An Update of Orthopoxvirus Molecular Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Retrieval of Genome Sequences and Alignment
2.2. Phylogenetic Tree Construction and Evolutionary Analyses
3. Results
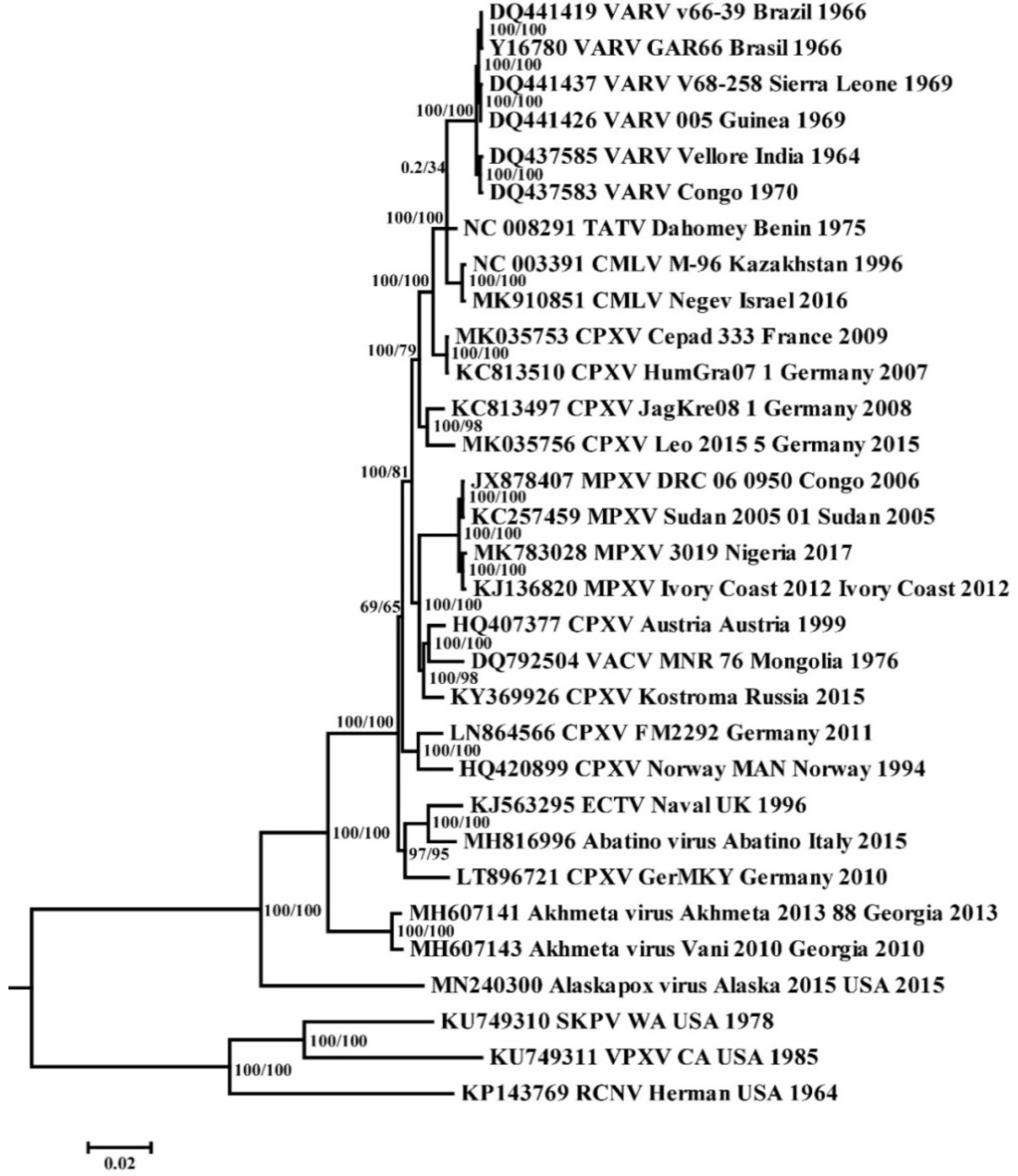
3.1. Phylogenetic Analysis of Orthopoxviruses
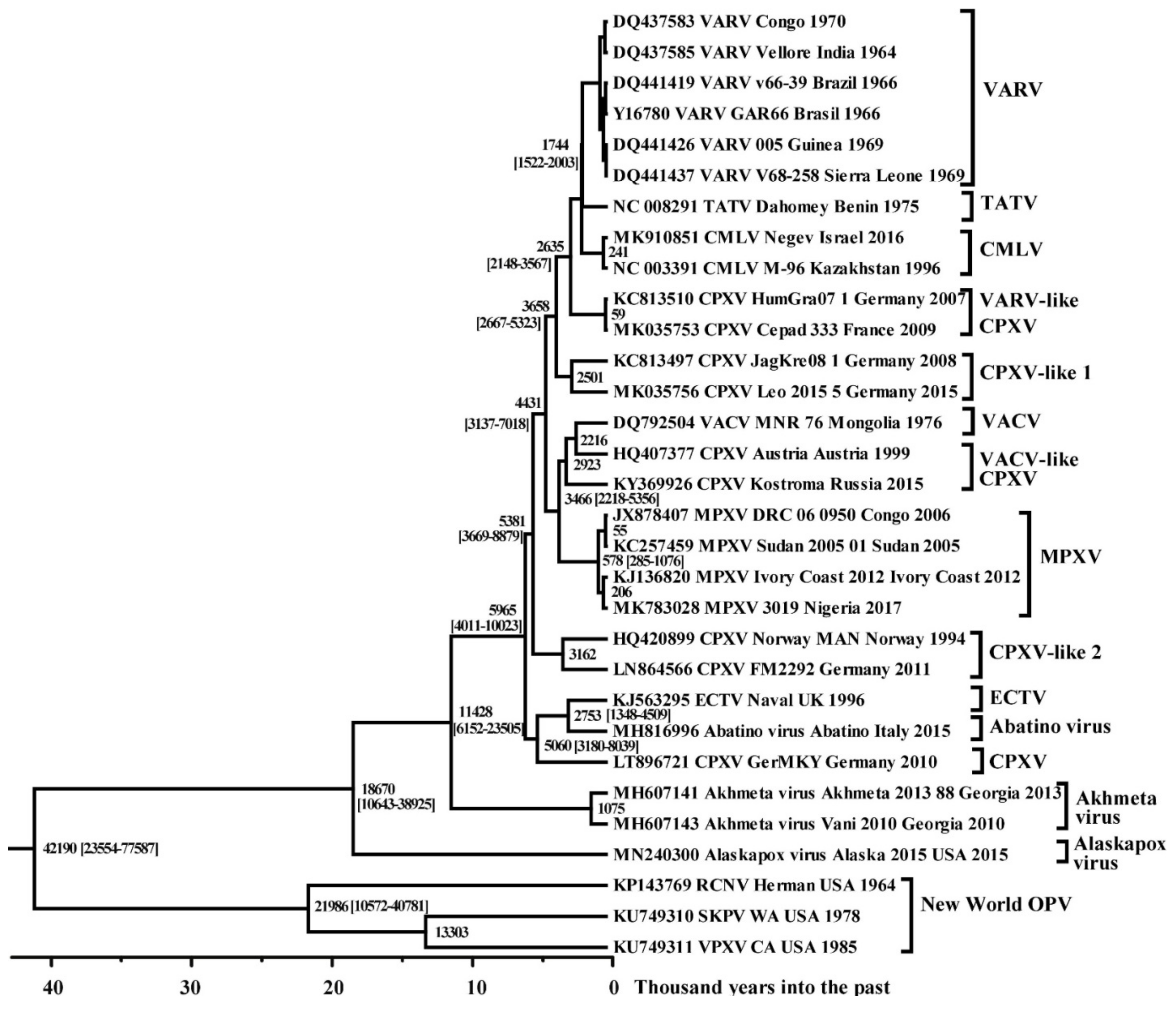
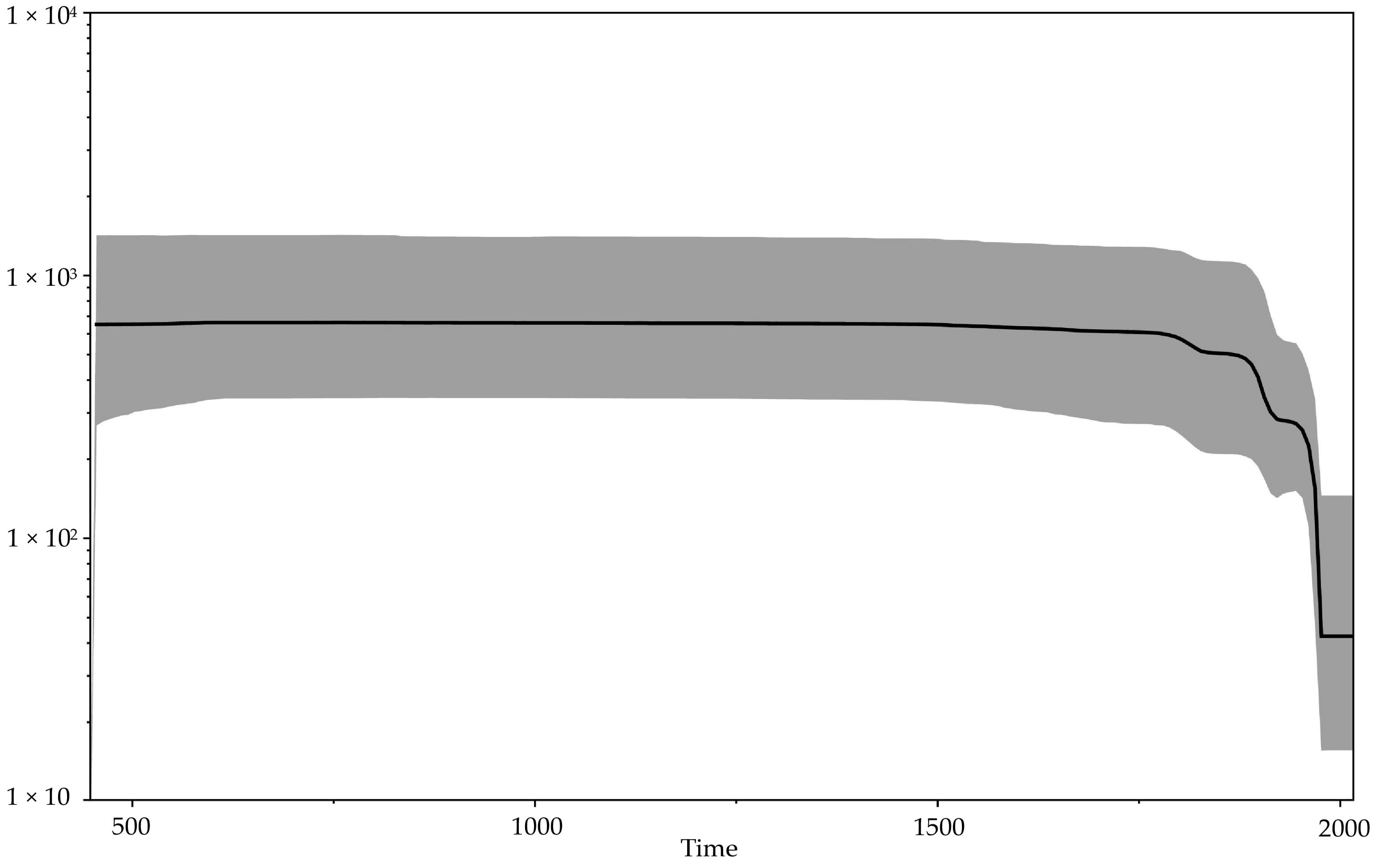
3.2. Molecular Evolution of VARV
3.3. Molecular Evolution of Orthopoxviruses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fenner, F.; Henderson, D.A.; Arita, I.; Jezek, Z.; Ladnyi, I.D. Smallpox and Its Eradication; WHO: Geneva, Switzerland, 1988. [Google Scholar]
- Mühlemann, B.; Vinner, L.; Margaryan, A.; Wilhelmson, H.; de la Fuente Castro, C.; Allentoft, M.E.; de Barros Damgaard, P.; Hansen, A.J.; Holtsmark Nielsen, S.; Strand, L.; et al. Diverse variola virus (smallpox) strains were widespread in northern Europe in the Viking Age. Science 2020, 369, eaaw8977. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N.; Marennikova, S.S.; Moyer, R.W. Orthopoxviruses Pathogenic for Humans; Springer: Berlin/Heildeberg Germany, 2005. [Google Scholar]
- Chen, N.; Bellone, C.J.; Schriewer, J.; Owens, G.; Fredrickson, T.; Parker, S.; Buller, R.M.L. Poxvirus interleukin-4 expression overcomes inherent resistance and vaccine-induced immunity: Pathogenesis, prophylaxis, and antiviral therapy. Virology 2011, 409, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N.; Totmenin, A.V.; Safronov, P.F.; Mikheev, M.V.; Gutorov, V.V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.V.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Analysis of the monkeypox virus genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef] [Green Version]
- Mauldin, M.R.; McCollum, A.M.; Nakazawa, Y.J.; Mandra, A.; Whitehouse, E.R.; Davidson, W.; Zhao, H.; Gao, J.; Li, Y.; Doty, J.; et al. Exportation of Monkeypox virus from the African continent. J. Infect. Dis. 2020, jiaa559. [Google Scholar] [CrossRef]
