Visualization of Retroviral Gag-Genomic RNA Cellular Interactions Leading to Genome Encapsidation and Viral Assembly: An Overview
Abstract
1. Introduction
2. Retroviral Gag Precursors
3. Gag Oligomerization
4. Retroviral Packaging Signals and gRNA Dimerization
5. Visualization of gRNA Dimer in Cell
6. Where Is the gRNA Recruited?
7. How Retroviral gRNA-Gag Complexes Are Trafficked to the PM?
8. Molecular Mechanisms Occurring at the PM
9. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.-C.; Vivet-Boudou, V.; Smyth, R. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses 2016, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Rein, A.; Datta, S.A.K.; Jones, C.P.; Musier-Forsyth, K. Diverse Interactions of Retroviral Gag Proteins with RNAs. Trends Biochem. Sci. 2011, S0968000411000545. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.D.; Musier-Forsyth, K. Retroviral Gag Protein-RNA Interactions: Implications for Specific Genomic RNA Packaging and Virion Assembly. Semin. Cell Dev. Biol. 2019, 86, 129–139. [Google Scholar] [CrossRef]
- Dubois, N.; Marquet, R.; Paillart, J.-C.; Bernacchi, S. Retroviral RNA Dimerization: From Structure to Functions. Front. Microbiol. 2018, 9, 527. [Google Scholar] [CrossRef]
- Lever, A.M.L. HIV RNA Packaging and Lentivirus-Based Vectors. In Advances in Pharmacology; Elsevier: Amsterdam, The Netherlands, 2000; Volume 48, pp. 1–28. ISBN 978-0-12-032949-6. [Google Scholar]
- Comas-Garcia, M.; Davis, S.; Rein, A. On the Selective Packaging of Genomic RNA by HIV-1. Viruses 2016, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Summers, M.F. How Retroviruses Select Their Genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, T.A.; Schmidt, R.D.; Lew, K.A. Mason–Pfizer Monkey Virus (MPMV) Constitutive Transport Element (CTE) Functions in a Position-Dependent Manner. Virology 1997, 236, 118–129. [Google Scholar] [CrossRef]
- Jouvenet, N.; Neil, S.J.D.; Bess, C.; Johnson, M.C.; Virgen, C.A.; Simon, S.M.; Bieniasz, P.D. Plasma Membrane Is the Site of Productive HIV-1 Particle Assembly. PLoS Biol. 2006, 4, e435. [Google Scholar] [CrossRef]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the Interaction of HIV-1 Genomes and Gag during Assembly of Individual Viral Particles. PNAS 2009, 106, 19114–19119. [Google Scholar] [CrossRef]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the Biogenesis of Individual HIV-1 Virions in Live Cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA Genome Dimerizes on the Plasma Membrane in the Presence of Gag Protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef]
- Ivanchenko, S.; Godinez, W.J.; Lampe, M.; Kräusslich, H.-G.; Eils, R.; Rohr, K.; Bräuchle, C.; Müller, B.; Lamb, D.C. Dynamics of HIV-1 Assembly and Release. PLoS Pathog. 2009, 5, e1000652. [Google Scholar] [CrossRef]
- Hendrix, J.; Baumgärtel, V.; Schrimpf, W.; Ivanchenko, S.; Digman, M.A.; Gratton, E.; Kräusslich, H.-G.; Müller, B.; Lamb, D.C. Live-Cell Observation of Cytosolic HIV-1 Assembly Onset Reveals RNA-Interacting Gag Oligomers. J. Cell Biol. 2015, 210, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Sardo, L.; Hatch, S.C.; Chen, J.; Nikolaitchik, O.; Burdick, R.C.; Chen, D.; Westlake, C.J.; Lockett, S.; Pathak, V.K.; Hu, W.-S. Dynamics of HIV-1 RNA Near the Plasma Membrane during Virus Assembly. J. Virol. 2015, 89, 10832–10840. [Google Scholar] [CrossRef] [PubMed]
- Hubner, W.; Chen, P.; Portillo, A.D.; Liu, Y.; Gordon, R.E.; Chen, B.K. Sequence of Human Immunodeficiency Virus Type 1 (HIV-1) Gag Localization and Oligomerization Monitored with Live Confocal Imaging of a Replication-Competent, Fluorescently Tagged HIV-1. J. Virol. 2007, 81, 12596–12607. [Google Scholar] [CrossRef]
- Larson, D.R.; Ma, Y.M.; Vogt, V.M.; Webb, W.W. Direct Measurement of Gag–Gag Interaction during Retrovirus Assembly with FRET and Fluorescence Correlation Spectroscopy. J. Cell. Biol. 2003, 162, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Arhel, N.; Genovesio, A.; Kim, K.-A.; Miko, S.; Perret, E.; Olivo-Marin, J.-C.; Shorte, S.; Charneau, P. Quantitative Four-Dimensional Tracking of Cytoplasmic and Nuclear HIV-1 Complexes. Nat. Methods 2006, 3, 817–824. [Google Scholar] [CrossRef]
- McDonald, D. The inside Track on HIV. Nat. Methods 2006, 3, 782–783. [Google Scholar] [CrossRef]
- Rudner, L.; Nydegger, S.; Coren, L.V.; Nagashima, K.; Thali, M.; Ott, D.E. Dynamic Fluorescent Imaging of Human Immunodeficiency Virus Type 1 Gag in Live Cells by Biarsenical Labeling. J. Virol. 2005, 79, 4055–4065. [Google Scholar] [CrossRef]
- Boutant, E.; Bonzi, J.; Anton, H.; Nasim, M.B.; Cathagne, R.; Réal, E.; Dujardin, D.; Carl, P.; Didier, P.; Paillart, J.-C.; et al. Zinc Fingers in HIV-1 Gag Precursor Are Not Equivalent for GRNA Recruitment at the Plasma Membrane. Biophys. J. 2020, 119, 419–433. [Google Scholar] [CrossRef]
- Chen, J.; Grunwald, D.; Sardo, L.; Galli, A.; Plisov, S.; Nikolaitchik, O.A.; Chen, D.; Lockett, S.; Larson, D.R.; Pathak, V.K.; et al. Cytoplasmic HIV-1 RNA Is Mainly Transported by Diffusion in the Presence or Absence of Gag Protein. Proc. Natl. Acad. Sci. USA 2014, 111, E5205–E5213. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Clerté, C.; Chamontin, C.; Basyuk, E.; Lainé, S.; Hottin, J.; Bertrand, E.; Margeat, E.; Mougel, M. Imaging HIV-1 RNA Dimerization in Cells by Multicolor Super-Resolution and Fluctuation Microscopies. Nucleic Acids Res. 2016, 44, 7922–7934. [Google Scholar] [CrossRef]
