Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Viral DNA Extraction
2.2. Virus Detection
2.3. Virus Amplification and Sequence Determination of Porcine Circovirus Type 2 (PCV2) ORF2
2.4. Bioinformatic Analysis
3. Results
3.1. Sample Screening and Co-Infection Status of PCV2
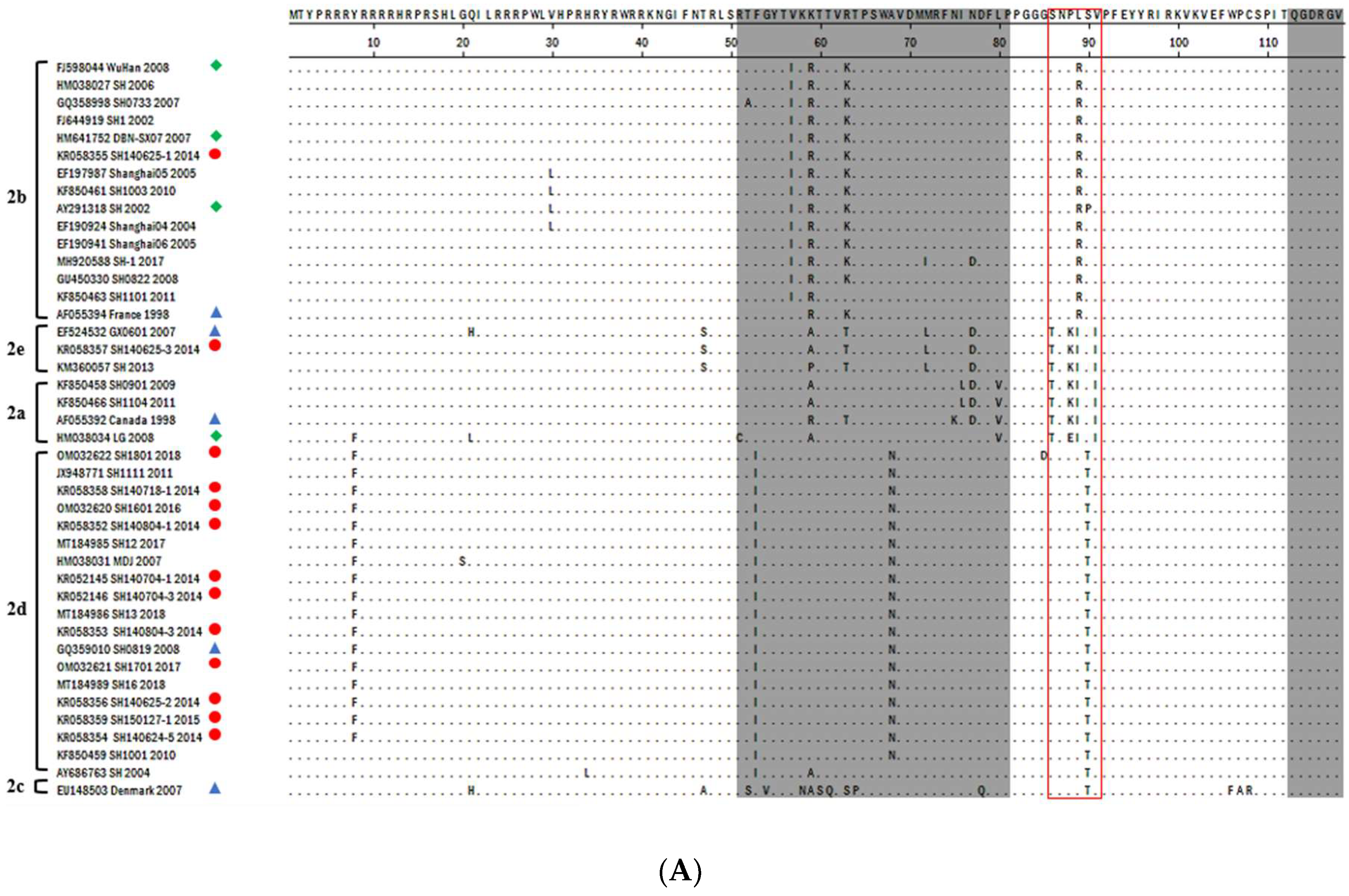
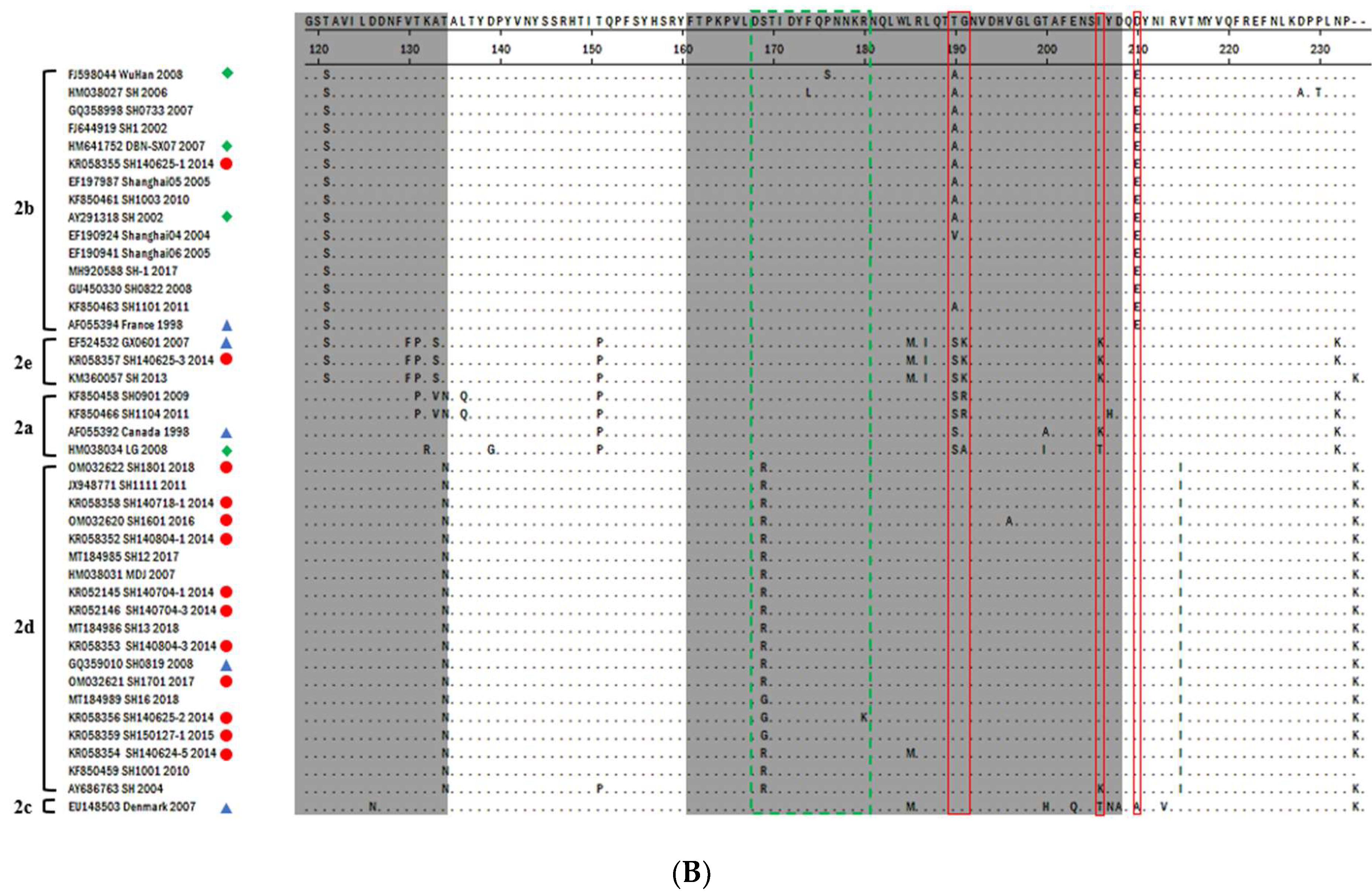
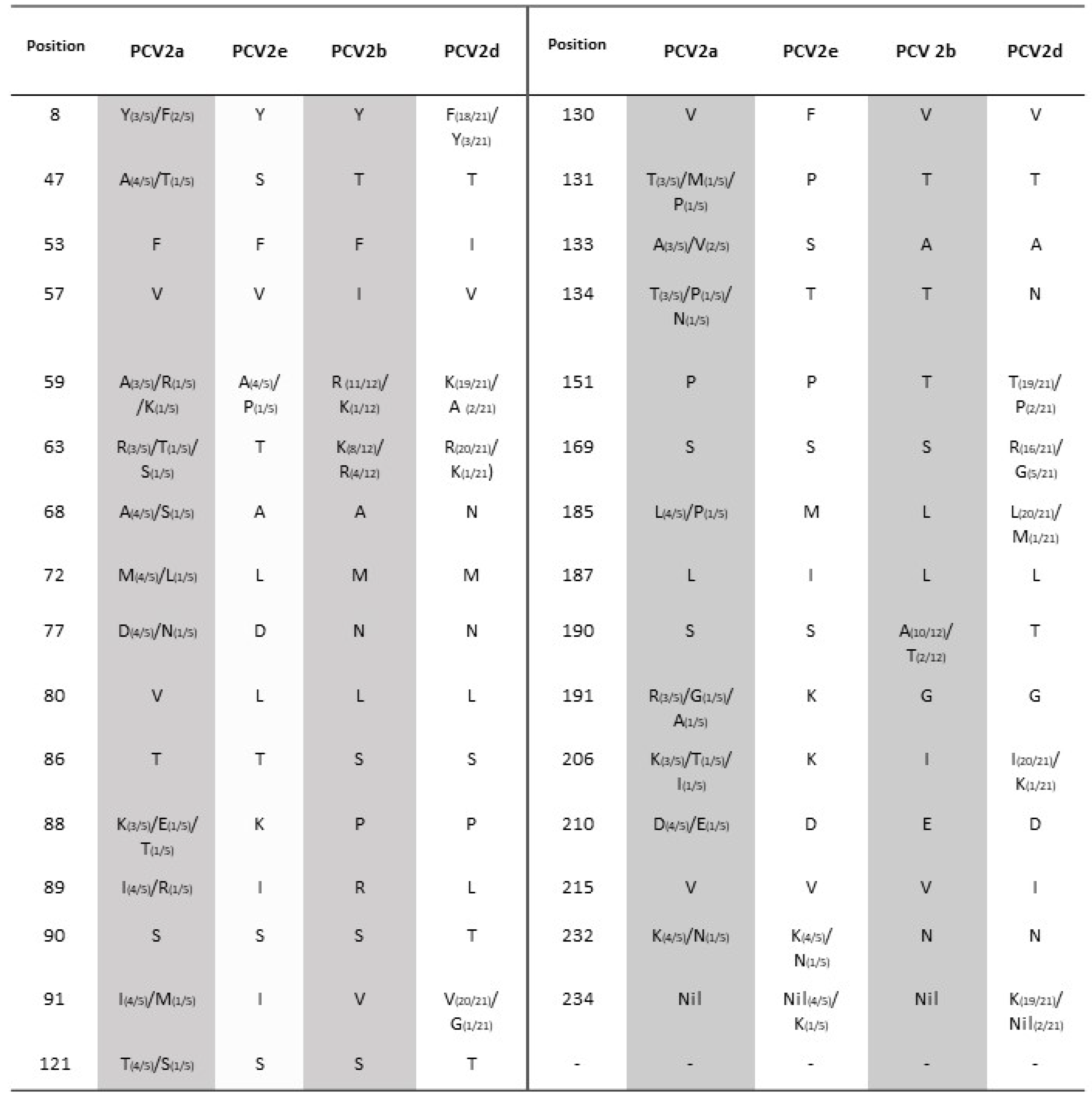
3.2. PCV2-ORF2 Sequencing and Analysis
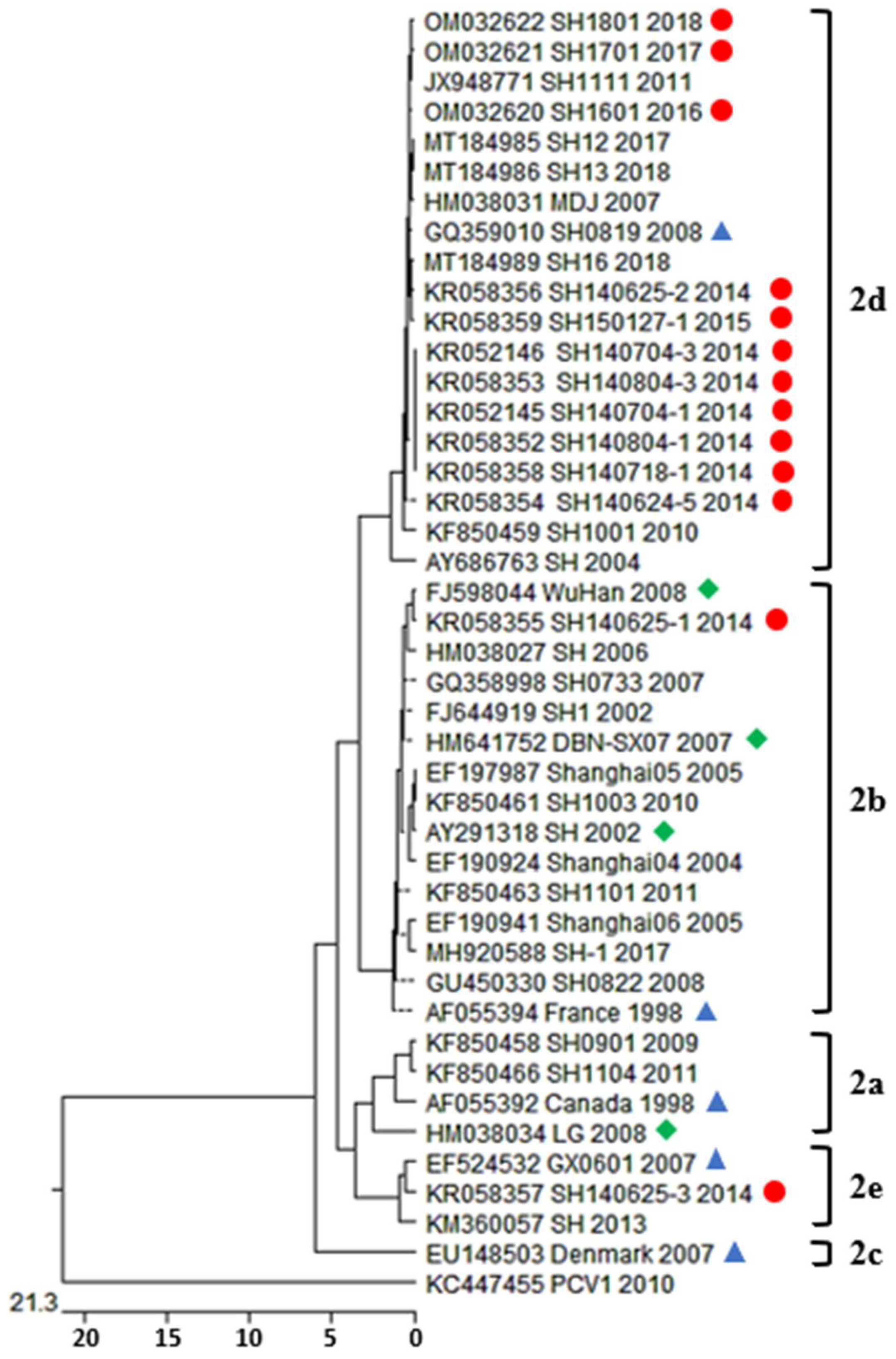
3.3. Phylogenetic Analysis and Genotype Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zenibl. Bukt. Orig A 1974, 226, 153–167. [Google Scholar]
- Hamel, A.L.; Lin, L.L.; Nayar, G.P. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 1998, 72, 5262–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meehan, B.M.; McNeilly, F.; Todd, D.; Kennedy, S.; Jewhurst, V.A.; Ellis, J.A.; Hassard, L.E.; Clark, E.G.; Haines, D.M.; Allan, G.M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 1998, 79, 2171–2179. [Google Scholar] [CrossRef]
- Cheung, A.K.; Lager, K.M.; Kohutyuk, O.I.; Vincent, A.L.; Henry, S.C.; Baker, R.B.; Rowland, R.R.; Dunham, A.G. Detection of two porcine circovirus type 2 genotypic groups in United States swine herds. Arch. Virol. 2007, 152, 1035–1044. [Google Scholar] [CrossRef]
- Harding, J.C. Postweaning multisystemic wasting syndrome: Epidemiology and clinical presentation. J. Swine Health Prod. 1998, 6, 249–254. [Google Scholar]
- Mankertz, A.; Domingo, M.; Folch, J.M.; LeCann, P.; Jestin, A.; Segalés, J.; Chmielewicz, B.; Plana-Durán, J.; Soike, D. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res. 2000, 66, 65–77. [Google Scholar] [CrossRef]
- Trible, B.R.; Suddith, A.W.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Hesse, R.A.; Rowland, R.R. Recognition of the different structural forms of the capsid protein determines the outcome following infection with porcine circovirus type 2. J. Virol. 2012, 86, 13508–13514. [Google Scholar] [CrossRef] [Green Version]
- Chae, C. A review of porcine circovirus 2-associated syndromes and diseases. Vet. J. 2005, 169, 326–336. [Google Scholar] [CrossRef]
- Cadar, D.; Cságola, A.; Lőrincz, M.; Tombácz, K.; Spînu, M.; Tuboly, T. Detection of natural inter- and intra-genotype recombination events revealed by cap gene analysis and decreasing prevalence of PCV2 in wild boars. Infect. Genet. Evol. 2012, 12, 420–427. [Google Scholar] [CrossRef]
