Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Image Processing of the Qβ gRNA
2.2. Modeling of Wild-Type gRNA
2.3. Purification of Qβ and VLPs
2.4. Western Blots
2.5. Electrophoretic Mobility Shift Assay
2.6. Cryo-EM Specimen Preparation and Data Collection
2.7. Cryo-EM Data Processing
2.8. Modeling of VLP Capsids
3. Results
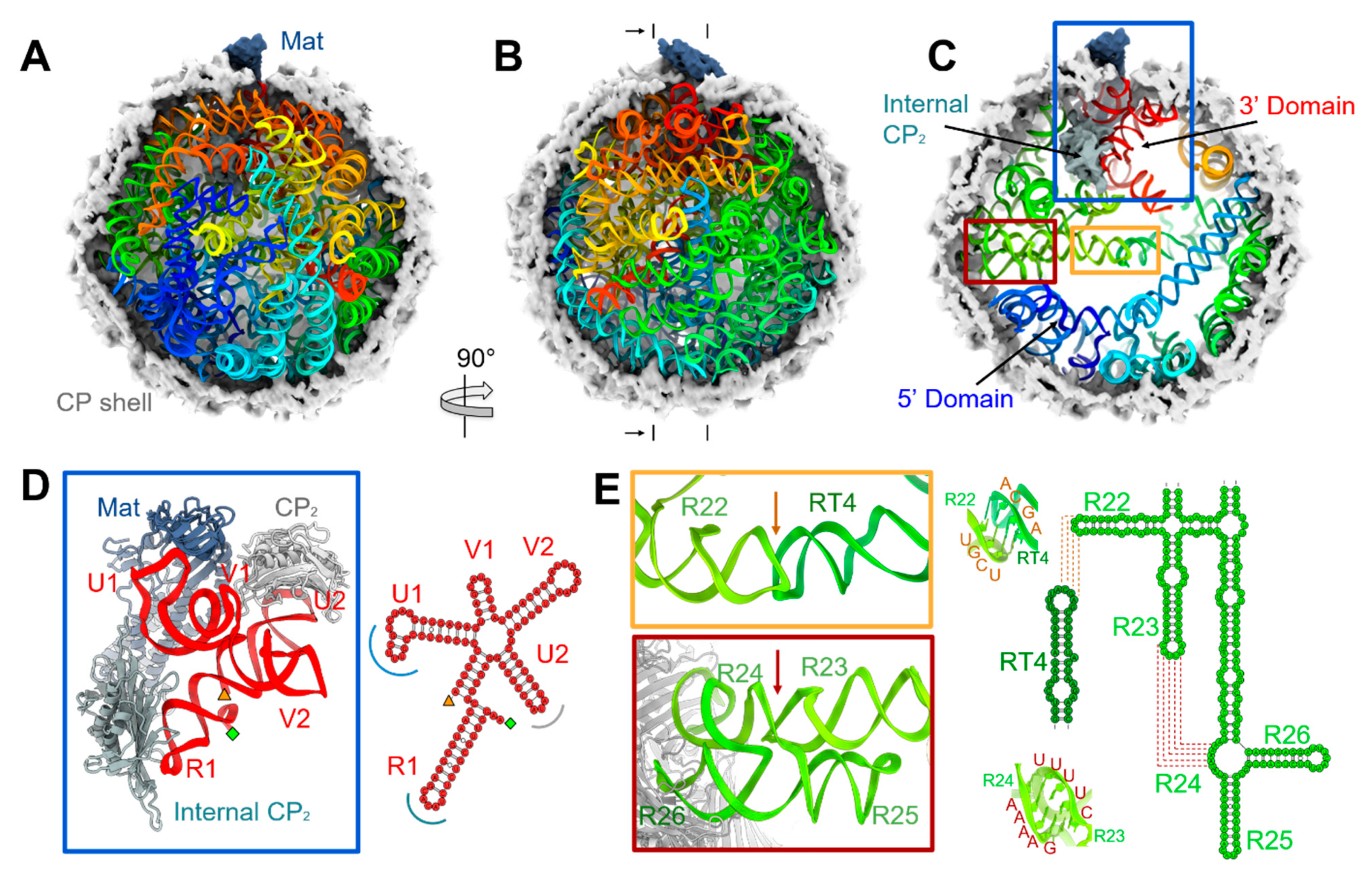
3.1. The Complete gRNA Model inside the Qβ Virion
3.2. Operator-like RNA Stem-Loops Support the Formation of CP Pentamers
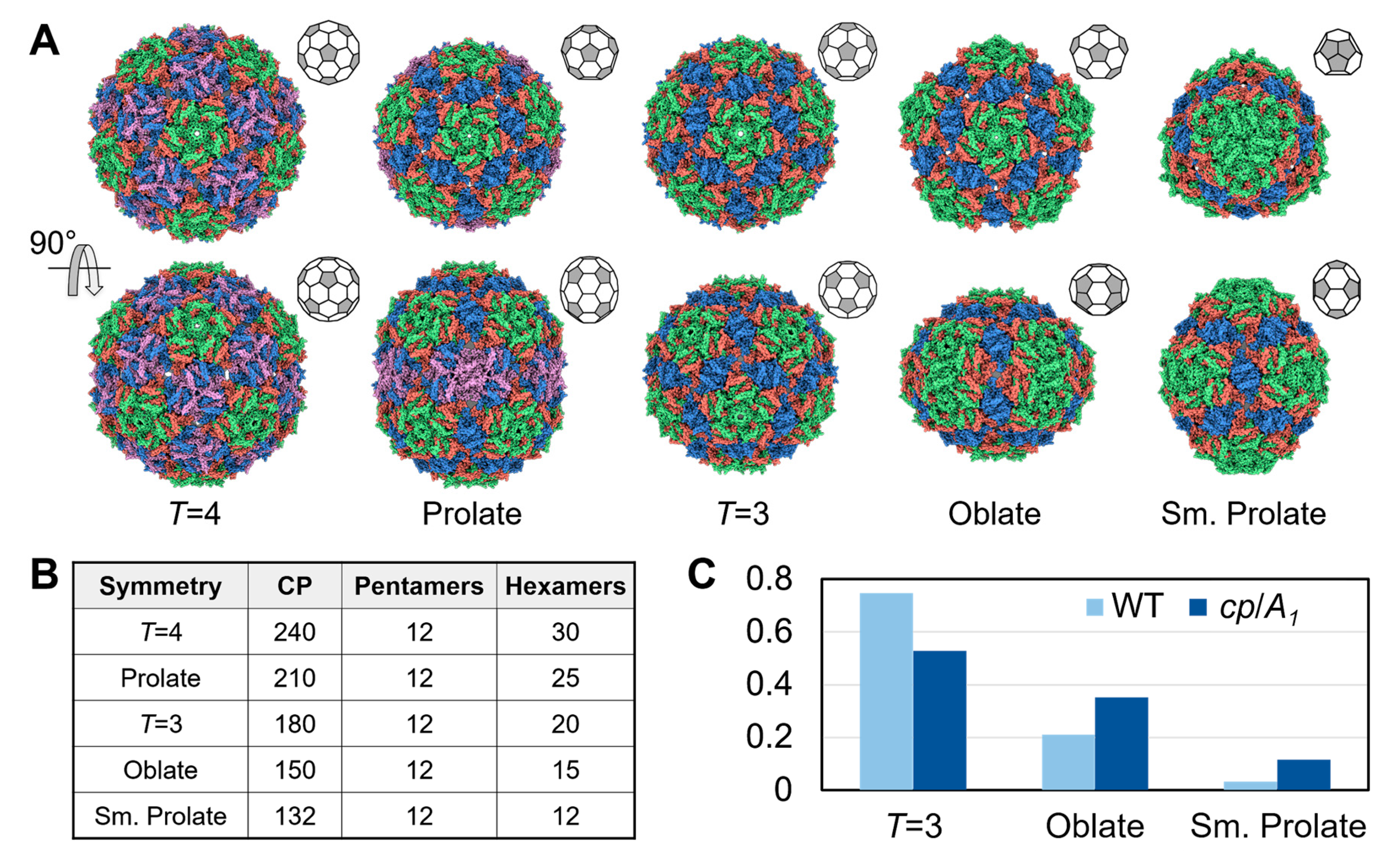
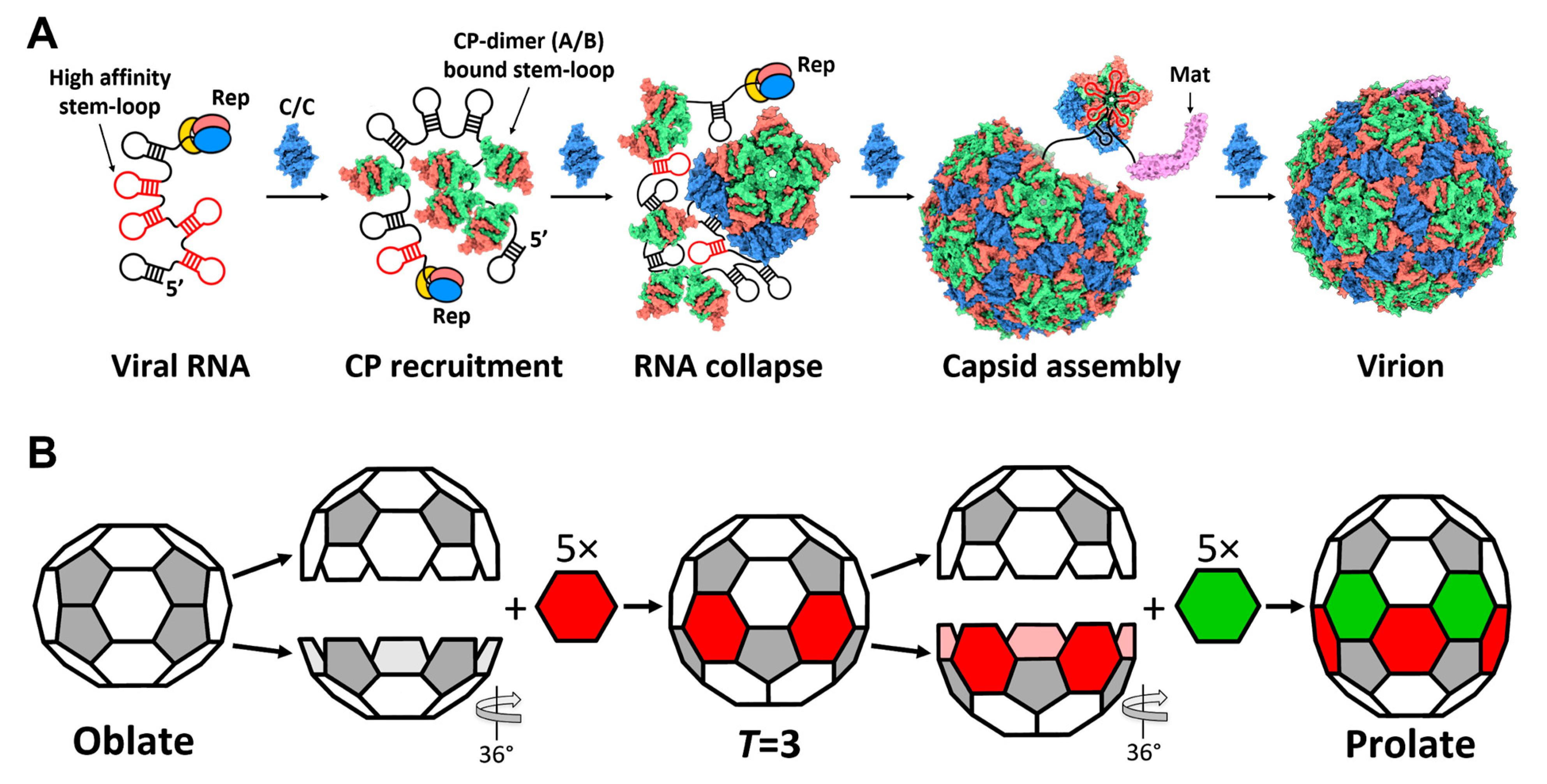
3.3. Qβ VLPs of Different Forms Are Made along with Infectious Virions
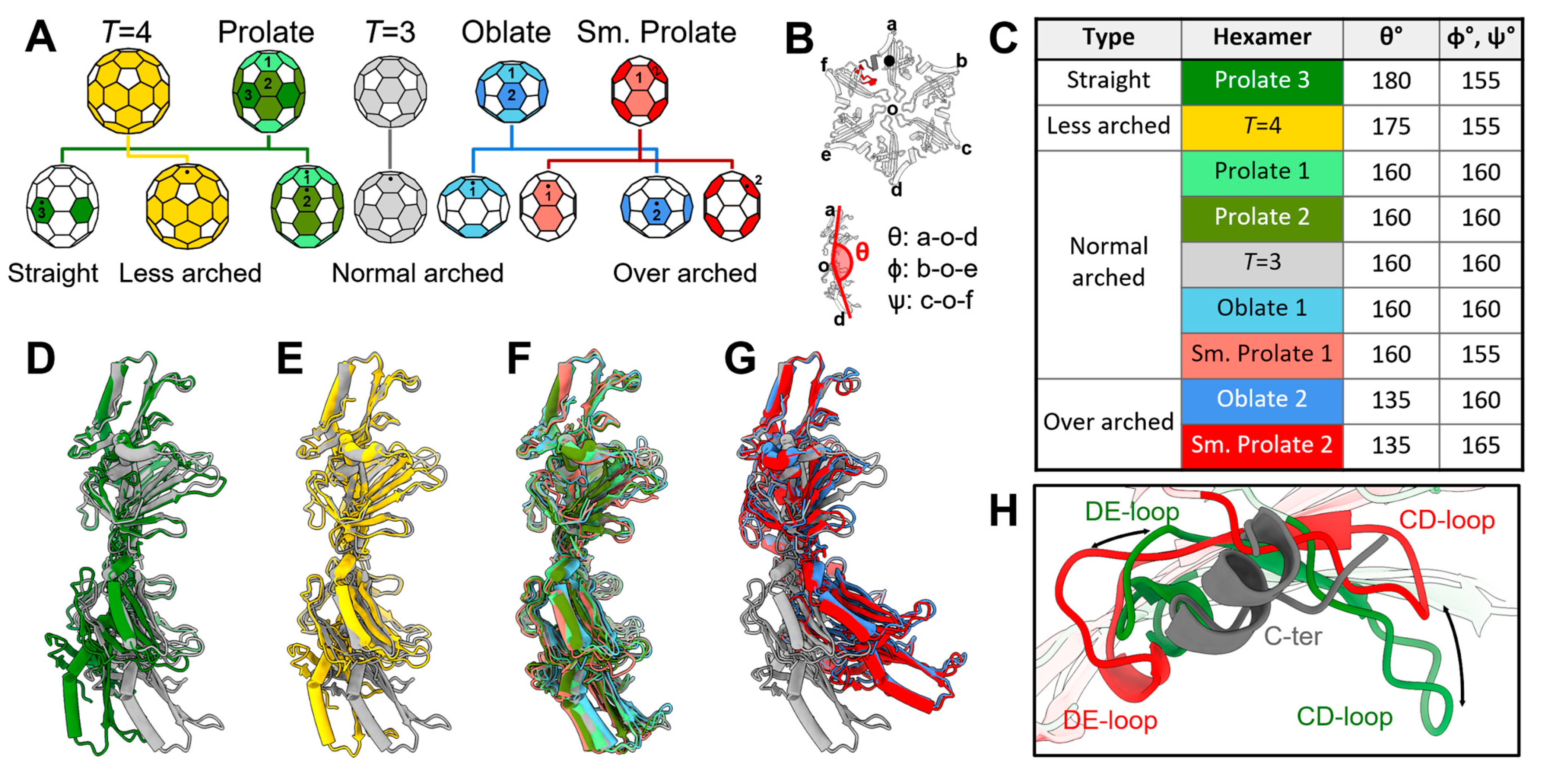
3.4. The Plasticity in the Hexamers Supports the Formation of Different VLPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Duin, J.; Tsareva, N. Single-stranded RNA phages. In The Bacteriophages, 2nd ed.; Calendar, R., Ed.; Oxford University Press: New York, NY, USA, 2006; pp. 175–196. [Google Scholar]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Zhou, Z.H.; Sun, R. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 2017, 541, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorzelnik, K.V.; Cui, Z.; Reed, C.A.; Jakana, J.; Young, R.; Zhang, J. Asymmetric cryo-EM structure of the canonical Allolevivirus Qbeta reveals a single maturation protein and the genomic ssRNA in situ. Proc. Natl. Acad. Sci. USA 2016, 113, 11519–11524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tars, K.; Fridborg, K.; Bundule, M.; Liljas, L. The