- Franke, A.; Pfaff, F.; Jenckel, M.; Hoffmann, B.; Höper, D.; Antwerpen, M.; Meyer, H.; Beer, M.; Hoffmann, D. Classification of cowpox viruses into several distinct clades and identification of a novel lineage. Viruses 2017, 9, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, D.S.; Emerson, G.L.; Li, Y.; Sammons, S.; Olson, V.; Frace, M.; Nakazawa, Y.; Czerny, C.P.; Tryland, M.; Kolodziejek, J.; et al. Chasing Jenner’s vaccine: Revisiting cowpox virus classification. PLoS ONE 2011, 6, e23086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Gigante, C.; Khmaladze, E.; Liu, P.; Tang, S.; Wilkins, K.; Zhao, K.; Davidson, W.; Nakazawa, Y.; Maghlakelidze, G.; et al. Genome sequences of Akhmeta virus, an early divergent Old World Orthopoxvirus. Viruses 2018, 10, 252. [Google Scholar] [CrossRef] [Green Version]
- Springer, Y.P.; Hsu, C.H.; Werle, Z.R.; Olson, L.E.; Cooper, M.P.; Castrodale, L.J.; Fowler, N.; McCollum, A.M.; Goldsmith, C.S.; Emerson, G.L.; et al. Novel orthopoxvirus infection in an Alaska resident. Clin. Infect. Dis. 2017, 64, 1737–1741. [Google Scholar] [CrossRef]
- Gruber, C.; Giombini, E.; Selleri, M.; Tausch, S.H.; Andrusch, A.; Tyshaieva, A.; Cardeti, G.; Lorenzetti, R.; De Marco, L.; Carletti, F.; et al. Whole genome characterization of Orthopoxvirus (OPV) Abatino, a zoonotic virus representing a putative novel clade of Old World orthopoxviruses. Viruses 2018, 10, 546. [Google Scholar] [CrossRef] [Green Version]
- Duggan, A.T.; Perdomo, M.F.; Piombino-Mascali, D.; Marciniak, S.; Poinar, D.; Emery, M.V.; Buchmann, J.P.; Duchêne, S.; Jankauskas, R.; Humphreys, M.; et al. 17th century variola virus reveals the recent history of smallpox. Curr. Biol. 2016, 26, 3407–3412. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, G.; Neukamm, J.; Baalsrud, H.T.; Breidenstein, A.M.; Ravinet, M.; Phillips, C.; Rühli, F.; Bouwman, A.; Schuenemann, V.J. Variola virus genome sequenced from an eighteenth-century museum specimen supports the recent origin of smallpox. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190572. [Google Scholar] [CrossRef] [PubMed]
