Characterisation of an Anti-Vaccinia Virus F13 Single Chain Fragment Variable from a Human Anti-Vaccinia Virus-Specific Recombinant Immunoglobulin Library
Abstract
:1. Introduction
2. Material and Methods
2.1. Cells and Viruses
2.2. Monoclonal and Polyclonal Antibodies
2.3. Immunisation, Lymphocyte Preparation and Library Construction
2.4. Construction and Purification of F13
2.5. Selection of Purified Recombinant F13 Protein
2.6. Plasmid Isolation and Sequencing of Positive Colonies
2.7. Upscale Production and Purification of Selected scFv
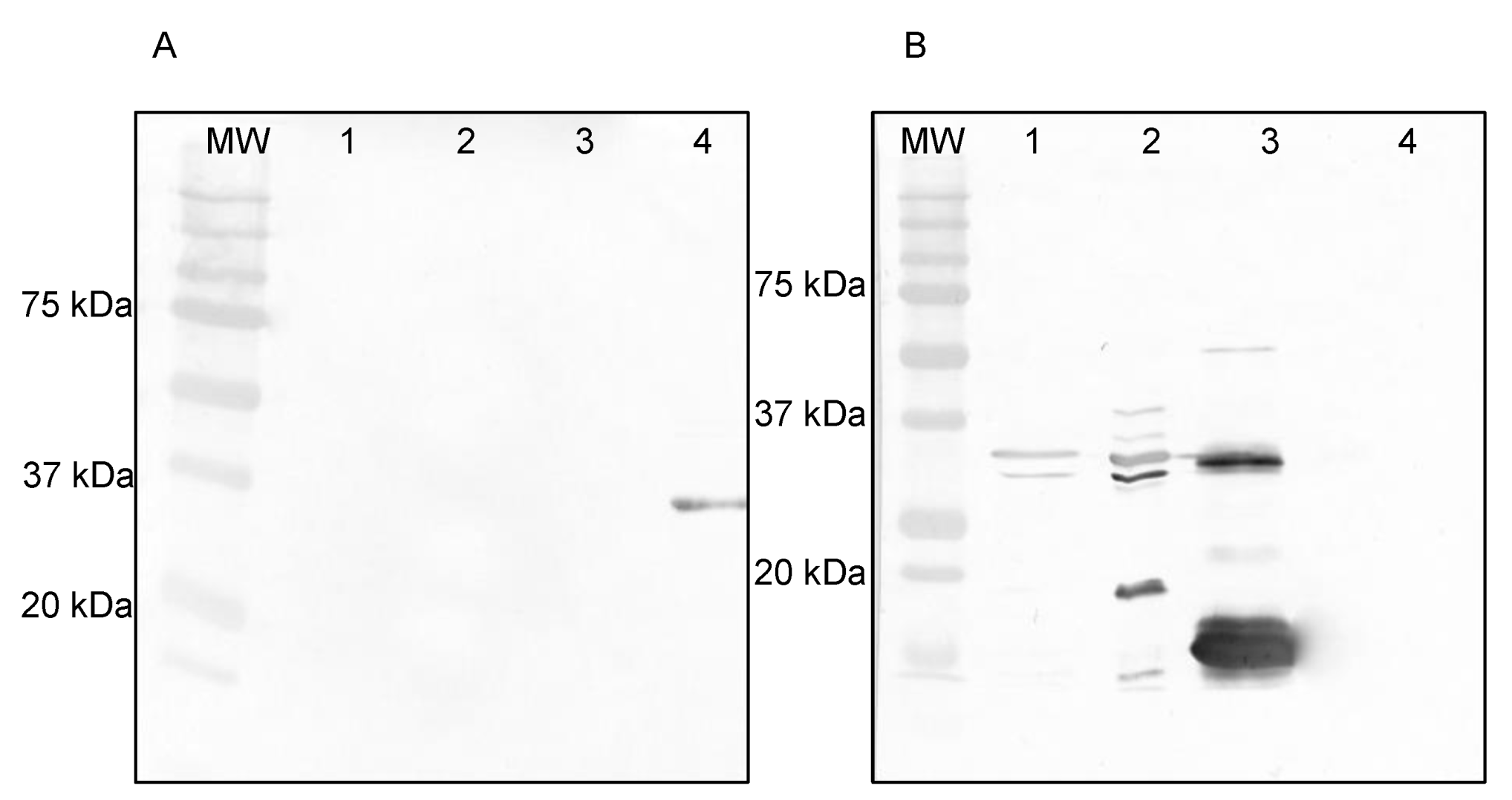
