3: 512. https://doi.org/10.3390/v14030512
APA StyleMa, R., Shao, S., Wei, S., Ye, J., Yang, Y., Jiao, N., & Zhang, R. (2022). A Novel Phage Infecting the Marine Photoheterotrophic Bacterium Citromicrobium bathyomarinum. Viruses, 14(3), 512. https://doi.org/10.3390/v14030512