- Mankertz, A.; Mankertz, J.; Wolf, K.; Buhk, H.J. Identification of a protein essential for replication of porcine circovirus. J. Gen. Virol. 1998, 79, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Finsterbusch, T.; Mankertz, A. Porcine circoviruses—Small but powerful. Virus Res. 2009, 143, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, I.; Kwang, J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 2005, 79, 8262–8274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chen, I.; Du, Q.; Chua, H.; Kwang, J. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J. Virol. 2006, 80, 5065–5073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grau-Roma, L.; Crisci, E.; Sibila, M.; López-Soria, S.; Nofrarias, M.; Cortey, M.; Fraile, L.; Olvera, A.; Segalés, J. A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence. Vet. Microbiol. 2008, 128, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Olvera, A.; Cortey, M.; Segalés, J. Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality. Virology 2007, 357, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Guo, X.; Yang, H. Genotyping of porcine circovirus type 2 from a variety of clinical conditions in China. Vet. Microbiol. 2005, 110, 141–146. [Google Scholar] [CrossRef]
- Hamel, A.L.; Lin, L.L.; Sachvie, C.; Grudeski, E.; Nayar, G.P. PCR detection and characterization of type-2 porcine circovirus. Can. J. Vet. Res. 2000, 64, 44–52. [Google Scholar]
- Martins Gomes de Castro, A.M.; Cortez, A.; Heinemann, M.B.; Brandão, P.E.; Richtzenhain, L.J. Genetic diversity of Brazilian strains of porcine circovirus type 2 (PCV-2) revealed by analysis of the cap gene (ORF-2). Arch. Virol. 2007, 152, 1435–1445. [Google Scholar] [CrossRef]
- De Boisséson, C.; Béven, V.; Bigarré, L.; Thiéry, R.; Rose, N.; Eveno, E.; Madec, F.; Jestin, A. Molecular characterization of Porcine circovirus type 2 isolates from post-weaning multisystemic wasting syndrome-affected and non-affected pigs. J. Gen. Virol. 2004, 85, 293–304. [Google Scholar] [CrossRef]
- Timmusk, S.; Wallgren, P.; Belák, K.; Berg, M.; Fossum, C. In Genetic analysis of PCV2 capsid protein sequences reveals two main groups of Swedish isolates. In Proceedings of the International Conference on Animal Circoviruses and Associated Diseases, Belfast, Ireland, 11–13 July 2005. [Google Scholar]
- Carman, S.; McEwen, B.; DeLay, J.; van Dreumel, T.; Lusis, P.; Cai, H.; Fairles, J. Porcine circovirus-2 associated disease in swine in Ontario (2004 to 2005). Can. Vet. J. 2006, 47, 761–762. [Google Scholar] [PubMed]
- Segalés, J.; Olvera, A.; Grau-Roma, L.; Charreyre, C.; Nauwynck, H.; Larsen, L.; Dupont, K.; McCullough, K.; Ellis, J.; Krakowka, S.; et al. PCV-2 genotype definition and nomenclature. Vet. Rec. 2008, 162, 867–868. [Google Scholar] [CrossRef] [PubMed]
