Conserved Residues Adjacent to ß-Barrel and Loop Intersection among Enterovirus VP1 Affect Viral Replication: Potential Target for Anti-Enteroviral Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Alignment of GenBank Sequences
2.2. Predicting Consensus Sequences and Protein Structure Modeling
2.3. Preparation of Reverse Genetics VP1 Variants
2.4. Plaque Forming Assay
2.5. Immunofluorescence Stain of rgVP1 Variants
2.6. Real-Time RT-PCR
2.7. One-Step Growth Curves and Temperature Sensitivity Assay
2.8. Thermostability
2.9. Binding Assay
2.10. Statistics
3. Results
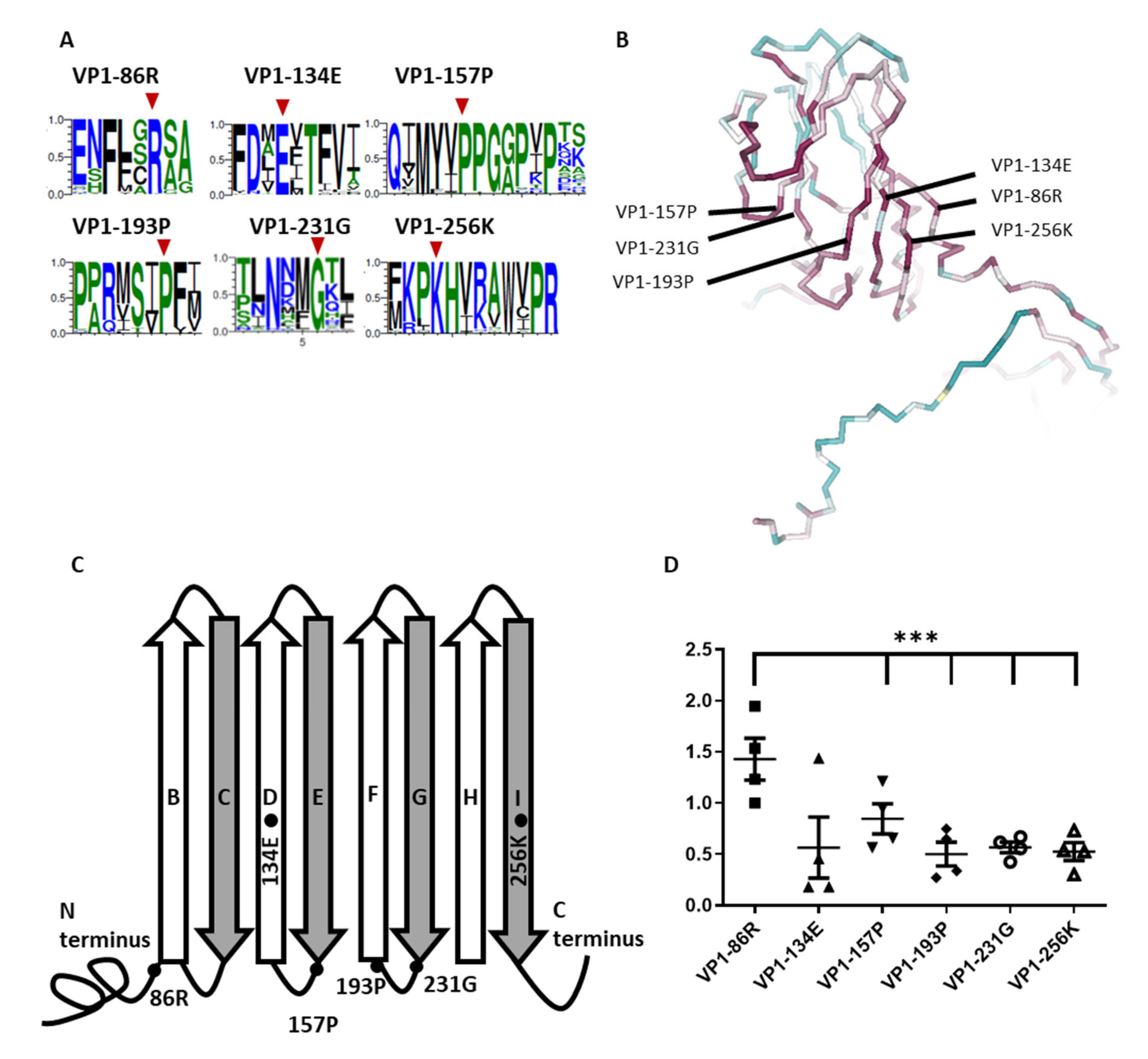
3.1. Identification of Critical Highly Conserved Residues
3.2. Conserved Residues Adjacent to β-Barrel and Loop Intersection Affect Viral Replication
3.3. Conserved Residues Adjacent to β-Barrel and Loop Intersection Affect RNA Level and Viral Infectious Titer
3.4. The Viral Properties of the rgVP1 Variants
3.5. The Viral Temperature Sensitivity and Stabilty of the rgVP1-P157A Variant of VP1
3.6. Effect of Amino Acid Interactions after Alanine Substution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; Garcia, M.L.; et al. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2021). Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef]
- Zell, R. Picornaviridae—The ever-growing virus family. Arch. Virol. 2018, 163, 299–317. [Google Scholar] [CrossRef]
- Chan, L.G.; Parashar, U.D.; Lye, M.S.; Ong, F.G.; Zaki, S.R.; Alexander, J.P.; Ho, K.K.; Han, L.L.; Pallansch, M.A.; Suleiman, A.B.; et al. Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: Clinical and pathological characteristics of the disease. For the outbreak study group. Clin. Infect. Dis. 2000, 31, 678–683. [Google Scholar] [CrossRef]
- Anasir, M.I.; Zarif, F.; Poh, C.L. Antivirals blocking entry of enteroviruses and therapeutic potential. J. Biomed. Sci. 2021, 28, 10. [Google Scholar] [CrossRef]
- Lin, J.Y.; Kung, Y.A.; Shih, S.R. Antivirals and vaccines for enterovirus a71. J. Biomed. Sci. 2019, 26, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Wang, X.; Hu, Z.; Gao, Q.; Sun, Y.; Li, X.; Porta, C.; Walter, T.S.; Gilbert, R.J.; Zhao, Y.; et al. Picornavirus uncoating intermediate captured in atomic detail. Nat. Commun. 2013, 4, 1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strauss, M.; Filman, D.J.; Belnap, D.M.; Cheng, N.; Noel, R.T.; Hogle, J.M. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J. Virol. 2015, 89, 4143–4157. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.F.; Chou, C.T.; Lei, H.Y.; Liu, C.C.; Wang, S.M.; Yan, J.J.; Su, I.J.; Wang, J.R.; Yeh, T.M.; Chen, S.H.; et al. A mouse-adapted enterovirus 71 strain causes neurological disease in mice after oral infection. J. Virol. 