- Carlson, L.-A.; Briggs, J.A.G.; Glass, B.; Riches, J.D.; Simon, M.N.; Johnson, M.C.; Müller, B.; Grünewald, K.; Kräusslich, H.-G. Three-Dimensional Analysis of Budding Sites and Released Virus Suggests a Revised Model for HIV-1 Morphogenesis. Cell Host Microbe 2008, 4, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global Changes in the RNA Binding Specificity of HIV-1 Gag Regulate Virion Genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Bieniasz, P.D. Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging. PLoS Pathog. 2010, 6, e1001200. [Google Scholar] [CrossRef] [PubMed]
- Bernacchi, S.; Abd El-Wahab, E.W.; Dubois, N.; Hijnen, M.; Smyth, R.P.; Mak, J.; Marquet, R.; Paillart, J.-C. HIV-1 Pr55 Gag Binds Genomic and Spliced RNAs with Different Affinity and Stoichiometry. RNA Biol. 2017, 14, 90–103. [Google Scholar] [CrossRef]
- Briggs, J.A.G.; Simon, M.N.; Gross, I.; Kräusslich, H.-G.; Fuller, S.D.; Vogt, V.M.; Johnson, M.C. The Stoichiometry of Gag Protein in HIV-1. Nat. Struct. Mol. Biol. 2004, 11, 672–675. [Google Scholar] [CrossRef]
- Freed, E.O. HIV-1 Assembly, Release and Maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef]
- Bussienne, C.; Marquet, R.; Paillart, J.-C.; Bernacchi, S. Post-Translational Modifications of Retroviral HIV-1 Gag Precursors: An Overview of Their Biological Role. IJMS 2021, 22, 2871. [Google Scholar] [CrossRef]
- Zhou, W.; Resh, M.D. Differential Membrane Binding of the Human Immunodeficiency Virus Type 1 Matrix Protein. J. Virol. 1996, 70, 8540–8548. [Google Scholar] [CrossRef]
- Paillart, J.-C.; Göttlinger, H.G. Opposing Effects of Human Immunodeficiency Virus Type 1 Matrix Mutations Support a Myristyl Switch Model of Gag Membrane Targeting. J. Virol. 1999, 73, 2604–2612. [Google Scholar] [CrossRef]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic Switch Regulates Myristate Exposure in the HIV-1 Matrix Protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Sandefur, S.; Smith, R.M.; Varthakavi, V.; Spearman, P. Mapping and Characterization of the N-Terminal I Domain of Human Immunodeficiency Virus Type 1 Pr55 Gag. J. Virol. 2000, 74, 7238–7249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ono, A.; Freed, E.O. Cell-Type-Dependent Targeting of Human Immunodeficiency Virus Type 1 Assembly to the Plasma Membrane and the Multivesicular Body. J. Virol. 2004, 78, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, V.; Ono, A. Molecular Determinants That Regulate Plasma Membrane Association of HIV-1 Gag. J. Mol. Biol. 2011, 410, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, V.; Hogue, I.B.; Boyko, V.; Hu, W.-S.; Ono, A. Interaction between the Human Immunodeficiency Virus Type 1 Gag Matrix Domain and Phosphatidylinositol-(4,5)-Bisphosphate Is Essential for Efficient Gag Membrane Binding. J. Virol. 2008, 82, 2405–2417. [Google Scholar] [CrossRef]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural Basis for Targeting HIV-1 Gag Proteins to the Plasma Membrane for Virus Assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef]
- Jones, C.P.; Datta, S.A.K.; Rein, A.; Rouzina, I.; Musier-Forsyth, K. Matrix Domain Modulates HIV-1 Gag’s Nucleic Acid Chaperone Activity via Inositol Phosphate Binding. J. Virol. 2011, 85, 1594–1603. [Google Scholar] [CrossRef]
- Todd, G.C.; Duchon, A.; Inlora, J.; Olson, E.D.; Musier-Forsyth, K.; Ono, A. Inhibition of HIV-1 Gag–Membrane Interactions by Specific RNAs. RNA 2017, 23, 395–405. [Google Scholar] [CrossRef]
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing Mechanisms Involving RNA and Lipids Regulate HIV-1 Gag Membrane Binding through the Highly Basic Region of the Matrix Domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605. [Google Scholar] [CrossRef]
- Bieniasz, P.; Telesnitsky, A. Multiple, Switchable Protein:RNA Interactions Regulate Human Immunodeficiency Virus Type 1 Assembly. Annu. Rev. Virol. 2018, 5, 165–183. [Google Scholar] [CrossRef]
- Munro, J.B.; Nath, A.; Färber, M.; Datta, S.A.K.; Rein, A.; Rhoades, E.; Mothes, W. A Conformational Transition Observed in Single HIV-1 Gag Molecules during In Vitro Assembly of Virus-Like Particles. J. Virol. 2014, 88, 3577–3585. [Google Scholar] [CrossRef] [PubMed]
- Obr, M.; Ricana, C.L.; Nikulin, N.; Feathers, J.-P.R.; Klanschnig, M.; Thader, A.; Johnson, M.C.; Vogt, V.M.; Schur, F.K.M.; Dick, R.A. Structure of the Mature Rous Sarcoma Virus Lattice Reveals a Role for IP6 in the Formation of the Capsid Hexamer. Nat. Commun. 2021, 12, 3226. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.M.; Dick, R.A.; Hagen, W.J.H.; Vogt, V.M.; Briggs, J.A.G. The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the P10 Domain in Assembly. J. Virol. 2015, 89, 10294–10302. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.R.; Yoo, S.; Vajdos, F.F.; von Schwedler, U.K.; Worthylake, D.K.; Wang, H.; McCutcheon, J.P.; Sundquist, W.I.; Hill, C.P. Structure of the Carboxyl-Terminal Dimerization Domain of the HIV-1 Capsid Protein. Science 1997, 278, 849–853. [Google Scholar] [CrossRef]
- Von Schwedler, U.K.; Stray, K.M.; Garrus, J.E.; Sundquist, W.I. Functional Surfaces of the Human Immunodeficiency Virus Type 1 Capsid Protein. JVI 2003, 77, 5439–5450. [Google Scholar] [CrossRef]
- Dorfman, T.; Bukovsky, A.; Ohagen, A.; Höglund, S.; Göttlinger, H.G. Functional Domains of the Capsid Protein of Human Immunodeficiency Virus Type 1. J. Virol. 1994, 68, 8180–8187. [Google Scholar] [CrossRef]