- Dupont, K.; Hjulsager, C.K.; Kristensen, C.S.; Baekbo, P.; Larsen, L.E. Transmission of different variants of PCV2 and viral dynamics in a research facility with pigs mingled from PMWS-affected herds and non-affected herds. Vet. Microbiol. 2009, 139, 219–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupont, K.; Nielsen, E.O.; Baekbo, P.; Larsen, L.E. Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet. Microbiol. 2008, 128, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Jantafong, T.; Boonsoongnern, A.; Poolperm, P.; Urairong, K.; Lekcharoensuk, C.; Lekcharoensuk, P. Genetic characterization of porcine circovirus type 2 in piglets from PMWS-affected and -negative farms in Thailand. Virol. J. 2011, 8, 88. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.J.; Lu, Y.H.; Wei, Y.W.; Huang, L.P.; Liu, C.M. Porcine circovirus type 2 (PCV2): Genetic variation and newly emerging genotypes in China. Virol. J. 2010, 7, 273. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, C.A.; Tremblay, D.; Tijssen, P.; Venne, M.H.; Houde, A.; Elahi, S.M. The emergence of porcine circovirus 2b genotype (PCV-2b) in swine in Canada. Can. Vet. J. 2007, 48, 811–819. [Google Scholar]
- Karuppannan, A.K.; Opriessnig, T. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Liu, M.; Liu, Z.; Meng, F.; Wang, H.; Cao, L.; Li, Y.; Jiao, Q.; Han, Z.; Liu, S. Epidemiological investigation of porcine circovirus type 2 and its coinfection rate in Shandong province in China from 2015 to 2018. BMC Vet. Res. 2021, 17, 17. [Google Scholar] [CrossRef]
- Chen, N.; Huang, Y.; Ye, M.; Li, S.; Xiao, Y.; Cui, B.; Zhu, J. Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infect. Genet. Evol. 2019, 68, 127–135. [Google Scholar] [CrossRef]
- Guo, Z.; Ruan, H.; Qiao, S.; Deng, R.; Zhang, G. Co-infection status of porcine circoviruses (PCV2 and PCV3) and porcine epidemic diarrhea virus (PEDV) in pigs with watery diarrhea in Henan province, central China. Microb. Pathog. 2020, 142, 104047. [Google Scholar] [CrossRef] [PubMed]
- Ssemadaali, M.A.; Ilha, M.; Ramamoorthy, S. Genetic diversity of porcine circovirus type 2 and implications for detection and control. Res. Vet. Sci. 2015, 103, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Kim, J.; Ha, Y.; Choi, C.; Chae, C. The effects of transplacental porcine circovirus type 2 infection on porcine epidemic diarrhoea virus-induced enteritis in preweaning piglets. Vet. J. 2006, 171, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Allan, G.M.; McNeilly, F.; Kennedy, S.; Daft, B.; Clarke, E.G.; Ellis, J.A.; Haines, D.M.; Meehan, B.M.; Adair, B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 1998, 10, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Yuan, W.; Guo, H.; Ma, Z.; Song, Q.; Wang, X.; Li, H. Prevalence and genetic variation of porcine circovirus type 2 in Hebei, China from 2004 to 2014. Gene 2016, 586, 222–227. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, M.; Chen, Y.; Xie, J.; Huang, Z.; Zhu, W.; Xu, T.; Cao, Z.; Zhou, P.; Su, S.; et al. Genetic evolution and phylogenetic analysis of porcine circovirus type 2 infections in southern China from 2011 to 2012. Infect. Genet. Evol. 2013, 17, 87–92. [Google Scholar] [CrossRef]