- Pajer, P.; Dresler, J.; Kabíckova, H.; Písa, L.; Aganov, P.; Fucik, K.; Elleder, D.; Hron, T.; Kuzelka, V.; Velemínsky, P.; et al. Characterization of two historic smallpox specimens from a Czech museum. Viruses 2017, 9, 200. [Google Scholar] [CrossRef] [PubMed]
- Smithson, C.; Imbery, J.; Upton, C. Re-assembly and analysis of an ancient variola virus genome. Viruses 2017, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.F.; Duggan, A.T.; Poinar, H.N.; Holmes, E.C. Comment: Characterization of two historic smallpox specimens from a Czech museum. Viruses 2017, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.L.; Irausquina, S.; Friedman, R. The evolutionary biology of poxviruses. Infect. Genet. Evol. 2010, 10, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Babkin, I.V.; Shchelkunov, S.N. The time scale in poxvirus evolution. Mol. Biol. Mosk. 2006, 40, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Babkin, I.V.; Babkina, I.N. Molecular dating in the evolution of vertebrate poxviruses. Intervirology 2011, 54, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Babkin, I.V.; Babkina, I.N. A retrospective study of the orthopoxvirus molecular evolution. Infect. Genet. Evol. 2012, 12, 1597–1604. [Google Scholar] [CrossRef]
- Firth, C.; Kitchen, A.; Shapiro, B.; Suchard, M.A.; Holmes, E.C.; Rambaut, A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol. Biol. Evol. 2010, 27, 2038–2051. [Google Scholar] [CrossRef] [Green Version]
- Esposito, J.J.; Sammons, S.A.; Frace, A.M.; Osborne, J.D.; Olsen-Rasmussen, M.; Zhang, M.; Govil, D.; Damon, I.K.; Kline, R.; Laker, M.; et al. Genome sequence diversity and clues to the evolution of variola smallpox virus. Science 2006, 313, 807–812. [Google Scholar] [CrossRef] [Green Version]