- Accola, M.A.; Strack, B.; Göttlinger, H.G. Efficient Particle Production by Minimal Gag Constructs Which Retain the Carboxy-Terminal Domain of Human Immunodeficiency Virus Type 1 Capsid-P2 and a Late Assembly Domain. J. Virol. 2000, 74, 5395–5402. [Google Scholar] [CrossRef]
- Ma, Y.M.; Vogt, V.M. Rous Sarcoma Virus Gag Protein-Oligonucleotide Interaction Suggests a Critical Role for Protein Dimer Formation in Assembly. J. Virol. 2002, 76, 5452–5462. [Google Scholar] [CrossRef]
- Fisher, R.J. Complex Interactions of HIV-1 Nucleocapsid Protein with Oligonucleotides. Nucleic Acids Res. 2006, 34, 472–484. [Google Scholar] [CrossRef]
- Houzet, L.; Morichaud, Z.; Didierlaurent, L.; Muriaux, D.; Darlix, J.-L.; Mougel, M. Nucleocapsid Mutations Turn HIV-1 into a DNA-Containing Virus. Nucleic Acids Res. 2008, 36, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Didierlaurent, L.; Houzet, L.; Morichaud, Z.; Darlix, J.-L.; Mougel, M. The Conserved N-Terminal Basic Residues and Zinc-Finger Motifs of HIV-1 Nucleocapsid Restrict the Viral CDNA Synthesis during Virus Formation and Maturation. Nucleic Acids Res. 2008, 36, 4745–4753. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Grigsby, I.F.; Gorelick, R.J.; Mansky, L.M.; Musier-Forsyth, K. Retrovirus-Specific Differences in Matrix and Nucleocapsid Protein-Nucleic Acid Interactions: Implications for Genomic RNA Packaging. J. Virol. 2014, 88, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.A.; Datta, S.A.K.; Nanda, H.; Fang, X.; Wen, Y.; Barros, M.; Wang, Y.-X.; Rein, A.; Vogt, V.M. Hydrodynamic and Membrane Binding Properties of Purified Rous Sarcoma Virus Gag Protein. J. Virol. 2015, 89, 10371–10382. [Google Scholar] [CrossRef]
- Zábranský, A.; Hoboth, P.; Hadravová, R.; Štokrová, J.; Sakalian, M.; Pichová, I. The Noncanonical Gag Domains P8 and n Are Critical for Assembly and Release of Mouse Mammary Tumor Virus. J. Virol. 2010, 84, 11555–11559. [Google Scholar] [CrossRef][Green Version]
- Abdusetir Cerfoglio, J.C.; González, S.A.; Affranchino, J.L. Structural Elements in the Gag Polyprotein of Feline Immunodeficiency Virus Involved in Gag Self-Association and Assembly. J. Gen. Virol. 2014, 95, 2050–2059. [Google Scholar] [CrossRef]
- Parent, L.J.; Bennett, R.P.; Craven, R.C.; Nelle, T.D.; Krishna, N.K.; Bowzard, J.B.; Wilson, C.B.; Puffer, B.A.; Montelaro, R.C.; Wills, J.W. Positionally Independent and Exchangeable Late Budding Functions of the Rous Sarcoma Virus and Human Immunodeficiency Virus Gag Proteins. J. Virol. 1995, 69, 5455–5460. [Google Scholar] [CrossRef]
- Yasuda, J.; Hunter, E. A Proline-Rich Motif (PPPY) in the Gag Polyprotein of Mason-Pfizer Monkey Virus Plays a Maturation-Independent Role in Virion Release. J. Virol. 1998, 72, 4095–4103. [Google Scholar] [CrossRef]
- Welker, L.; Paillart, J.-C.; Bernacchi, S. Importance of Viral Late Domains in Budding and Release of Enveloped RNA Viruses. Viruses 2021, 13, 1559. [Google Scholar] [CrossRef]
- Mendonça, L.; Sun, D.; Ning, J.; Liu, J.; Kotecha, A.; Olek, M.; Frosio, T.; Fu, X.; Himes, B.A.; Kleinpeter, A.B.; et al. CryoET Structures of Immature HIV Gag Reveal Six-Helix Bundle. Commun. Biol. 2021, 4, 481. [Google Scholar] [CrossRef]
- Wiegers, K.; Rutter, G.; Kottler, H.; Tessmer, U.; Hohenberg, H.; Kräusslich, H.-G. Sequential Steps in Human Immunodeficiency Virus Particle Maturation Revealed by Alterations of Individual Gag Polyprotein Cleavage Sites. J. Virol. 1998, 72, 2846–2854. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.B.; Russell, R.S.; Turner, D.; Liang, C. The T12I Mutation within the SP1 Region of Gag Restricts Packaging of Spliced Viral RNA into Human Immunodeficiency Virus Type 1 with Mutated RNA Packaging Signals and Mutated Nucleocapsid Sequence. Virology 2006, 344, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.S.; Roldan, A.; Detorio, M.; Hu, J.; Wainberg, M.A.; Liang, C. Effects of a Single Amino Acid Substitution within Thep2 Region of Human Immunodeficiency Virus Type 1 on Packagingof Spliced ViralRNA. J. Virol. 2003, 77, 12986–12995. [Google Scholar] [CrossRef] [PubMed]
- Wanaguru, M.; Barry, D.J.; Benton, D.J.; O’Reilly, N.J.; Bishop, K.N. Murine Leukemia Virus P12 Tethers the Capsid-Containing Pre-Integration Complex to Chromatin by Binding Directly to Host Nucleosomes in Mitosis. PLoS Pathog. 2018, 14, e1007117. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, H.S.; Khoo, K.K.; Garvey, M.; Waddington, L.; Leis, A.; Hijnen, M.; Velkov, T.; Dumsday, G.J.; McKinstry, W.J.; Mak, J. The Thermodynamics of Pr55Gag-RNA Interaction Regulate the Assembly of HIV. PLoS Pathog. 2017, 13, e1006221. [Google Scholar] [CrossRef]
- Dubois, N.; Khoo, K.K.; Ghossein, S.; Seissler, T.; Wolff, P.; McKinstry, W.J.; Mak, J.; Paillart, J.-C.; Marquet, R.; Bernacchi, S. The C-Terminal P6 Domain of the HIV-1 Pr55 Gag Precursor Is Required for Specific Binding to the Genomic RNA. RNA Biol. 2018, 15, 923–936. [Google Scholar] [CrossRef]
- Müller, B.; Daecke, J.; Fackler, O.T.; Dittmar, M.T.; Zentgraf, H.; Kräusslich, H.-G. Construction and Characterization of a Fluorescently Labeled Infectious Human Immunodeficiency Virus Type 1 Derivative. J. Virol. 2004, 78, 10803–10813. [Google Scholar] [CrossRef]