- An, D.J.; Roh, I.S.; Song, D.S.; Park, C.K.; Park, B.K. Phylogenetic characterization of porcine circovirus type 2 in PMWS and PDNS Korean pigs between 1999 and 2006. Virus Res. 2007, 129, 115–122. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT, Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J. Gen. Virol. 2015, 96, 1830–1841. [Google Scholar] [CrossRef]
- Cheung, A.K. Homologous recombination within the capsid gene of porcine circovirus type 2 subgroup viruses via natural co-infection. Arch. Virol. 2009, 154, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wei, C.; Dai, A.; Lin, Z.; Fan, K.; Fan, J.; Liu, J.; Luo, M.; Yang, X. Detection of PCV2e strains in Southeast China. PeerJ 2018, 6, e4476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trible, B.R.; Kerrigan, M.; Crossland, N.; Potter, M.; Faaberg, K.; Hesse, R.; Rowland, R.R. Antibody recognition of porcine circovirus type 2 capsid protein epitopes after vaccination, infection, and disease. Clin. Vaccine Immunol. 2011, 18, 749–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, S.-L.; Chen, S.-N.; Xu, Z.-H.; Tang, M.-H.; Wang, F.-G.; Li, X.-J.; Sun, B.-B.; Deng, S.-F.; Hu, J.; Lv, D.-H.; et al. Porcine circovirus type 2 in China: An update on and insights to its prevalence and control. Virol. J. 2014, 11, 88. [Google Scholar] [CrossRef] [Green Version]
- Krakowka, S.; Allan, G.; Ellis, J.; Hamberg, A.; Charreyre, C.; Kaufmann, E.; Brooks, C.; Meehan, B. A nine-base nucleotide sequence in the porcine circovirus type 2 (PCV2) nucleocapsid gene determines viral replication and virulence. Virus Res. 2012, 164, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Trible, B.R.; Rowland, R.R. Genetic variation of porcine circovirus type 2 (PCV2) and its relevance to vaccination, pathogenesis and diagnosis. Virus Res. 2012, 164, 68–77. [Google Scholar] [CrossRef]
- Zheng, G.; Lu, Q.; Wang, F.; Xing, G.; Feng, H.; Jin, Q.; Guo, Z.; Teng, M.; Hao, H.; Li, D.; et al. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Vet. Res. 2020, 16, 6. [Google Scholar] [CrossRef]
- Xia, D.; Huang, L.; Xie, Y.; Zhang, X.; Wei, Y.; Liu, D.; Zhu, H.; Bian, H.; Feng, L.; Liu, C. The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Arch. Virol. 2019, 164, 2435–2449. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Chen, S.-N.; Zhang, J.-W.; Wei, Z.-Z.; Long, J.-X.; Yuan, S.-S.; Wei, W.-K.; Chen, Q.-L.; Xuan, H.; Wu, D.-C. Dissection of the possible routes on porcine circoviruses infecting human. J. Anim. Vet. Adv. 2012, 11, 1281–1286. [Google Scholar]