- Esteban, D.J.; Hutchinson, A.P. Genes in the terminal regions of orthopoxvirus genomes experience adaptive molecular evolution. BMC Genom. 2011, 12, e261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archard, L.C.; Mackett, M. Restriction endonuclease analysis of red cowpox virus and its white pock variant. J. Gen. Virol. 1979, 45, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Delhon, G.; Afonso, C.L.; Lu, Z.; Zsak, L.; Sandybaev, N.T.; Kerembekova, U.Z.; Zaitsev, V.L.; Kutish, G.F.; Rock, D.L. Genome of horsepox virus. J. Virol. 2006, 80, 9244–9258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, B.; Rambaut, A.; Drummond, A.J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006, 23, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Lam, T.T.; Carvalho, L.M.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [Green Version]
- Lefkowitz, E.J.; Wang, C.; Upton, C. Poxviruses: Past, present and future. Virus Res. 2006, 117, 105–118. [Google Scholar] [CrossRef]
- Upton, C.; Slack, S.; Hunter, A.L.; Ehlers, A.; Roper, R.L. Poxvirus orthologous clusters: Toward defining the minimum essential poxvirus genome. J. Virol. 2003, 77, 7590–7600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babkin, I.V.; Babkina, I.N. The origin of the variola virus. Viruses 2015, 7, 1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrickson, R.C.; Wang, C.; Hatcher, E.L.; Lefkowitz, E.J. Orthopoxvirus genome evolution: The role of gene loss. Viruses 2010, 2, 1933. [Google Scholar] [CrossRef] [PubMed]
- McLysaght, A.; Baldi, P.F.; Gaut, B.S. Extensive gene gain associated with adaptive evolution of poxviruses. Proc. Natl. Acad. Sci. USA 2003, 100, 15655–15660. [Google Scholar] [CrossRef] [Green Version]
- Babkina, I.N.; Babkin, I.V.; Le, U.; Ropp, S.; Kline, R.; Damon, I.; Esposito, J.; Sandakhchiev, L.S.; Shchelkunov, S.N. Phylogenetic comparison of the genomes of different strains of variola virus. Dokl. Biochem. Biophys. 2004, 398, 316–319. [Google Scholar] [CrossRef]