- Pornillos, O.; Higginson, D.S.; Stray, K.M.; Fisher, R.D.; Garrus, J.E.; Payne, M.; He, G.-P.; Wang, H.E.; Morham, S.G.; Sundquist, W.I. HIV Gag Mimics the Tsg101-Recruiting Activity of the Human Hrs Protein. J. Cell Biol. 2003, 162, 425–434. [Google Scholar] [CrossRef]
- Larson, D.R.; Johnson, M.C.; Webb, W.W.; Vogt, V.M. Visualization of Retrovirus Budding with Correlated Light and Electron Microscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 15453–15458. [Google Scholar] [CrossRef]
- Wachter, R. Photoconvertible Fluorescent Proteins and the Role of Dynamics in Protein Evolution. IJMS 2017, 18, 1792. [Google Scholar] [CrossRef]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A Guide to Choosing Fluorescent Proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, J.; Oswald, F.; Nienhaus, G.U. Fluorescent Proteins for Live Cell Imaging: Opportunities, Limitations, and Challenges. IUBMB Life 2009, 61, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.A.; Adams, S.R.; Tsien, R.Y. Specific Covalent Labeling of Recombinant Protein Molecules Inside Live Cells. Science 1998, 281, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-M.; Yu, X.-F. Identification and Characterization of Virus Assembly Intermediate Complexes in HIV-1-Infected CD4+T Cells. Virology 1998, 243, 78–93. [Google Scholar] [CrossRef][Green Version]
- Nermut, M.V.; Fassati, A. Structural Analyses of Purified Human Immunodeficiency Virus Type 1 Intracellular Reverse Transcription Complexes. J. Virol. 2003, 77, 8196–8206. [Google Scholar] [CrossRef][Green Version]
- Gomez, C.Y.; Hope, T.J. Mobility of Human Immunodeficiency Virus Type 1 Pr55 Gag in Living Cells. J. Virol. 2006, 80, 8796–8806. [Google Scholar] [CrossRef]
- Hogue, I.B.; Hoppe, A.; Ono, A. Quantitative Fluorescence Resonance Energy Transfer Microscopy Analysis of the Human Immunodeficiency Virus Type 1 Gag-Gag Interaction: Relative Contributions of the CA and NC Domains and Membrane Binding. J. Virol. 2009, 83, 7322–7336. [Google Scholar] [CrossRef]
- Alfadhli, A.; Dhenub, T.C.; Still, A.; Barklis, E. Analysis of Human Immunodeficiency Virus Type 1 Gag Dimerization-Induced Assembly. J. Virol. 2005, 79, 14498–14506. [Google Scholar] [CrossRef]
- Johnson, M.C.; Scobie, H.M.; Ma, Y.M.; Vogt, V.M. Nucleic Acid-Independent Retrovirus Assembly Can Be Driven by Dimerization. J. Virol. 2002, 76, 11177–11185. [Google Scholar] [CrossRef]
- Ma, Y.M.; Vogt, V.M. Nucleic Acid Binding-Induced Gag Dimerization in the Assembly of Rous Sarcoma Virus Particles In Vitro. J. Virol. 2004, 78, 52–60. [Google Scholar] [CrossRef]
- Roldan, A.; Russell, R.S.; Marchand, B.; Götte, M.; Liang, C.; Wainberg, M.A. In Vitro Identification and Characterization of an Early Complex Linking HIV-1 Genomic RNA Recognition and Pr55Gag Multimerization. J. Biol. Chem. 2004, 279, 39886–39894. [Google Scholar] [CrossRef]
- Guo, X.; Roy, B.B.; Hu, J.; Roldan, A.; Wainberg, M.A.; Liang, C. The R362A Mutation at the C-Terminus of CA Inhibits Packaging of Human Immunodeficiency Virus Type 1 RNA. Virology 2005, 343, 190–200. [Google Scholar] [CrossRef]
- Kaye, J.F.; Lever, A.M.L. Nonreciprocal Packaging of Human Immunodeficiency Virus Type 1 and Type 2 RNA: A Possible Role for the P2 Domain of Gag in RNA Encapsidation. J. Virol. 1998, 72, 5877–5885. [Google Scholar] [CrossRef]
- Carlson, L.-A.; Bai, Y.; Keane, S.C.; Doudna, J.A.; Hurley, J.H. Reconstitution of Selective HIV-1 RNA Packaging in Vitro by Membrane-Bound Gag Assemblies. eLife 2016, 5, e14663. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, K.H.; Chen, Y.; Grigsby, I.F.; Macdonald, P.J.; Smith, E.M.; Johnson, J.L.; Rawson, J.M.; Mansky, L.M.; Mueller, J.D. Characterization of Cytoplasmic Gag-Gag Interactions by Dual-Color Z-Scan Fluorescence Fluctuation Spectroscopy. Biophys. J. 2011, 100, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Pillai, V.N.; Ali, L.M.; Prabhu, S.G.; Krishnan, A.; Chameettachal, A.; Pitchai, F.N.N.; Mustafa, F.; Rizvi, T.A. A Stretch of Unpaired Purines in the Leader Region of Simian Immunodeficiency Virus (SIV) Genomic RNA Is Critical for Its Packaging into Virions. J. Mol. Biol. 2021, 433, 167293. [Google Scholar] [CrossRef] [PubMed]
- Ali, L.M.; Pitchai, F.N.N.; Vivet-Boudou, V.; Chameettachal, A.; Jabeen, A.; Pillai, V.N.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Role of Purine-Rich Regions in Mason-Pfizer Monkey Virus (MPMV) Genomic RNA Packaging and Propagation. Front. Microbiol. 2020, 11, 595410. [Google Scholar] [CrossRef] [PubMed]
- Jaballah, S.A.; Aktar, S.J.; Ali, J.; Phillip, P.S.; Al Dhaheri, N.S.; Jabeen, A.; Rizvi, T.A. A G–C-Rich Palindromic Structural Motif and a Stretch of Single-Stranded Purines Are Required for Optimal Packaging of Mason–Pfizer Monkey Virus (MPMV) Genomic RNA. J. Mol. Biol. 2010, 401, 996–1014. [Google Scholar] [CrossRef]
- Houzet, L.; Paillart, J.C.; Smagulova, F.; Maurel, S.; Morichaud, Z.; Marquet, R.; Mougel, M. HIV Controls the Selective Packaging of Genomic, Spliced Viral and Cellular RNAs into Virions through Different Mechanisms. Nucleic Acids Res. 2007, 35, 2695–2704. [Google Scholar] [CrossRef]
- Smyth, R.P.; Despons, L.; Huili, G.; Bernacchi, S.; Hijnen, M.; Mak, J.; Jossinet, F.; Weixi, L.; Paillart, J.-C.; von Kleist, M.; et al. Mutational Interference Mapping Experiment (MIME) for Studying RNA Structure and Function. Nat. Methods 2015, 12, 866–872. [Google Scholar] [CrossRef]