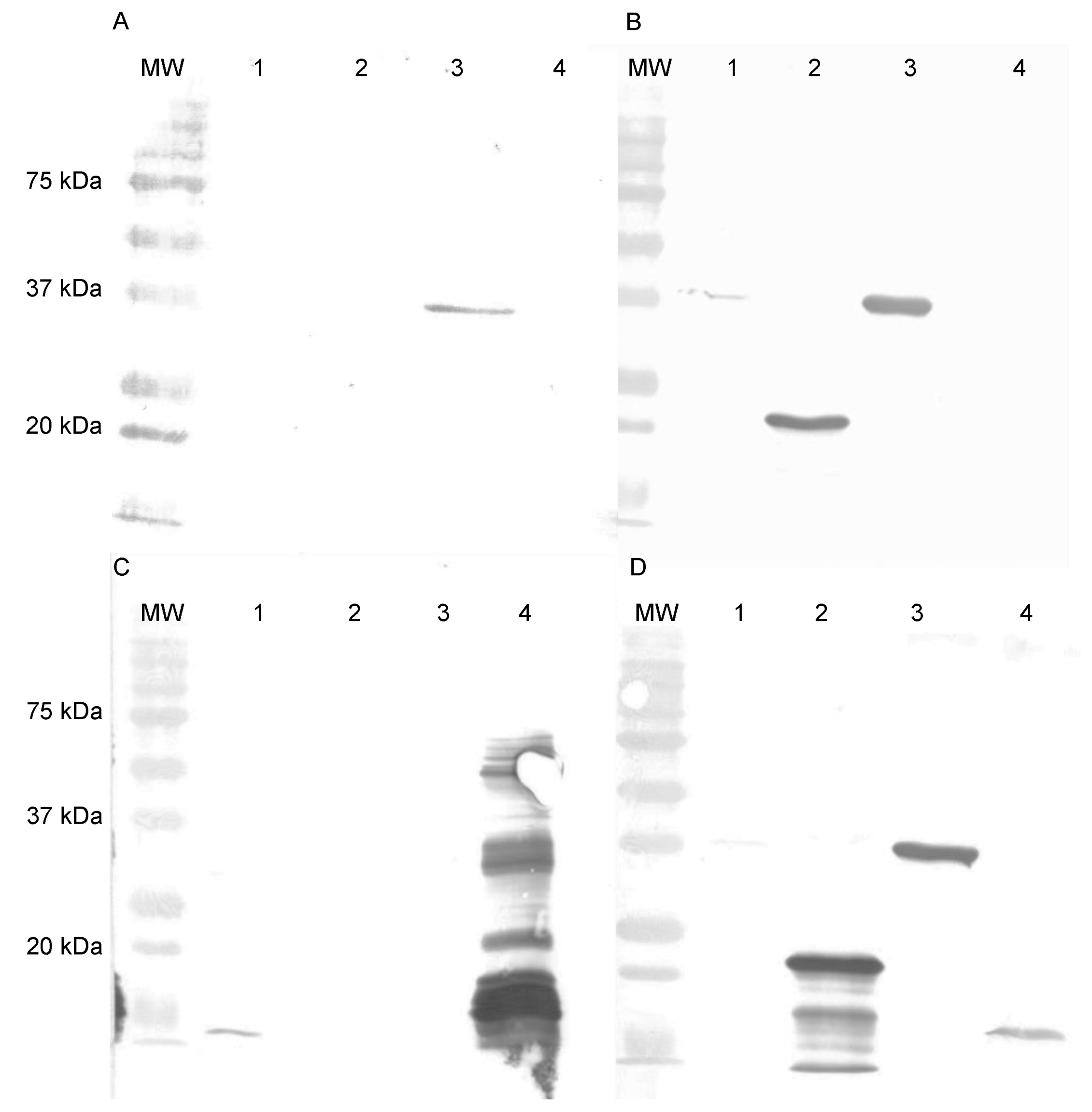
2.8. SDS-PAGE and Western Blotting
2.9. Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. Epitope Mapping by SPOT Synthesis on Nitrocellulose Membranes
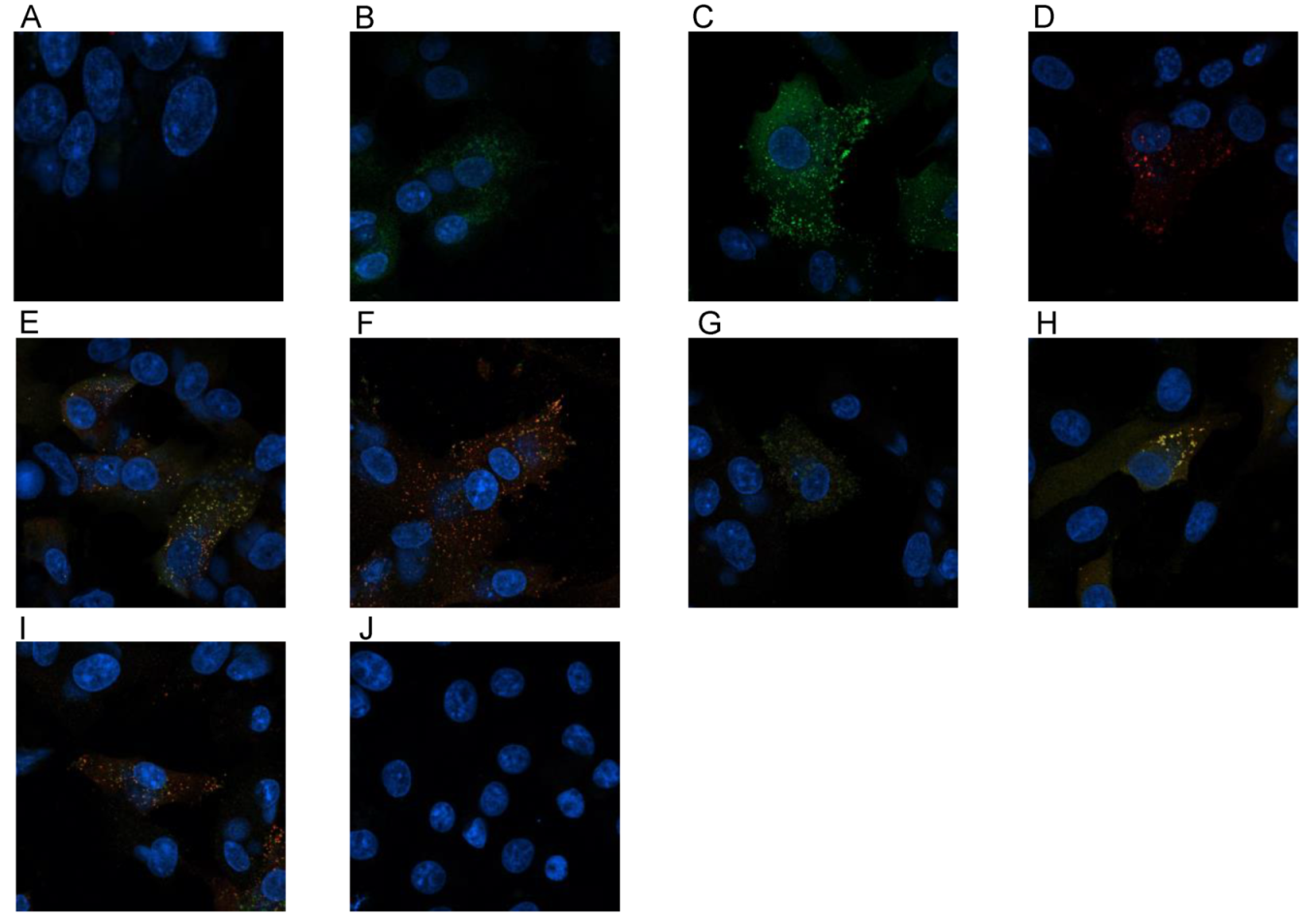
2.11. Cell Infection and Confocal Microscopy
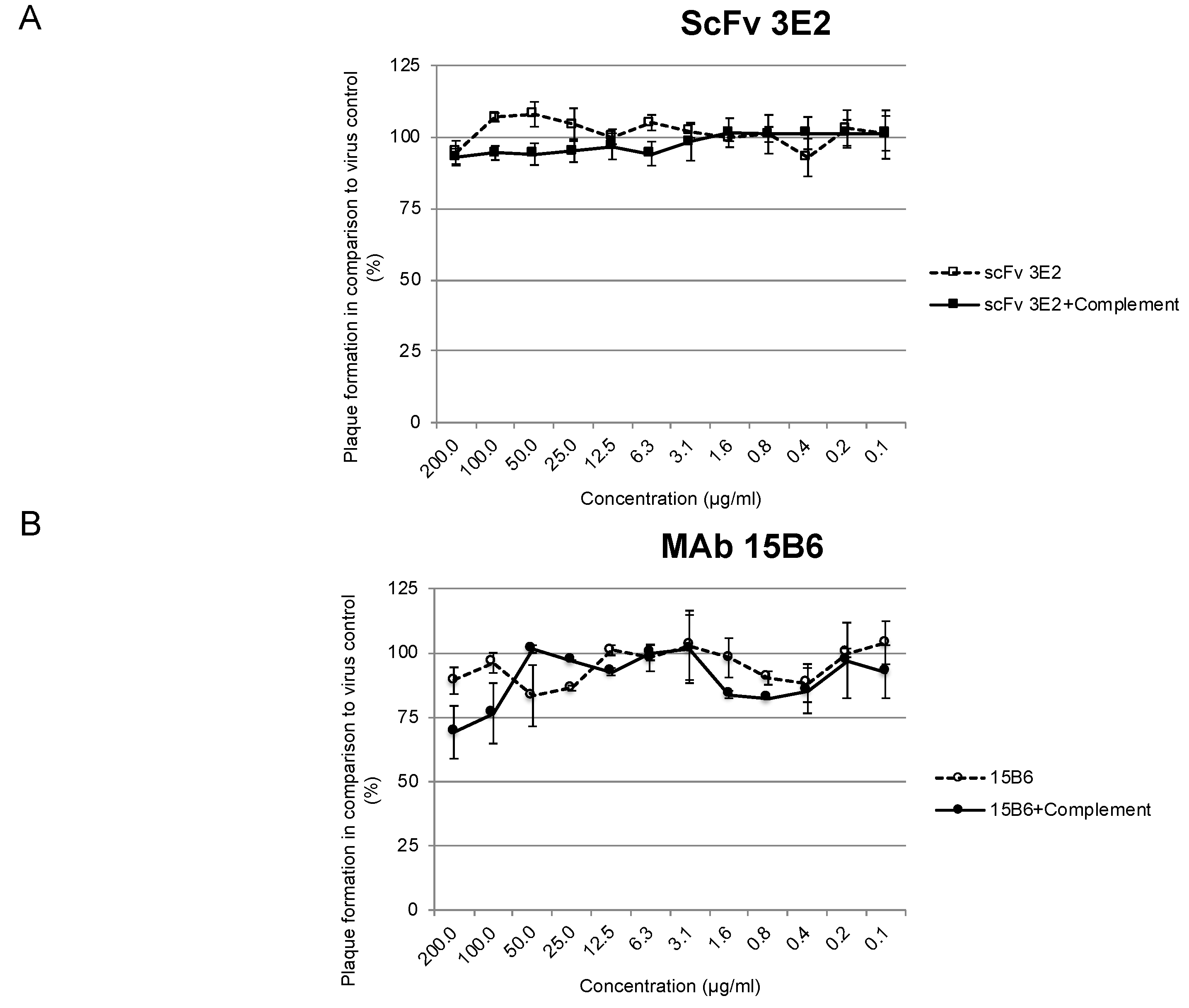