- Wang, F.; Guo, X.; Ge, X.; Wang, Z.; Chen, Y.; Cha, Z.; Yang, H. Genetic variation analysis of Chinese strains of porcine circovirus type 2. Virus Res. 2009, 145, 151–156. [Google Scholar] [CrossRef]
- Segales, J.; Domingo, M.; Latimer, K. In Porcine circovirus is present in cases of porcine dermatitis and nephropathy syndrome (PDNS). In Proceedings of the 15th International Pig Veterinary Society Congress, Birmingham, UK, 5–9 July 1998; p. 215. [Google Scholar]
- Segalés, J.; Domingo, M. Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet. Q. 2002, 24, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Lipej, Z.; Segalés, J.; Jemersić, L.; Olvera, A.; Roić, B.; Novosel, D.; Mihaljević, Z.; Manojlović, L. First description of postweaning multisystemic wasting syndrome (PMWS) in wild boar (Sus scrofa) in Croatia and phylogenetic analysis of partial PCV2 sequences. Acta Vet. Hung. 2007, 55, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Sofia, M.; Billinis, C.; Psychas, V.; Birtsas, P.; Sofianidis, G.; Leontides, L.; Knowles, N.; Spyrou, V. Detection and genetic characterization of porcine circovirus 2 isolates from the first cases of postweaning multisystemic and wasting syndrome in wild boars in Greece. J. Wildl. Dis. 2008, 44, 864–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H. Epidemical characterizations and control strategies of swine immunosuppressive diseases (PRRS and PMWS). Chin. J. Anim. Husband Vet. Med. 2004, 31, 41–43. [Google Scholar]
- Sun, J.; Huang, L.; Wei, Y.; Wang, Y.; Chen, D.; Du, W.; Wu, H.; Liu, C. Prevalence of emerging porcine parvoviruses and their co-infections with porcine circovirus type 2 in China. Arch. Virol. 2015, 160, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Wang, X.; Dvorak, C.M.T.; Marthaler, D.; Murtaugh, M.P. Diagnostic phylogenetics reveals a new Porcine circovirus 2 cluster. Virus Res. 2016, 217, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, F.X.; Zhu, H.W.; Sun, N.; Wu, H. Phylogenetic analysis of porcine circovirus type 2 (PCV2) isolates from China with high homology to PCV2c. Arch. Virol. 2016, 161, 1591–1599. [Google Scholar] [CrossRef]
- Jiang, C.-G.; Wang, G.; Tu, Y.-B.; Liu, Y.-G.; Wang, S.-J.; Cai, X.-H.; An, T.-Q. Genetic analysis of porcine circovirus type 2 in China. Arch. Virol. 2017, 162, 2715–2726. [Google Scholar] [CrossRef]
- Franzo, G.; Cortey, M.; de Castro, A.M.; Piovezan, U.; Szabo, M.P.; Drigo, M.; Segalés, J.; Richtzenhain, L.J. Genetic characterisation of Porcine circovirus type 2 (PCV2) strains from feral pigs in the Brazilian Pantanal: An opportunity to reconstruct the history of PCV2 evolution. Vet. Microbiol. 2015, 178, 158–162. [Google Scholar] [CrossRef]