- Tounekti, N.; Mougel, M.; Roy, C.; Marquet, R.; Darlix, J.-L.; Paoletti, J.; Ehresmann, B.; Ehresmann, C. Effect of Dimerization on the Conformation of the Encapsidation Psi Domain of Moloney Murine Leukemia Virus RNA. J. Mol. Biol. 1992, 223, 205–220. [Google Scholar] [CrossRef]
- Mougel, M.; Barklis, E. A Role for Two Hairpin Structures as a Core RNA Encapsidation Signal in Murine Leukemia Virus Virions. J. Virol. 1997, 71, 8061–8065. [Google Scholar] [CrossRef] [PubMed]
- Oroudjev, E.M.; Kang, P.C.E.; Kohlstaedt, L.A. An Additional Dimer Linkage Structure in Moloney Murine Leukemia Virus RNA 1 1Edited by D. E. Draper. J. Mol. Biol. 1999, 291, 603–613. [Google Scholar] [CrossRef]
- Ly, H.; Parslow, T.G. Bipartite Signal for Genomic RNA Dimerization in Moloney Murine Leukemia Virus. J. Virol. 2002, 76, 3135–3144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, S.; Kaddis Maldonado, R.; Rye-McCurdy, T.; Binkley, C.; Bah, A.; Chen, E.C.; Rice, B.L.; Parent, L.J.; Musier-Forsyth, K. Rous Sarcoma Virus Genomic RNA Dimerization Capability In Vitro Is Not a Prerequisite for Viral Infectivity. Viruses 2020, 12, 568. [Google Scholar] [CrossRef]
- Lever, A.; Gottlinger, H.; Haseltine, W.; Sodroski, J. Identification of a Sequence Required for Efficient Packaging of Human Immunodeficiency Virus Type 1 RNA into Virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [CrossRef]
- Clever, J.; Sassetti, C.; Parslow, T.G. RNA Secondary Structure and Binding Sites for Gag Gene Products in the 5’ Packaging Signal of Human Immunodeficiency Virus Type 1. J. Virol. 1995, 69, 2101–2109. [Google Scholar] [CrossRef]
- Laughrea, M.; Jetté, L.; Mak, J.; Kleiman, L.; Liang, C.; Wainberg, M.A. Mutations in the Kissing-Loop Hairpin of Human Immunodeficiency Virus Type 1 Reduce Viral Infectivity as Well as Genomic RNA Packaging and Dimerization. J. Virol. 1997, 71, 3397–3406. [Google Scholar] [CrossRef]
- Ennifar, E.; Walter, P.; Ehresmann, B.; Ehresmann, C.; Dumas, P. Crystal Structures of Coaxially Stacked Kissing Complexes of the HIV-1 RNA Dimerization Iniziatin Site. Nat. Struct. Biol. 2001, 8, 1064–1068. [Google Scholar] [CrossRef]
- Ennifar, E.; Carpentier, P.; Ferrer, J.L.; Walter, P.; Dumas, P. X-Ray-Induced Debromination of Nucleic Acids at the Br K Absorption Edge and Implications for MAD Phasing. Acta Cryst. D Biol. Cryst. 2002, 58, 1262–1268. [Google Scholar] [CrossRef]
- Skripkin, E.; Paillart, J.C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. Identification of the Primary Site of the Human Immunodeficiency Virus Type 1 RNA Dimerization in Vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 4945–4949. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. A Loop-Loop “Kissing” Complex Is the Essential Part of the Dimer Linkage of Genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 5572–5577. [Google Scholar] [CrossRef] [PubMed]
- Weixlbaumer, A. Determination of Thermodynamic Parameters for HIV DIS Type Loop-Loop Kissing Complexes. Nucleic Acids Res. 2004, 32, 5126–5133. [Google Scholar] [CrossRef] [PubMed]
- Chameettachal, A.; Vivet-Boudou, V.; Pitchai, F.N.N.; Pillai, V.N.; Ali, L.M.; Krishnan, A.; Bernacchi, S.; Mustafa, F.; Marquet, R.; Rizvi, T.A. A Purine Loop and the Primer Binding Site Are Critical for the Selective Encapsidation of Mouse Mammary Tumor Virus Genomic RNA by Pr77Gag. Nucleic Acids Res. 2021, 49, 4668–4688. [Google Scholar] [CrossRef] [PubMed]
- Muriaux, D.; Fossé, P.; Paoletti, J. A Kissing Complex Together with a Stable Dimer Is Involved in the HIV-1 Lai RNA Dimerization Process in Vitro. Biochemistry 1996, 35, 5075–5082. [Google Scholar] [CrossRef]
- Bernacchi, S.; Ennifar, E.; Tóth, K.; Walter, P.; Langowski, J.; Dumas, P. Mechanism of Hairpin-Duplex Conversion for the HIV-1 Dimerization Initiation Site. J. Biol. Chem. 2005, 280, 40112–40121. [Google Scholar] [CrossRef]
- Levin, J.G.; Guo, J.; Rouzina, I.; Musier-Forsyth, K. Nucleic Acid Chaperone Activity of HIV-1 Nucleocapsid Protein: Critical Role in Reverse Transcription and Molecular Mechanism. In Progress in Nucleic Acid Research and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2005; Volume 80, pp. 217–286. ISBN 978-0-12-540080-0. [Google Scholar]
- Rist, M.J.; Marino, J.P. Mechanism of Nucleocapsid Protein Catalyzed Structural Isomerization of the Dimerization Initiation Site of HIV-1. Biochemistry 2002, 41, 14762–14770. [Google Scholar] [CrossRef]
- Brigham, B.S.; Kitzrow, J.P.; Reyes, J.-P.C.; Musier-Forsyth, K.; Munro, J.B. Intrinsic Conformational Dynamics of the HIV-1 Genomic RNA 5′UTR. Proc. Natl. Acad. Sci. USA 2019, 116, 10372–10381. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR Detection of Structures in the HIV-1 5’-Leader RNA That Regulate Genome Packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef]
- Tran, T.; Liu, Y.; Marchant, J.; Monti, S.; Seu, M.; Zaki, J.; Yang, A.L.; Bohn, J.; Ramakrishnan, V.; Singh, R.; et al. Conserved Determinants of Lentiviral Genome Dimerization. Retrovirology 2015, 12, 83. [Google Scholar] [CrossRef]
- Kalloush, R.M.; Vivet-Boudou, V.; Ali, L.M.; Pillai, V.N.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Stabilizing Role of Structural Elements within the 5´ Untranslated Region (UTR) and Gag Sequences in Mason-Pfizer Monkey Virus (MPMV) Genomic RNA Packaging. RNA Biol. 2019, 16, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Kemler, I.; Meehan, A.; Poeschla, E.M. Live-Cell Coimaging of the Genomic RNAs and Gag Proteins of Two Lentiviruses. J. Virol. 2010, 84, 6352–6366. [Google Scholar] [CrossRef] [PubMed]