three-dimensional structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7-A resolution. Virology 2000, 272, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klovins, J.; Overbeek, G.P.; van den Worm, S.H.E.; Ackermann, H.W.; van Duin, J. Nucleotide sequence of a ssRNA phage from Acinetobacter: Kinship to coliphages. J. Gen. Virol. 2002, 83, 1523–1533. [Google Scholar] [CrossRef]
- Cui, Z.; Gorzelnik, K.V.; Chang, J.Y.; Langlais, C.; Jakana, J.; Young, R.; Zhang, J. Structures of Qbeta virions, virus-like particles, and the Qbeta-MurA complex reveal internal coat proteins and the mechanism of host lysis. Proc. Natl. Acad. Sci. USA 2017, 114, 11697–11702. [Google Scholar] [CrossRef] [Green Version]
- Meng, R.; Jiang, M.; Cui, Z.; Chang, J.Y.; Yang, K.; Jakana, J.; Yu, X.; Wang, Z.; Hu, B.; Zhang, J. Structural basis for the adsorption of a single-stranded RNA bacteriophage. Nat. Commun. 2019, 10, 3130. [Google Scholar] [CrossRef] [Green Version]
- Krahn, P.M.; O’Callaghan, R.J.; Paranchych, W. Stages in phage R17 infection. VI. Injection of A protein and RNA into the host cell. Virology 1972, 47, 628–637. [Google Scholar] [CrossRef]
- Borodavka, A.; Tuma, R.; Stockley, P.G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. USA 2012, 109, 15769–15774. [Google Scholar] [CrossRef] [Green Version]
- Zeltins, A. Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef]