- Molini, U.; Franzo, G.; Gous, L.; Moller, S.; Hemberger, Y.M.; Chiwome, B.; Marruchella, G.; Khaiseb, S.; Cattoli, G.; Dundon, W.G. Three different genotypes of porcine circovirus 2 (PCV-2) identified in pigs and warthogs in Namibia. Arch. Virol. 2021, 166, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Beach, N.M.; Ramamoorthy, S.; Opriessnig, T.; Wu, S.Q.; Meng, X.J. Novel chimeric porcine circovirus (PCV) with the capsid gene of the emerging PCV2b subtype cloned in the genomic backbone of the non-pathogenic PCV1 is attenuated in vivo and induces protective and cross-protective immunity against PCV2b and PCV2a subtypes in pigs. Vaccine 2010, 29, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, X.; Ma, T.; Feng, Z.; Li, Y.; Jiang, P. Genetic analysis of porcine circovirus type 2 (PCV2) strains isolated between 2001 and 2009: Genotype PCV2b predominate in postweaning multisystemic wasting syndrome occurrences in eastern China. Virus Genes 2010, 40, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Lee, D.U.; Yoo, S.J.; Je, S.H.; Shin, J.Y.; Lyoo, Y.S. Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res. 2017, 228, 24–29. [Google Scholar] [CrossRef]
- Yang, S.; Yin, S.; Shang, Y.; Liu, B.; Yuan, L.; Zafar Khan, M.U.; Liu, X.; Cai, J. Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transbound. Emerg. Dis. 2018, 65, e383–e392. [Google Scholar] [CrossRef]
- Mahé, D.; Blanchard, P.; Truong, C.; Arnauld, C.; Le Cann, P.; Cariolet, R.; Madec, F.; Albina, E.; Jestin, A. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 2000, 81, 1815–1824. [Google Scholar] [CrossRef]
- Fachinger, V.; Bischoff, R.; Jedidia, S.B.; Saalmüller, A.; Elbers, K. The effect of vaccination against porcine circovirus type 2 in pigs suffering from porcine respiratory disease complex. Vaccine 2008, 26, 1488–1499. [Google Scholar] [CrossRef]
- Fort, M.; Sibila, M.; Allepuz, A.; Mateu, E.; Roerink, F.; Segalés, J. Porcine circovirus type 2 (PCV2) vaccination of conventional pigs prevents viremia against PCV2 isolates of different genotypes and geographic origins. Vaccine 2008, 26, 1063–1071. [Google Scholar] [CrossRef]
- Khayat, R.; Brunn, N.; Speir, J.A.; Hardham, J.M.; Ankenbauer, R.G.; Schneemann, A.; Johnson, J.E. The 2.3-angstrom structure of porcine circovirus 2. J. Virol. 2011, 85, 7856–7862. [Google Scholar] [CrossRef] [Green Version]
- Lekcharoensuk, P.; Morozov, I.; Paul, P.S.; Thangthumniyom, N.; Wajjawalku, W.; Meng, X.J. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 2004, 78, 8135–8145. [Google Scholar] [CrossRef] [Green Version]