2004, 78, 7916–7924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.W.; Wang, Y.F.; Yu, C.K.; Su, I.J.; Wang, J.R. Mutations in VP2 and VP1 capsid proteins increase infectivity and mouse lethality of enterovirus 71 by virus binding and RNA accumulation enhancement. Virology 2012, 422, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.W.; Cheng, D.; Wang, J.R. Enterovirus A71: Virulence, antigenicity, and genetic evolution over the years. J. Biomed. Sci. 2019, 26, 81. [Google Scholar] [CrossRef]
- Laursen, N.S.; Wilson, I.A. Broadly neutralizing antibodies against influenza viruses. Antivir. Res. 2013, 98, 476–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stejskal, L.; Lees, W.D.; Moss, D.S.; Palor, M.; Bingham, R.J.; Shepherd, A.J.; Grove, J. Flexibility and intrinsic disorder are conserved features of hepatitis C virus E2 glycoprotein. PLoS Comput. Biol. 2020, 16, e1007710. [Google Scholar] [CrossRef]
- Ma, C.; Su, S.; Wang, J.; Wei, L.; Du, L.; Jiang, S. From Sars-CoV to Sars-CoV-2: Safety and broad-spectrum are important for coronavirus vaccine development. Microbes Infect. 2020, 22, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new endscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. Consurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. Consurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. Weblogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Kuriata, A.; Gierut, A.M.; Oleniecki, T.; Ciemny, M.P.; Kolinski, A.; Kurcinski, M.; Kmiecik, S. Cabs-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018, 46, W338–W343. [Google Scholar] [CrossRef] [Green Version]
- Plevka, P.; Perera, R.; Cardosa, J.; Kuhn, R.J.; Rossmann, M.G. Crystal structure of human enterovirus 71. Science 2012, 336, 1274. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, M.; Chen, E.R.; Hsu, K.H.; Twu, S.J.; Chen, K.T.; Tsai, S.F.; Wang, J.R.; Shih, S.R. An epidemic of enterovirus 71 infection in Taiwan. Taiwan enterovirus epidemic working group. N. Engl. J. Med. 1999, 341, 929–935. [Google Scholar] [CrossRef]
- La Monica, N.; Meriam, C.; Racaniello, V.R. Mapping of sequences required for mouse neurovirulence of poliovirus type 2 lansing. J. Virol. 1986, 57, 515–525. [Google Scholar] [CrossRef] [Green Version]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Tan, E.L.; Yong, L.L.; Quak, S.H.; Yeo, W.C.; Chow, V.T.; Poh, C.L. Rapid detection of enterovirus 71 by real-time TaqMan RT-PCR. J. Clin. Virol. 2008, 42, 203–206. [Google Scholar] [CrossRef]