- Asensio, M.A.; Morella, N.M.; Jakobson, C.M.; Hartman, E.C.; Glasgow, J.E.; Sankaran, B.; Zwart, P.H.; Tullman-Ercek, D. A Selection for Assembly Reveals That a Single Amino Acid Mutant of the Bacteriophage MS2 Coat Protein Forms a Smaller Virus-like Particle. Nano Lett. 2016, 16, 5944–5950. [Google Scholar] [CrossRef] [Green Version]
- De Martin Garrido, N.; Crone, M.A.; Ramlaul, K.; Simpson, P.A.; Freemont, P.S.; Aylett, C.H.S. Bacteriophage MS2 displays unreported capsid variability assembling T = 4 and mixed capsids. Mol. Microbiol. 2020, 113, 143–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Kopylov, M.; Potter, C.S.; Carragher, B.; Finn, M.G. Engineering the PP7 Virus Capsid as a Peptide Display Platform. ACS Nano 2019, 13, 4443–4454. [Google Scholar] [CrossRef] [PubMed]
- Lieknina, I.; Kalnins, G.; Akopjana, I.; Bogans, J.; Sisovs, M.; Jansons, J.; Rumnieks, J.; Tars, K. Production and characterization of novel ssRNA bacteriophage virus-like particles from metagenomic sequencing data. J. Nanobiotechnol. 2019, 17, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rūmnieks, J.; Liekniņa, I.; Kalniņš, G.; Šišovs, M.; Akopjana, I.; Bogans, J.; Tārs, K. Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes. Sci. Adv. 2020, 6, eabc0023. [Google Scholar] [CrossRef]
- Cielens, I.; Ose, V.; Petrovskis, I.; Strelnikova, A.; Renhofa, R.; Kozlovska, T.; Pumpens, P. Mutilation of RNA phage Qbeta virus-like particles: From icosahedrons to rods. FEBS Lett. 2000, 482, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Yi, H.; Kim, W.J.; Kang, K.; Yun, D.S.; Strano, M.S.; Ceder, G.; Belcher, A.M. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 2009, 324, 1051–1055. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.T.; Kim, D.W.; Yoo, P.J.; Chiang, C.Y.; Meethong, N.; Hammond, P.T.; Chiang, Y.M.; Belcher, A.M. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science 2006, 312, 885–888. [Google Scholar] [CrossRef] [Green Version]
- Bao, Q.; Li, X.; Han, G.; Zhu, Y.; Mao, C.; Yang, M. Phage-based vaccines. Adv. Drug Deliv. Rev. 2019, 145, 40–56. [Google Scholar] [CrossRef]
- Frietze, K.M.; Peabody, D.S.; Chackerian, B. Engineering virus-like particles as vaccine platforms. Curr. Opin. Virol. 2016, 18, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Karimi, M.; Mirshekari, H.; Moosavi Basri, S.M.; Bahrami, S.; Moghoofei, M.; Hamblin, M.R. Bacteriophages and phage-inspired nanocarriers for targeted delivery of therapeutic cargos. Adv. Drug Deliv. Rev. 2016, 106, 45–62. [Google Scholar] [CrossRef] [Green Version]
- Rohovie, M.J.; Nagasawa, M.; Swartz, J.R. Virus-like particles: Next-generation nanoparticles for targeted therapeutic delivery. Bioeng. Transl. Med. 2017, 2, 43–57. [Google Scholar] [CrossRef]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beekwilder, M.J.; Nieuwenhuizen, R.; van Duin, J. Secondary structure model for the last two domains of single-stranded RNA phage Q beta. J. Mol. Biol. 1995, 247, 903–917. [Google Scholar] [CrossRef] [PubMed]