- Wertheim, J.O. Viral evolution: Mummy virus challenges presumed history of smallpox. Curr. Biol. 2017, 27, R119–R120. [Google Scholar] [CrossRef]
- Pomeroy, L.W.; Bjornstad, O.N.; Holmes, E.C. The evolutionary and epidemiological dynamics of the paramyxoviridae. J. Mol. Evol. 2008, 66, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Morgan, K. Slavery and the British Empire: From Africa to America; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Hopkins, D.R. The Greatest Killer: Smallpox in History; University of Chicago Press: Chicago, IL, USA, 2002. [Google Scholar]
- Biagini, P.; Theves, C.; Balaresque, P.; Geraut, A.; Cannet, C.; Keyser, C.; Nikolaeva, D.; Gerard, P.; Duchesne, S.; Orlando, L.; et al. Variola virus in a 300-year-old Siberian mummy. N. Engl. J. Med. 2012, 367, 2057–2059. [Google Scholar] [CrossRef] [Green Version]
- Hu, A.; Meehl, G.A.; Otto-Bliesner, B.L.; Waelbroeck, C.; Han, W.; Loutre, M.F.; Lambeck, K.; Mitrovica, J.X.; Rosenbloom, N. Influence of Bering Strait flow and North Atlantic circulation on glacial sea-level changes. Nature Geosci. 2010, 3, 118–121. [Google Scholar] [CrossRef]
- Doty, J.B.; Maghlakelidze, G.; Sikharulidze, I.; Tu, S.L.; Morgan, C.N.; Mauldin, M.R.; Parkadze, O.; Kartskhia, N.; Turmanidze, M.; Matheny, A.M.; et al. Isolation and characterization of Akhmeta virus from wild-caught rodents (Apodemus spp.) in Georgia. J. Virol. 2019, 93, e00966–e01019. [Google Scholar] [CrossRef] [Green Version]
- Vora, N.M.; Li, Y.; Geleishvili, M.; Emerson, G.L.; Khmaladze, E.; Maghlakelidze, G.; Navdarashvili, A.; Zakhashvili, K.; Kokhreidze, M.; Endeladze, M.; et al. Human infection with a zoonotic orthopoxvirus in the country of Georgia. N. Engl. J. Med. 2015, 372, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y.; Tong, K.J.; Foster, C.S.; Ritchie, A.M.; Lo, N.; Crisp, M.D. Biogeographic calibrations for the molecular clock. Biol. Lett. 2015, 11, 20150194. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No | Species | Strain Name | Country of Isolation | Year of Isolation | Accession Number |