- Arita, M.; Shimizu, H.; Nagata, N.; Ami, Y.; Suzaki, Y.; Sata, T.; Iwasaki, T.; Miyamura, T. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 2005, 86, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, W.; Ren, J.; Hu, Z.; Xu, J.; Lou, Z.; Li, X.; Yin, W.; Shen, X.; Porta, C.; et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 2012, 19, 424–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, S.; Li, G.; Wang, Y.; Gao, Q.; Wang, Y.; Cui, R.; Altmeyer, R.; Zou, G. Identification of positively charged residues in enterovirus 71 capsid protein VP1 essential for production of infectious particles. J. Virol. 2016, 90, 741–752. [Google Scholar] [CrossRef] [Green Version]
- Baggen, J.; Thibaut, H.J.; Strating, J.R.; van Kuppeveld, F.J. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef]
- Morgan, A.A.; Rubenstein, E. Proline: The distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS ONE 2013, 8, e53785. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Kishore, A.; Maity, D.; Sunanda, P.; Krishnarjuna, B.; Vappala, S.; Raghothama, S.; Kenyon, L.C.; Pal, D.; Das Sarma, J. A proline insertion-deletion in the spike glycoprotein fusion peptide of mouse hepatitis virus strongly alters neuropathology. J. Biol. Chem. 2019, 294, 8064–8087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rout, S.S.; Singh, M.; Shindler, K.S.; Das Sarma, J. One proline deletion in the fusion peptide of neurotropic mouse hepatitis virus (MHV) restricts retrograde axonal transport and neurodegeneration. J. Biol. Chem. 2020, 295, 6926–6935. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Zhao, Y.; Kotecha, A.; Fry, E.E.; Kelly, J.T.; Wang, X.; Rao, Z.; Rowlands, D.J.; Ren, J.; Stuart, D.I. Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. Nat. Microbiol. 2019, 4, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, P.; Gui, X.; Zhou, L.; Ge, X.; Guo, X.; Wills, J.W.; Han, J.; Yang, H. Induction of rod-shaped structures by herpes simplex virus glycoprotein I. J. Virol. 2020, 94, e00231-20. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion Mers-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puenpa, J.; Wanlapakorn, N.; Vongpunsawad, S.; Poovorawan, Y. The history of enterovirus A71 outbreaks and molecular epidemiology in the asia-pacific region. J. Biomed. Sci. 2019, 26, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, S.; Yan, D.; Liu, G.; Bai, R.; Wang, D.; Chen, L.; Zhu, H.; An, H.; Kew, O. Natural type 3/type 2 intertypic vaccine-related poliovirus recombinants with the first crossover sites within the VP1 capsid coding region. PLoS ONE 2010, 5, e15300. [Google Scholar] [CrossRef] [Green Version]