- Beekwilder, J. Secondary Structure of the RNA Genome of Bacteriophage Qβ; University of Leiden: Leiden, Netherlands, 1996. [Google Scholar]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Chou, F.C.; Das, R. Modeling complex RNA tertiary folds with Rosetta. Methods Enzymol. 2015, 553, 35–64. [Google Scholar] [PubMed]
- Chang, J.Y.; Cui, Z.; Yang, K.; Huang, J.; Minary, P.; Zhang, J. Hierarchical Natural Move Monte Carlo Refines Flexible RNA Structures into Cryo-EM Densities. RNA 2020, 26, 1755–1766. [Google Scholar] [CrossRef]
- Singharoy, A.; Teo, I.; McGreevy, R.; Stone, J.E.; Zhao, J.; Schulten, K. Molecular dynamics-based refinement and validation for sub-5 A cryo-electron microscopy maps. eLife 2016, 5, e16105. [Google Scholar] [CrossRef]
- Wriggers, W. Conventions and workflows for using Situs. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 344–351. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Reed, C.A.; Langlais, C.; Wang, I.N.; Young, R. A2 expression and assembly regulates lysis in Qbeta infections. Microbiology 2013, 159, 507–514. [Google Scholar] [CrossRef] [Green Version]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.Y.; Song, Y.; Barad, B.A.; Cheng, Y.; Fraser, J.S.; DiMaio, F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 2016, 5, e17219. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Suzuki, Y. Localization of A protein in the RNA-A protein complex of RNA phage MS2. Biochim. Biophys. Acta 1981, 654, 249–255. [Google Scholar] [CrossRef]