|---|---|---|---|---|---|
| 1. | Abatino-virus | Abatino | Italy | 2015 | MH816996 |
| 2. | Akhmeta-virus | Akhmeta-88 | Georgia | 2013 | MH607141 |
| 3. | Akhmeta-virus | Vani-2010 | Georgia | 2010 | MH607143 |
| 4. | Alaskapox-virus | Alaska-2015 | The USA ** | 2015 | MN240300 |
| 5. | CMLV | M-96 | Kazakhstan | 1996 | NC003391 |
| 6. | CMLV | Negev | Israel | 2016 | MK910851 |
| 7. | CPXV | Austria | Austria | 1999 | HQ407377 |
| 8. | CPXV | Cepad-333 | France | 2009 | MK035753 |
| 9. | CPXV | FM2292 | Germany | 2011 | LN864566 |
| 10. | CPXV | GerMKY | Germany | 2010 | LT896721 |
| 11. | CPXV | HumGra07-1 | Germany | 2007 | KC813510 |
| 12. | CPXV | JagKre08-1 | Germany | 2008 | KC813497 |
| 13. | CPXV | Kostroma | Russia | 2015 | KY369926 |
| 14. | CPXV | Leo-2015-5 | Germany | 2015 | MK035756 |
| 15. | CPXV | Norway-MAN | Norway | 1994 | HQ420899 |
| 16. | ECTV | Naval | The UK | 1996 | KJ563295 |
| 17. | MPXV | 3019 | Nigeria | 2017 | MK783028 |
| 18. | MPXV | DRC-6-0950 | Congo | 2006 | JX878407 |
| 19. | MPXV | IvoryCoast2012 | Ivory Coast | 2012 | KJ136820 |
| 20. | MPXV | Sudan-2005-01 | Sudan | 2005 | KC257459 |
| 21. | RCNV | Herman | The USA | 1964 | KP143769 |
| 22. | SKPV | WA | The USA | 1978 | KU749310 |
| 23. | TATV | Dahomey | Benin | 1975 | NC008291 |
| 24. | VACV | MNR-76 | Mongolia | 1976 | DQ792504 |
| 25. | VARV | 001 | Nigeria | 1969 | DQ441434 |
| 26. | VARV | 005 | Guinea | 1969 | DQ441426 |
| 27. | VARV | 102 | South-Africa | 1965 | DQ441435 |
| 28. | VARV | 103 | South-Africa | 1965 | DQ441436 |
| 29. | VARV | 7124-Vellore | India | 1964 | DQ437585 |
| 30. | VARV | 7125-Vellore | India | 1964 | DQ437586 |
| 31. | VARV | BSH75 | Bangladesh | 1975 | L22579 |
| 32. | VARV | Butler | The UK * | 1952 | DQ441447 |
| 33. | VARV | China Horn | China | 1948 | DQ437582 |
| 34. | VARV | Congo-9 | Congo | 1970 | DQ437583 |