- Saha, D.; Lefebvre, D.J.; Ooms, K.; Huang, L.; Delputte, P.L.; Van Doorsselaere, J.; Nauwynck, H.J. Single amino acid mutations in the capsid switch the neutralization phenotype of porcine circovirus 2. J. Gen. Virol. 2012, 93, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.P.; Lu, Y.H.; Wei, Y.W.; Guo, L.J.; Liu, C.M. Identification of one critical amino acid that determines a conformational neutralizing epitope in the capsid protein of porcine circovirus type 2. BMC Microbiol. 2011, 11, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.K.; Wei, C.H.; Yang, X.Y.; Dai, A.L.; Li, X.H. Multiplex PCR for the simultaneous detection of porcine reproductive and respiratory syndrome virus, classical swine fever virus, and porcine circovirus in pigs. Mol. Cell. Probes 2013, 27, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Wang, F.; Guo, X.; Yang, H. Porcine circovirus type 2 and its associated diseases in China. Virus Res. 2012, 164, 100–106. [Google Scholar] [CrossRef]
- Ellis, J.; Hassard, L.; Clark, E.; Harding, J.; Allan, G.; Willson, P.; Strokappe, J.; Martin, K.; McNeilly, F.; Meehan, B.; et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 1998, 39, 44–51. [Google Scholar] [PubMed]
- Pescador, C.A.; Bandarra, P.M.; Castro, L.A.; Antoniassi, N.A.B.; Ravazzolo, A.P.; Sonne, L.; Cruz, C.E.F.; Driemeier, D. Co-infection by porcine circovirus type 2 and porcine parvovirus in aborted fetuses and stillborn piglets in southern Brazil. Pesqui. Veterinária Bras. 2007, 27, 425–429. [Google Scholar] [CrossRef]
- Saekhow, P.; Kishizuka, S.; Sano, N.; Mitsui, H.; Akasaki, H.; Mawatari, T.; Ikeda, H. Coincidental detection of genomes of porcine parvoviruses and porcine circovirus type 2 infecting pigs in Japan. J. Vet. Med. Sci. 2016, 77, 1581–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, T.; Li, R.; Yan, M.; Luo, B.; Yang, T.; Yu, X. High prevalence of PCV2d in Hunan province, China: A retrospective analysis of samples collected from 2006 to 2016. Arch. Virol. 2018, 163, 1897–1906. [Google Scholar] [CrossRef]
| Primer Pairs | Primer Sequence | Amplified Length | Annealing Temperature | Virus Type |
|---|---|---|---|---|
| ORF5-F | 5′-AGGTGGGCAACTGTTTTAGC-3′ | 697 bp | 53.5 °C | PRRSV |
| ORF5-R | 5′-TTTGTGGAGCCGTGCTATCA-3′ | |||
| VP2-F | 5′-CACGCATCAAGACTCATA-3′ | 472 bp | 48.0 °C | PPV |
| VP2-R | 5′-TTGGTGGATTTAGGTTTC-3′ | |||
| PCV-F | 5′-CCGCGGGCTGGCTGAACTT-3′ | 1154 bp | 58.0 °C | PCV2 |
| PCV-R | 5′-ACCCCCGCCACCGCTACC-3′ |
| Samples’ ID | Farm/Area | Herd Size | Positive Samples/Total Samples | PCV2 Positive Rate (%) | PCV2 + PRRSV | PCV2 + PPV | Farm Vaccination Status |
|---|---|---|---|---|---|---|---|
| SH150127-1 | Fengxian | >500 | 6/10 | 60.0% | 0/6 | 6/6 | Yes |
| SH140624-5 | Pudong | >3000 | 13/21 | 61.9% | 0/13 | 13/13 | Yes |
| SH140718-1 | Pudong | >5000 | 10/15 | 66.7% | 0/10 | 10/10 | Yes |
| SH140704-1 | Jiading | >2000 | 16/27 | 59.3% | 0/16 | 16/16 | Yes |
| SH140704-3 | Jiading | >2000 | 9/15 | 60.0% | 0/9 | 9/9 | Yes |
| SH140804-1 | Fengxian | >2000 | 16/31 | 51.6% | 0/31 | 16/31 | Yes |
| SH140804-3 | Chongmei | >2000 | 13/20 | 65.0% | 0/13 | 13/13 | Yes |