- Pocock, G.M.; Becker, J.T.; Swanson, C.M.; Ahlquist, P.; Sherer, N.M. HIV-1 and M-PMV RNA Nuclear Export Elements Program Viral Genomes for Distinct Cytoplasmic Trafficking Behaviors. PLoS Pathog. 2016, 12, e1005565. [Google Scholar] [CrossRef]
- Becker, J.T.; Sherer, N.M. Subcellular Localization of HIV-1 Gag-Pol MRNAs Regulates Sites of Virion Assembly. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.-S. High Efficiency of HIV-1 Genomic RNA Packaging and Heterozygote Formation Revealed by Single Virion Analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef] [PubMed]
- St Johnston, D. Moving Messages: The Intracellular Localization of MRNAs. Nat. Rev. Mol. Cell Biol. 2005, 6, 363–375. [Google Scholar] [CrossRef]
- Fusco, D.; Bertrand, E.; Singer, R.H. Imaging of Single MRNAs in the Cytoplasm of Living Cells. In RNA Trafficking and Nuclear Structure Dynamics; Progress in Molecular and Subcellular Biology; Jeanteur, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 35, pp. 135–150. ISBN 978-3-540-74265-4. [Google Scholar]
- Sinck, L.; Richer, D.; Howard, J.; Alexander, M.; Purcell, D.F.J.; Marquet, R.; Paillart, J.-C. In Vitro Dimerization of Human Immunodeficiency Virus Type 1 (HIV-1) Spliced RNAs. RNA 2007, 13, 2141–2150. [Google Scholar] [CrossRef]
- Moore, M.D.; Nikolaitchik, O.A.; Chen, J.; Hammarskjöld, M.-L.; Rekosh, D.; Hu, W.-S. Probing the HIV-1 Genomic RNA Trafficking Pathway and Dimerization by Genetic Recombination and Single Virion Analyses. PLoS Pathog. 2009, 5, e1000627. [Google Scholar] [CrossRef]
- Maurel, S.; Houzet, L.; Garcia, E.L.; Telesnitsky, A.; Mougel, M. Characterization of a Natural Heterodimer between MLV Genomic RNA and the SD′ Retroelement Generated by Alternative Splicing. RNA 2007, 13, 2266–2276. [Google Scholar] [CrossRef]
- Maurel, S.; Mougel, M. Murine Leukemia Virus RNA Dimerization Is Coupled to Transcription and Splicing Processes. Retrovirology 2010, 7, 64. [Google Scholar] [CrossRef]
- Chen, E.C.; Maldonado, R.J.K.; Parent, L.J. Visualizing Rous Sarcoma Virus Genomic RNA Dimerization in the Nucleus, Cytoplasm, and at the Plasma Membrane. Viruses 2021, 13, 903. [Google Scholar] [CrossRef] [PubMed]
- Basyuk, E.; Boulon, S.; Skou Pedersen, F.; Bertrand, E.; Vestergaard Rasmussen, S. The Packaging Signal of MLV Is an Integrated Module That Mediates Intracellular Transport of Genomic RNAs. J. Mol. Biol. 2005, 354, 330–339. [Google Scholar] [CrossRef]
- Smagulova, F.; Maurel, S.; Morichaud, Z.; Devaux, C.; Mougel, M.; Houzet, L. The Highly Structured Encapsidation Signal of MuLV RNA Is Involved in the Nuclear Export of Its Unspliced RNA. J. Mol. Biol. 2005, 354, 1118–1128. [Google Scholar] [CrossRef]
- Abd El-Wahab, E.W.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.-C.; Marquet, R. Specific Recognition of the HIV-1 Genomic RNA by the Gag Precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef] [PubMed]
- Muriaux, D.; Darlix, J.-L. Properties and Functions of the Nucleocapsid Protein in Virus Assembly. RNA Biol. 2010, 7, 744–753. [Google Scholar] [CrossRef]
- Rein, A. Nucleic Acid Chaperone Activity of Retroviral Gag Proteins. RNA Biol. 2010, 7, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Rein, A.; Henderson, L.E.; Levin, J.G. Nucleic-Acid-Chaperone Activity of Retroviral Nucleocapsid Proteins: Significance for Viral Replication. Trends Biochem. Sci. 1998, 23, 297–301. [Google Scholar] [CrossRef]
- Paillart, J.C.; Berthoux, L.; Ottmann, M.; Darlix, J.L.; Marquet, R.; Ehresmann, B.; Ehresmann, C. A Dual Role of the Putative RNA Dimerization Initiation Site of Human Immunodeficiency Virus Type 1 in Genomic RNA Packaging and Proviral DNA Synthesis. J. Virol. 1996, 70, 8348–8354. [Google Scholar] [CrossRef]
- Berkhout, B.; van Wamel, J.L. Role of the DIS Hairpin in Replication of Human Immunodeficiency Virus Type 1. J. Virol. 1996, 70, 6723–6732. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Wu, B.; Nikolaitchik, O.A.; Mohan, P.R.; Chen, J.; Pathak, V.K.; Hu, W.-S. Visualizing the Translation and Packaging of HIV-1 Full-Length RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 6145–6155. [Google Scholar] [CrossRef]
- Parent, L.J. New Insights into the Nuclear Localization of Retroviral Gag Proteins. Nucleus 2011, 2, 92–97. [Google Scholar] [CrossRef]
- Garbitt-Hirst, R.; Kenney, S.P.; Parent, L.J. Genetic Evidence for a Connection between Rous Sarcoma Virus Gag Nuclear Trafficking and Genomic RNA Packaging. J. Virol. 2009, 83, 6790–6797. [Google Scholar] [CrossRef]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of Nucleocytoplasmic Transport of the Retroviral Gag Protein Depends on Sequential Binding of Karyopherins and Viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef]
- Kenney, S.P.; Lochmann, T.L.; Schmid, C.L.; Parent, L.J. Intermolecular Interactions between Retroviral Gag Proteins in the Nucleus. J. Virol. 2008, 82, 683–691. [Google Scholar] [CrossRef]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A Subset of Pr65gag Is Nucleus Associated in Murine Leukemia Virus-Infected Cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Schliephake, A.W.; Rethwilm, A. Nuclear Localization of Foamy Virus Gag Precursor Protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar] [CrossRef]