| 35. | VARV | Eth16 | Ethiopia | 1972 | DQ441424 |
| 36. | VARV | Eth17 | Ethiopia | 1972 | DQ441425 |
| 37. | VARV | GAR66 | Brazil | 1966 | Y16780 |
| 38. | VARV | Harper | Japan | 1951 | DQ441430 |
| 39. | VARV | Harvey | The UK | 1946 | DQ441444 |
| 40. | VARV | Heidelberg | Germany | 1958 | DQ437584 |
| 41. | VARV | Higgins | The UK | 1947 | DQ441446 |
| 42. | VARV | Hinden | The UK | 1946 | DQ441445 |
| 43. | VARV | Juba | Sudan | 1947 | DQ441440 |
| 44. | VARV | K1629 | Kuwait | 1967 | DQ441433 |
| 45. | VARV | KaliMuthu-M50 | India | 1953 | DQ441427 |
| 46. | VARV | Kembula | Tanzania | 1965 | DQ441443 |
| 47. | VARV | Lee | Korea | 1947 | DQ441432 |
| 48. | VARV | NewDelhi | India | 1953 | DQ441428 |
| 49. | VARV | nur-islam | Bangladesh | 1974 | DQ441420 |
| 50. | VARV | Rafig-Lahore | Pakistan | 1969 | DQ437589 |
| 51. | VARV | Rumbec | Sudan | 1947 | DQ441441 |
| 52. | VARV | Shahzaman | Bangladesh | 1974 | DQ441421 |
| 53. | VARV | Solaiman | Bangladesh | 1974 | DQ441422 |
| 54. | VARV | Stillwell | Japan | 1951 | DQ441431 |
| 55. | VARV | Tabriz | Iran | 1972 | DQ437587 |
| 56. | VARV | V1588 | Czechia | 1937 | LT706529 |
| 57. | VARV | V563 | Czechia | 1933 | LT706528 |
| 58. | VARV | v66-39 | Brazil | 1966 | DQ441419 |
| 59. | VARV | V68-258 | Sierra-Leone | 1969 | DQ441437 |
| 60. | VARV | v68-59 | Benin | 1968 | DQ441416 |
| 61. | VARV | V70-222 | Sumatra | 1970 | DQ437591 |
| 62. | VARV | V70-228 | Sumatra | 1970 | DQ441442 |
| 63. | VARV | v72-143 | Botswana | 1972 | DQ441417 |
| 64. | VARV | V72-164 | Yugoslavia | 1972 | DQ441448 |
| 65. | VARV | V72-199 | Syria | 1972 | DQ437592 |
| 66. | VARV | V73-175 | Nepal | 1973 | DQ437588 |
| 67. | VARV | v73-225 | Botswana | 1973 | DQ441418 |
| 68. | VARV | v74-227 | Congo | 1970 | DQ441423 |
| 69. | VARV | v75-550-Banu | Bangladesh | 1975 | DQ437581 |
| 70. | VARV | V77-1252 | Somalia | 1977 | DQ441438 |
| 71. | VARV | V77-1605 | Somalia | 1977 | DQ441439 |