- Qing, J.; Wang, Y.; Sun, Y.; Huang, J.; Yan, W.; Wang, J.; Su, D.; Ni, C.; Li, J.; Rao, Z.; et al. Cyclophilin A associates with enterovirus-71 virus capsid and plays an essential role in viral infection as an uncoating regulator. PLoS Pathog. 2014, 10, e1004422. [Google Scholar] [CrossRef] [Green Version]
- Kojima, Y.; Ryo, A. Pinning down viral proteins: A new prototype for virus-host cell interaction. Front Microbiol. 2010, 1, 107. [Google Scholar] [CrossRef] [Green Version]
- Tang, H. Cyclophilin inhibitors as a novel HCV therapy. Viruses 2010, 2, 1621–1634. [Google Scholar] [CrossRef] [Green Version]
| Variants | rgVP1- R86A | rgVP1- E134A | rgVP1- P157A | rgVP1- P193A | rgVP1- G231A | rgVP1- K256A | |
|---|---|---|---|---|---|---|---|
| P0 1 | CPE | 2/5 2 | 5/5 | 5/5 | 1/5 | 3/5 | 0/5 |
| IFA 1 | 3/5 | 5/5 | 5/5 | 1/5 | 3/5 | 1/5 | |
| P1 | CPE | 1/5 | 5/5 | 5/5 | — | 1/5 | — |
| IFA | 1/5 | 5/5 | 5/5 | — | 1/5 | — | |
| P2–P5 3 | CPE | — | 5/5 | 5/5 | — | — | — |
| IFA | — | 5/5 | 5/5 | — | — | — | |
| Viruses (MOI = 0.1) | Titer at: (log10 PFU/mL) | ∆log 1 | Phenotype | |
|---|---|---|---|---|
| 35 °C | 39.5 °C | |||
| rgVP1-E134A | 7.70 | 6.5 | 1.20 | TR 2 |
| rgVP1-P157A | 8.18 | 0 | 8.18 | TS 2 |
| rgVP1(WT) | 7.70 | 6.5 | 1.20 | TR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-L.; Huang, S.-W.; Shen, C.-Y.; Cheng, D.; Wang, J.-R. Conserved Residues Adjacent to ß-Barrel and Loop Intersection among Enterovirus VP1 Affect Viral Replication: Potential Target for Anti-Enteroviral Development. Viruses 2022, 14, 364. https://doi.org/10.3390/v14020364
Huang Y-L, Huang S-W, Shen C-Y, Cheng D, Wang J-R. Conserved Residues Adjacent to ß-Barrel and Loop Intersection among Enterovirus VP1 Affect Viral Replication: Potential Target for Anti-Enteroviral Development. Viruses. 2022; 14(2):364. https://doi.org/10.3390/v14020364
Chicago/Turabian StyleHuang, Ya-Ling, Sheng-Wen Huang, Chun-Yu Shen, Dayna Cheng, and Jen-Ren Wang. 2022. "Conserved Residues Adjacent to ß-Barrel and Loop Intersection among Enterovirus VP1 Affect Viral Replication: Potential Target for Anti-Enteroviral Development" Viruses 14, no. 2: 364. https://doi.org/10.3390/v14020364
APA StyleHuang, Y.-L., Huang, S.-W., Shen, C.-Y., Cheng, D., & Wang, J.-R. (2022). Conserved Residues Adjacent to ß-Barrel and Loop Intersection among Enterovirus VP1 Affect Viral Replication: Potential Target for Anti-Enteroviral Development. Viruses, 14(2), 364. https://doi.org/10.3390/v14020364