- Weiner, A.M.; Weber, K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat. New Biol. 1971, 234, 206–209. [Google Scholar] [CrossRef]
- Liu, D.; Geary, C.W.; Chen, G.; Shao, Y.; Li, M.; Mao, C.; Andersen, E.S.; Piccirilli, J.A.; Rothemund, P.W.K.; Weizmann, Y. Branched kissing loops for the construction of diverse RNA homooligomeric nanostructures. Nat. Chem. 2020, 12, 249–259. [Google Scholar] [CrossRef]
- Gytz, H.; Mohr, D.; Seweryn, P.; Yoshimura, Y.; Kutlubaeva, Z.; Dolman, F.; Chelchessa, B.; Chetverin, A.B.; Mulder, F.A.; Brodersen, D.E.; et al. Structural basis for RNA-genome recognition during bacteriophage Qbeta replication. Nucleic Acids Res. 2015, 43, 10893–10906. [Google Scholar] [CrossRef] [Green Version]
- Stockley, P.G.; Twarock, R.; Bakker, S.E.; Barker, A.M.; Borodavka, A.; Dykeman, E.; Ford, R.J.; Pearson, A.R.; Phillips, S.E.; Ranson, N.A.; et al. Packaging signals in single-stranded RNA viruses: Nature’s alternative to a purely electrostatic assembly mechanism. J. Biol. Phys. 2013, 39, 277–287. [Google Scholar] [CrossRef] [Green Version]
- Golmohammadi, R.; Valegard, K.; Fridborg, K.; Liljas, L. The refined structure of bacteriophage MS2 at 2.8 A resolution. J. Mol. Biol. 1993, 234, 620–639. [Google Scholar] [CrossRef]
- Golmohammadi, R.; Fridborg, K.; Bundule, M.; Valegard, K.; Liljas, L. The crystal structure of bacteriophage Q beta at 3.5 A resolution. Structure 1996, 4, 543–554. [Google Scholar] [CrossRef] [Green Version]