| 72. | VARV | V77-2479 | Somalia | 1977 | DQ437590 |
| 73. | VARV | Variolator4 | Afghanistan | 1970 | DQ437580 |
| 74. | VARV | VD21 | Lithuania | 1654 | BK010317 |
| 75. | VARV | VK281 | Denmark | 938 | LR800246 |
| 76. | VARV | VK382 | Sweden | 705 | LR800245 |
| 77. | VARV | VK388 | Norway | 628 | LR800244 |
| 78. | VARV | VK470 | Russia | 975 | LR800247 |
| 79. | VARV | Yamada-MS-2A | Japan | 1946 | DQ441429 |
| 80. | VPXV | CA | The USA | 1985 | KU749311 |
| Genome Region and Approach used for Timing | ||||||
|---|---|---|---|---|---|---|
| Complete VARV Genomes; Strict Clock [12] | Complete VARV Genomes; Relaxed Clock [14] | Complete VARV Genomes; Relaxed Clock [13] | Conservative Region of VARV Genomes; Strict Clock [15] | Complete VARV Genomes; Relaxed Clock [2] | Conservative Region of Orthopoxvirus Genomes; Relaxed Clock, this Study | |
| tMRCA–VARV/CMLV/TATV | - | - | - | - | - | 272 (13–494) |
| tMRCA–P1/P2 VARV | 1764 (1734–1793) | 1695 | 1809 (1797–1820) | 1623 (1579–1667) | 1705 (1588–1813) | 1694 (1580–1828) |
| tMRCA–P1 VARV | 1910 (1902–1917) | 1887 | 1911 (1908–1915) | 1881 (1861–1897) | - | 1908 (1888–1926) |
| tMRCA–P2 VARV | 1870 (1855–1885) | 1808 | 1886 (1877–1893) | 1794 (1754–1828) | - | 1878 (1825–1920) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babkin, I.V.; Babkina, I.N.; Tikunova, N.V. An Update of Orthopoxvirus Molecular Evolution. Viruses 2022, 14, 388. https://doi.org/10.3390/v14020388
Babkin IV, Babkina IN, Tikunova NV. An Update of Orthopoxvirus Molecular Evolution. Viruses. 2022; 14(2):388. https://doi.org/10.3390/v14020388
Chicago/Turabian StyleBabkin, Igor V., Irina N. Babkina, and Nina V. Tikunova. 2022. "An Update of Orthopoxvirus Molecular Evolution" Viruses 14, no. 2: 388. https://doi.org/10.3390/v14020388
APA StyleBabkin, I. V., Babkina, I. N., & Tikunova, N. V. (2022). An Update of Orthopoxvirus Molecular Evolution. Viruses, 14(2), 388. https://doi.org/10.3390/v14020388