| SH140625-2 | Pudong | >2000 | 4/7 | 57.1% | 4/4 | 4/4 | Yes |
| SH140625-1 | Fengxian | >500 | 4/8 | 50.0% | 2/4 | 4/4 | No |
| SH140625-3 | Chongmei | >2000 | 7/12 | 58.3% | 7/7 | 0/7 | Yes |
| SH1601 | Pudong | >2000 | 8/15 | 53.3% | 6/8 | 8/8 | Yes |
| SH1701 | Jiading | >2000 | 6/13 | 46.2% | 3/6 | 6/6 | Yes |
| SH1801 | Chongmei | >2000 | 3/5 | 60.0% | 3/3 | 3/3 | Yes |
| Samples’ ID | Accession Number (GenBank) | Year | PCV2 Genotype | History of Disease | ORF2 Size (bp) | Tissues |
|---|---|---|---|---|---|---|
| SH150127-1 | KR058359 | 2015 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph node |
| SH140624-5 | KR058354 | 2014 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph nodes |
| SH140718-1 | KR058358 | 2014 | 2d | PMWS/PNDS | 705 | Inguinal lymph nodes |
| SH140704-1 | KR052145 | 2014 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph nodes |
| SH140704-3 | KR052146 | 2014 | 2d | No signs | 705 | Lung, spleen, lymph nodes |
| SH140804-1 | KR058352 | 2014 | 2d | PMWS/PNDS | 705 | Inguinal lymph nodes |
| SH140804-3 | KR058353 | 2014 | 2d | PMWS/PNDS | 705 | Inguinal lymph nodes |
| SH140625-2 | KR058356 | 2014 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph nodes |
| SH140625-1 | KR058355 | 2014 | 2b | PMWS/PNDS | 702 | Inguinal lymph nodes |
| SH140625-3 | KR058357 | 2014 | 2e | No signs | 702 | Inguinal lymph nodes |
| SH1601 | OM032620 | 2016 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph nodes |
| SH1701 | OM032621 | 2017 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph nodes |
| SH1801 | OM032622 | 2018 | 2d | PMWS/PNDS | 705 | Lung, spleen, lymph nodes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, L.; Wahaab, A.; Shi, K.; Mustafa, B.E.; Zhang, Y.; Zhang, J.; Li, Z.; Qiu, Y.; Li, B.; Liu, K.; et al. Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China. Viruses 2022, 14, 289. https://doi.org/10.3390/v14020289
Kang L, Wahaab A, Shi K, Mustafa BE, Zhang Y, Zhang J, Li Z, Qiu Y, Li B, Liu K, et al. Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China. Viruses. 2022; 14(2):289. https://doi.org/10.3390/v14020289
Chicago/Turabian StyleKang, Le, Abdul Wahaab, Kun Shi, Bahar E Mustafa, Yan Zhang, Junjie Zhang, Zongjie Li, Yafeng Qiu, Beibei Li, Ke Liu, and et al. 2022. "Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China" Viruses 14, no. 2: 289. https://doi.org/10.3390/v14020289
APA StyleKang, L., Wahaab, A., Shi, K., Mustafa, B. E., Zhang, Y., Zhang, J., Li, Z., Qiu, Y., Li, B., Liu, K., Shao, D., Ma, Z., Zhong, D., & Wei, J. (2022). Molecular Epidemic Characteristics and Genetic Evolution of Porcine Circovirus Type 2 (PCV2) in Swine Herds of Shanghai, China. Viruses, 14(2), 289. https://doi.org/10.3390/v14020289