- Beyer, A.R.; Bann, D.V.; Rice, B.; Pultz, I.S.; Kane, M.; Goff, S.P.; Golovkina, T.V.; Parent, L.J. Nucleolar Trafficking of the Mouse Mammary Tumor Virus Gag Protein Induced by Interaction with Ribosomal Protein L9. J. Virol. 2013, 87, 1069–1082. [Google Scholar] [CrossRef]
- Cullen, B.R. Nuclear MRNA Export: Insights from Virology. Trends Biochem. Sci. 2003, 28, 419–424. [Google Scholar] [CrossRef]
- Butterfield-Gerson, K.L.; Scheifele, L.Z.; Ryan, E.P.; Hopper, A.K.; Parent, L.J. Importin-β Family Members Mediate Alpharetrovirus Gag Nuclear Entry via Interactions with Matrix and Nucleocapsid. J. Virol. 2006, 80, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Kaddis Maldonado, R.; Parent, L. Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors. Viruses 2016, 8, 257. [Google Scholar] [CrossRef]
- Yu, F.; Joshi, S.M.; Ma, Y.M.; Kingston, R.L.; Simon, M.N.; Vogt, V.M. Characterization of Rous Sarcoma Virus Gag Particles Assembled In Vitro. J. Virol. 2001, 75, 2753–2764. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Ryan, E.P.; Parent, L.J. Detailed Mapping of the Nuclear Export Signal in the Rous Sarcoma Virus Gag Protein. J. Virol. 2005, 79, 8732–8741. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear Entry and CRM1-Dependent Nuclear Export of the Rous Sarcoma Virus Gag Polyprotein. PNAS 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.J.K.; Rice, B.; Chen, E.C.; Tuffy, K.M.; Chiari, E.F.; Fahrbach, K.M.; Hope, T.J.; Parent, L.J. Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus. mBio 2020, 11. [Google Scholar] [CrossRef]
- Hernandez, F.P.; Sandri-Goldin, R.M. Bimolecular Fluorescence Complementation Analysis to Reveal Protein Interactions in Herpes Virus Infected Cells. Methods 2011, 55, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses 2020, 12, 1281. [Google Scholar] [CrossRef]
- Dupont, S.; Sharova, N.; DéHoratius, C.; Virbasius, C.-M.A.; Zhu, X.; Bukrinskaya, A.G.; Stevenson, M.; Green, M.R. A Novel Nuclear Export Activity in HIV-1 Matrix Protein Required for Viral Replication. Nature 1999, 402, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; Strappe, P.; Mok, H.-P.; Hicks, R.; Lever, A.M.L. HIV-1 Gag-RNA Interaction Occurs at a Perinuclear/Centrosomal Site; Analysis by Confocal Microscopy and FRET: HIV-1 Gag-RNA Interaction Occurs in a Perinuclear Region. Traffic 2005, 6, 741–755. [Google Scholar] [CrossRef]
- Baumgärtel, V.; Müller, B.; Lamb, D.C. Quantitative Live-Cell Imaging of Human Immunodeficiency Virus (HIV-1) Assembly. Viruses 2012, 4, 777–799. [Google Scholar] [CrossRef]
- Digman, M.A.; Stakic, M.; Gratton, E. Raster Image Correlation Spectroscopy and Number and Brightness Analysis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 518, pp. 121–144. ISBN 978-0-12-388422-0. [Google Scholar]
- Rhee, S.S.; Hunter, E. Myristylation Is Required for Intracellular Transport but Not for Assembly of D-Type Retrovirus Capsids. J. Virol. 1987, 61, 1045–1053. [Google Scholar] [CrossRef]
- Schultz, A.M.; Oroszlan, S. In Vivo Modification of Retroviral Gag Gene-Encoded Polyproteins by Myristic Acid. J. Virol. 1983, 46, 355–361. [Google Scholar] [CrossRef]
- Houzet, L.; Gay, B.; Morichaud, Z.; Briant, L.; Mougel, M. Intracellular Assembly and Budding of the Murine Leukemia Virus in Infected Cells. Retrovirology 2006, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Blot, V.; Perugi, F.; Gay, B.; Prévost, M.-C.; Briant, L.; Tangy, F.; Abriel, H.; Staub, O.; Dokhélar, M.-C.; Pique, C. Nedd4.1-Mediated Ubiquitination and Subsequent Recruitment of Tsg101 Ensure HTLV-1 Gag Trafficking towards the Multivesicular Body Pathway Prior to Virus Budding. J. Cell Sci. 2004, 117, 2357–2367. [Google Scholar] [CrossRef] [PubMed]
- Basyuk, E.; Galli, T.; Mougel, M.; Blanchard, J.-M.; Sitbon, M.; Bertrand, E. Retroviral Genomic RNAs Are Transported to the Plasma Membrane by Endosomal Vesicles. Dev. Cell 2003, 5, 161–174. [Google Scholar] [CrossRef]
- Jouvenet, N.; Lainé, S.; Pessel-Vivares, L.; Mougel, M. Cell Biology of Retroviral RNA Packaging. RNA Biol. 2011, 8, 572–580. [Google Scholar] [CrossRef]
- Welsch, S.; Keppler, O.T.; Habermann, A.; Allespach, I.; Krijnse-Locker, J.; Kräusslich, H.-G. HIV-1 Buds Predominantly at the Plasma Membrane of Primary Human Macrophages. PLoS Pathog. 2007, 3, e36. [Google Scholar] [CrossRef]
- Gladnikoff, M.; Rousso, I. Directly Monitoring Individual Retrovirus Budding Events Using Atomic Force Microscopy. Biophys. J. 2008, 94, 320–326. [Google Scholar] [CrossRef]
- Saffarian, S.; Kirchhausen, T. Differential Evanescence Nanometry: Live-Cell Fluorescence Measurements with 10-Nm Axial Resolution on the Plasma Membrane. Biophys. J. 2008, 94, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Ku, P.-I.; Miller, A.K.; Ballew, J.; Sandrin, V.; Adler, F.R.; Saffarian, S. Identification of Pauses during Formation of HIV-1 Virus Like Particles. Biophys. J. 2013, 105, 2262–2272. [Google Scholar] [CrossRef]
- Kemler, I.; Barraza, R.; Poeschla, E.M. Mapping the Encapsidation Determinants of Feline Immunodeficiency Virus. J. Virol. 2002, 76, 11889–11903. [Google Scholar] [CrossRef]
- Kerviel, A.; Thomas, A.; Chaloin, L.; Favard, C.; Muriaux, D. Virus Assembly and Plasma Membrane Domains: Which Came First? Virus Res. 2013, 171, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.R.; Göttlinger, H. The Role of Cellular Factors in Promoting HIV Budding. J. Mol. Biol. 2011, 410, 525–533. [Google Scholar] [CrossRef]