- Stockley, P.G.; Rolfsson, O.; Thompson, G.S.; Basnak, G.; Francese, S.; Stonehouse, N.J.; Homans, S.W.; Ashcroft, A.E. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T=3 viral capsid. J. Mol. Biol. 2007, 369, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Saxena, P.; Lomonossoff, G.P. Virus infection cycle events coupled to RNA replication. Annu. Rev. Phytopathol. 2014, 52, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Luque, A.; Reguera, D. The structure of elongated viral capsids. Biophys. J. 2010, 98, 2993–3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickett, G.G.; Peabody, D.S. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic. Acids Res. 1993, 21, 4621–4626. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, J.-Y.; Gorzelnik, K.V.; Thongchol, J.; Zhang, J. Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles. Viruses 2022, 14, 225. https://doi.org/10.3390/v14020225
Chang J-Y, Gorzelnik KV, Thongchol J, Zhang J. Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles. Viruses. 2022; 14(2):225. https://doi.org/10.3390/v14020225
Chicago/Turabian StyleChang, Jeng-Yih, Karl V. Gorzelnik, Jirapat Thongchol, and Junjie Zhang. 2022. "Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles" Viruses 14, no. 2: 225. https://doi.org/10.3390/v14020225
APA StyleChang, J.-Y., Gorzelnik, K. V., Thongchol, J., & Zhang, J. (2022). Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles. Viruses, 14(2), 225. https://doi.org/10.3390/v14020225