- Hurley, J.H.; Boura, E.; Carlson, L.-A.; Różycki, B. Membrane Budding. Cell 2010, 143, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Martin-Serrano, J.; Neil, S.J.D. Host Factors Involved in Retroviral Budding and Release. Nat. Rev. Microbiol. 2011, 9, 519–531. [Google Scholar] [CrossRef]
- Klingler, J.; Anton, H.; Réal, E.; Zeiger, M.; Moog, C.; Mély, Y.; Boutant, E. How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain. Viruses 2020, 12, 888. [Google Scholar] [CrossRef]
- Mouland, A.J.; Mercier, J.; Luo, M.; Bernier, L.; DesGroseillers, L.; Cohen, É.A. The Double-Stranded RNA-Binding Protein Staufen Is Incorporated in Human Immunodeficiency Virus Type 1: Evidence for a Role in Genomic RNA Encapsidation. J. Virol. 2000, 74, 5441–5451. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Clément, J.-F.; Martel, C.; Bériault, V.; Gatignol, A.; DesGroseillers, L.; Mouland, A.J. Identification of Staufen in the Human Immunodeficiency Virus Type 1 Gag Ribonucleoprotein Complex and a Role in Generating Infectious Viral Particles. Mol. Cell Biol. 2004, 24, 2637–2648. [Google Scholar] [CrossRef]
- Itano, M.S.; Arnion, H.; Wolin, S.L.; Simon, S.M. Recruitment of 7SL RNA to Assembling HIV-1 Virus-like Particles. Traffic 2018, 19, 36–43. [Google Scholar] [CrossRef]
- Strebel, K.; Khan, M.A. APOBEC3G Encapsidation into HIV-1 Virions: Which RNA Is It? Retrovirology 2008, 5, 55. [Google Scholar] [CrossRef]
- Hell, S.W.; Wichmann, J. Breaking the Diffraction Resolution Limit by Stimulated Emission: Stimulated-Emission-Depletion Fluorescence Microscopy. Opt. Lett. 1994, 19, 780. [Google Scholar] [CrossRef]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 2006, 3, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, X.; Xu, Z.; Zhang, X.; Gao, Y. Super-Resolution Fluorescence Microscopy for Single Cell Imaging. In Single Cell Biomedicine; Advances in Experimental Medicine and Biology; Gu, J., Wang, X., Eds.; Springer: Singapore, 2018; Volume 1068, pp. 59–71. ISBN 9789811305016. [Google Scholar]
- Han, R.; Li, Z.; Fan, Y.; Jiang, Y. Recent Advances in Super-Resolution Fluorescence Imaging and Its Applications in Biology. J. Genet. Genom. 2013, 40, 583–595. [Google Scholar] [CrossRef] [PubMed]

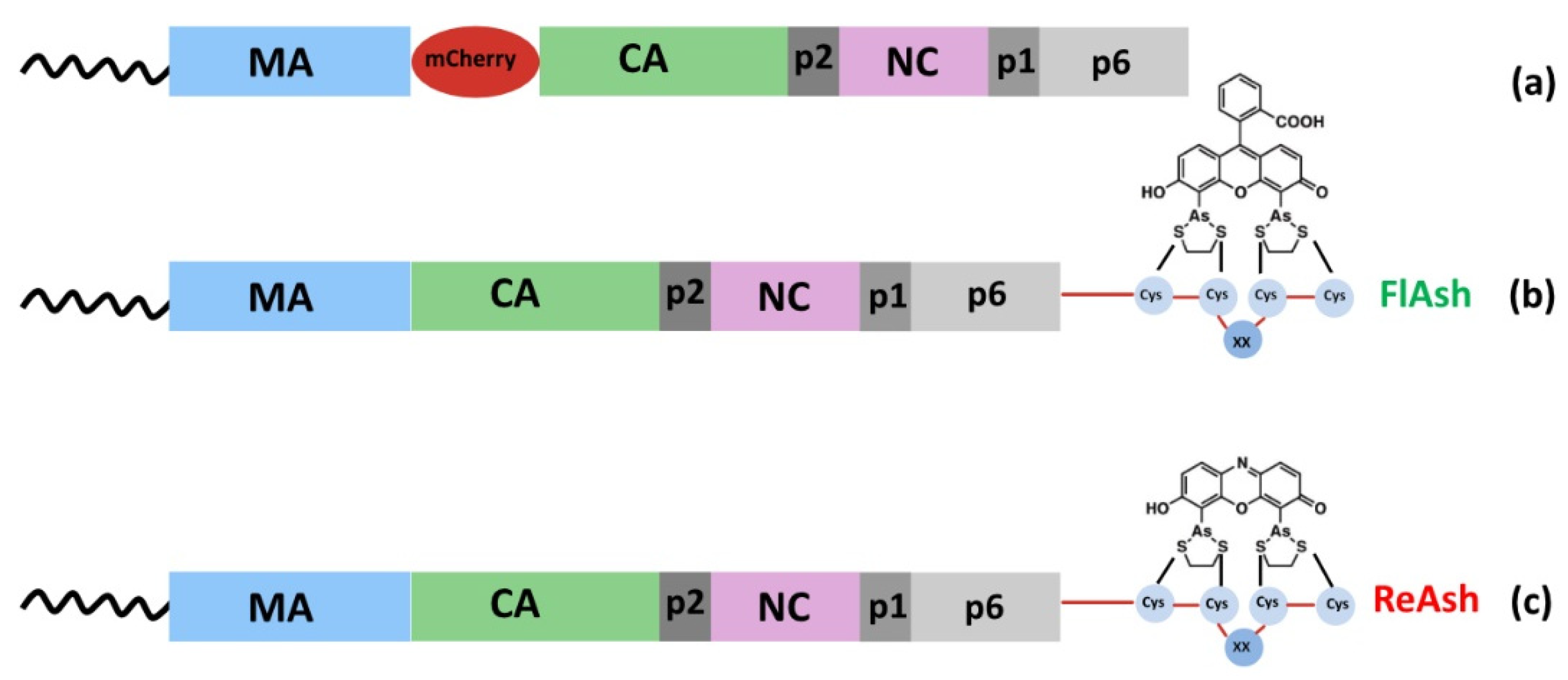
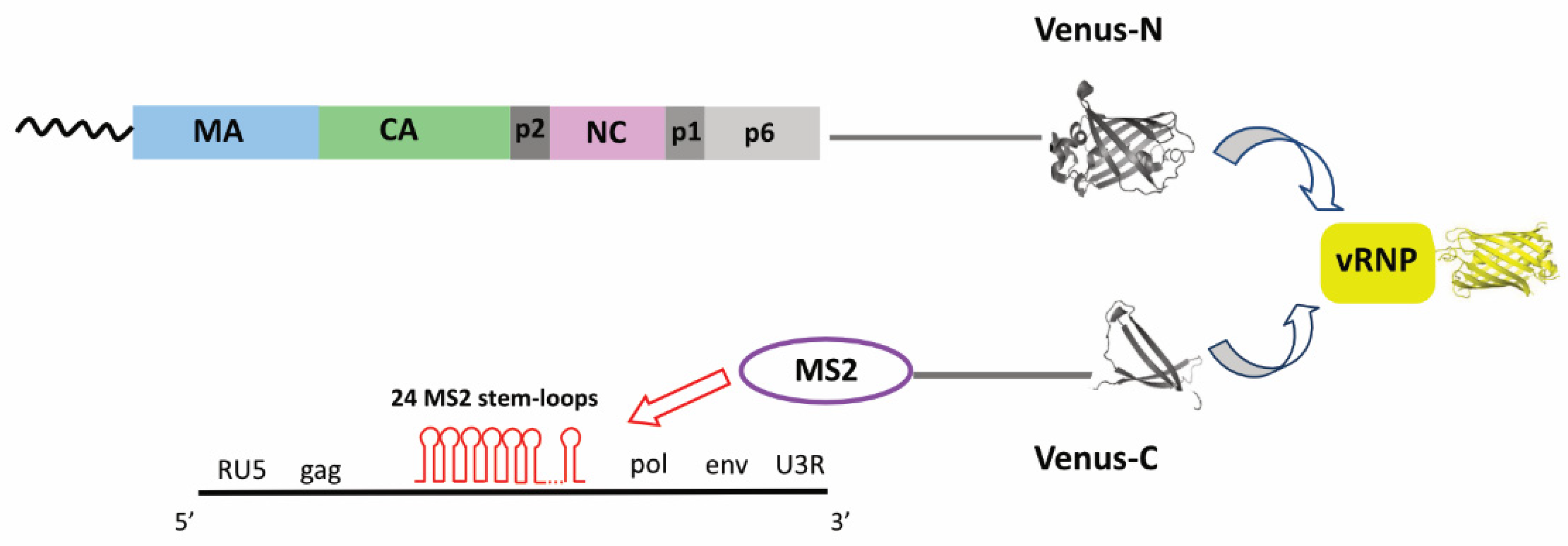
| Method | Advantages | Disadvantages |
|---|---|---|
| Atomic Force Microscopy (AM) | Appropriate to measure the distortion of the membrane (resolution <1 nm up to 1µm). | Technically limited time resolution of several minutes per frame. |
| Electron Microscopy (EM) | Resolution of finer spatial detail compared to the classical microscopy (0.2 nm). | Cells are fixed, and temporal resolution is lost. |
| Confocal Microscopy | Suitable method to investigate processes that are not limited to the PM (resolution 180 nm laterally and 500 nm axially). | Low signal-to-noise ratio could require stronger illumination resulting in photobleached samples. |
| FLIM-FRET | Only the measurement of donor lifetime is required and acceptors with poor quantum yield can be used. Less excitation is required because of wider emission filters. | Careful measure of lifetime for the donor without the acceptor is required for accurate calibration. |
| Fluorescence Fluctuation Spectroscopy (FFS) | Ensemble of microscopy tools (e.g., FCCS and RICS) appropriate to analyze biomolecular dynamics, interactions, and structural changes in living cells. | Highly stable light source is necessary. Cumulative effects of photobleaching are possible. Analysis of molecules with different diffusive properties, as it can be the case of Gag, is complicated by the relative excitation intensities, different diffusion times, and the number of diffusing molecules for each population. |
| FRAP | Suitable method for determining the kinetics of diffusion in cells to study cellular membrane diffusion and membrane anchoring. | The estimation of intrinsic photobleaching. The precise identification of several mobile species corresponding to various degrees of oligomerization or having different interactions with the membranes, can be difficult. |
| TIRF | Accurate determination of axial position within ∼200 nm of the specimen surface. Proper to study events near to the PM as the retroviral assembly sites. It displays good signal to noise ratio to allow quantification of the assembly of individual HIV-1 particles. | The fluorescence signal can be affected by azimuthal movement of the VLP. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernacchi, S. Visualization of Retroviral Gag-Genomic RNA Cellular Interactions Leading to Genome Encapsidation and Viral Assembly: An Overview. Viruses 2022, 14, 324. https://doi.org/10.3390/v14020324
Bernacchi S. Visualization of Retroviral Gag-Genomic RNA Cellular Interactions Leading to Genome Encapsidation and Viral Assembly: An Overview. Viruses. 2022; 14(2):324. https://doi.org/10.3390/v14020324
Chicago/Turabian StyleBernacchi, Serena. 2022. "Visualization of Retroviral Gag-Genomic RNA Cellular Interactions Leading to Genome Encapsidation and Viral Assembly: An Overview" Viruses 14, no. 2: 324. https://doi.org/10.3390/v14020324
APA StyleBernacchi, S. (2022). Visualization of Retroviral Gag-Genomic RNA Cellular Interactions Leading to Genome Encapsidation and Viral Assembly: An Overview. Viruses, 14(2), 324. https://doi.org/10.3390/v14020324