2.12. In Vitro Plaque Reduction Neutralisation Test (PRT)
3. Results
3.1. Selection of an Anti-F13 Specific scFv Antibody
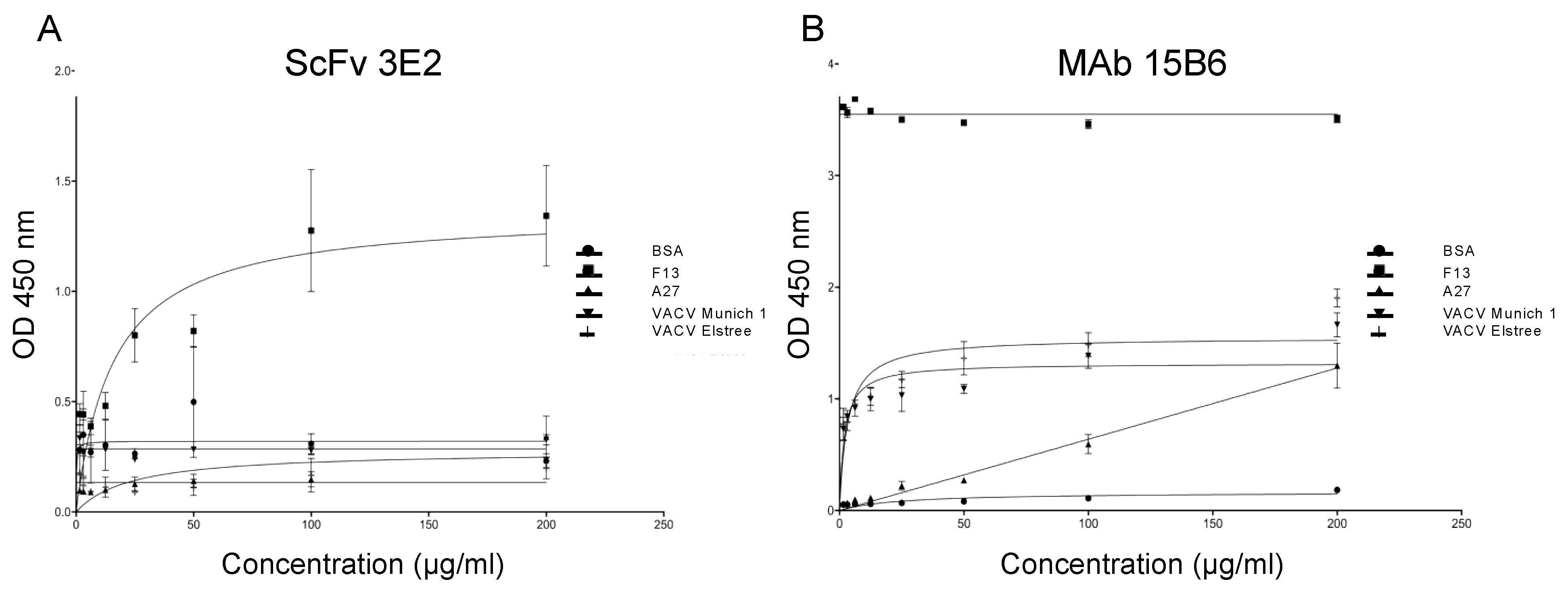
3.2. Specificity and Binding Affinity Studies
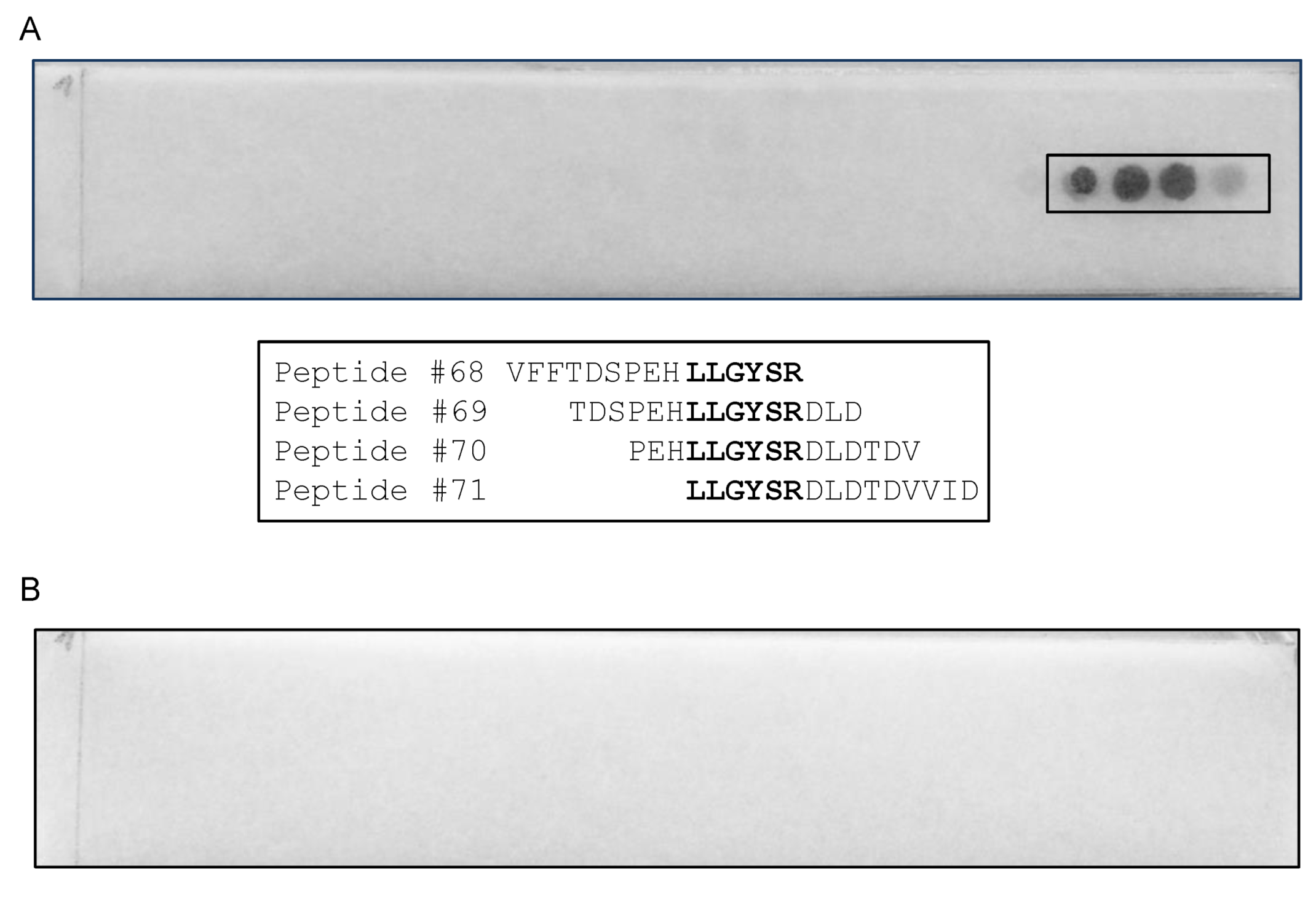
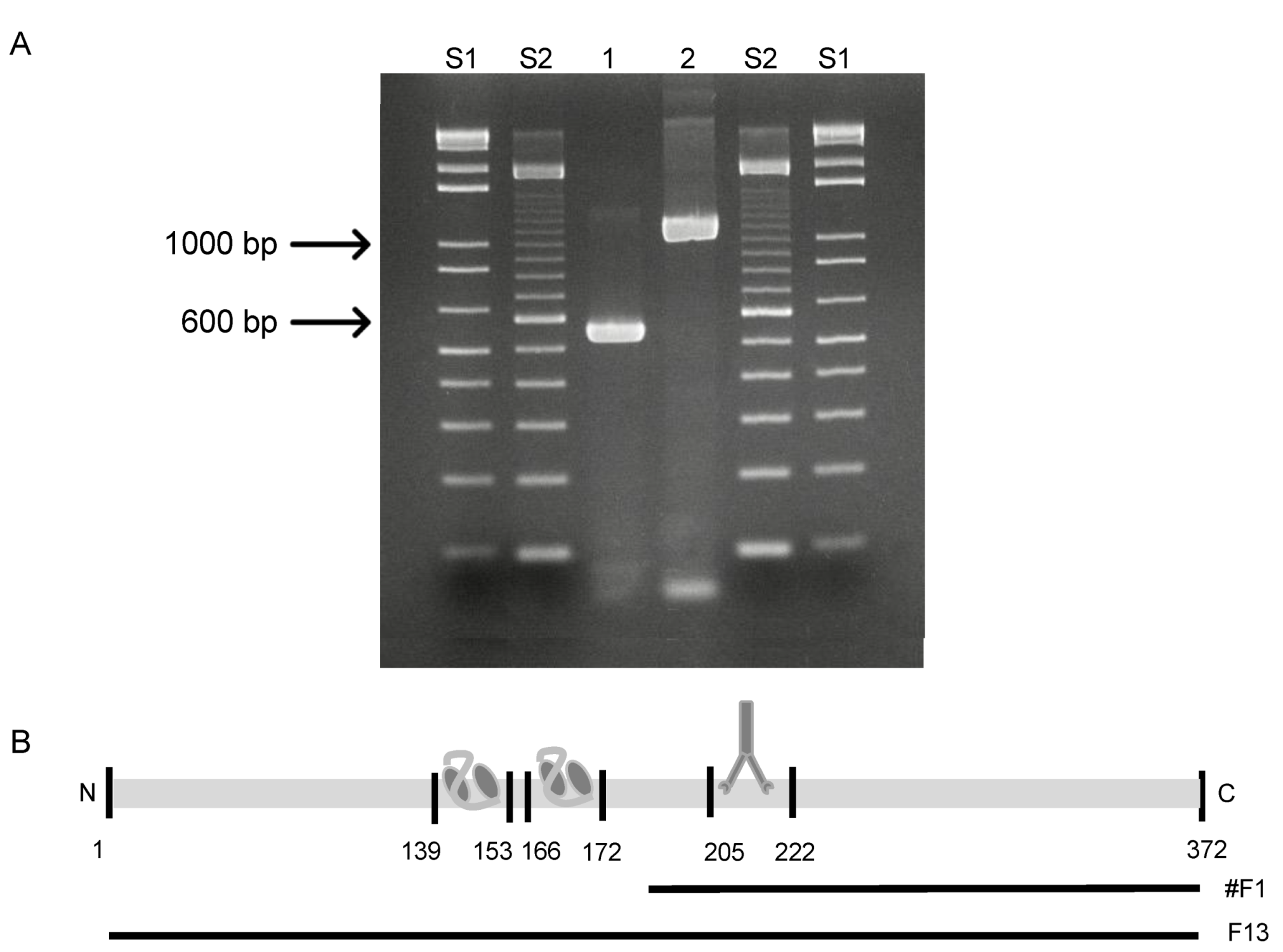
3.3. Epitope Mapping Using a Peptide Membrane and Truncated Recombinant F13 Fragment
3.4. Confocal Microscopy
3.5. Neutralisation Abilities In Vitro
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moss, B. Poxvirus entry and membrane fusion. Virology 2006, 344, 48–54. [Google Scholar] [CrossRef] [Green Version]
- Fenner, F.; Henderson, D.A.; Arita, I.; Jezek, Z.; Ladnyi, I.D. Smallpox and its Eradication; World Health Organization: Geneva, Switzerland, 1988; pp. 1–1460. [Google Scholar]
- Kurth, A.; Wibbelt, G.; Gerber, H.P.; Petschaelis, A.; Pauli, G.; Nitsche, A. Rat-to-elephant-to-human transmission of cowpox virus. Emerg. Infect. Dis. 2008, 14, 670–671. [Google Scholar] [CrossRef]
- Becker, C.; Kurth, A.; Hessler, F.; Kramp, H.; Gokel, M.; Hoffmann, R.; Kuczka, A.; Nitsche, A. Cowpox virus infection in pet rat owners: Not always immediately recognized. Dtsch. Ärzteblatt Int. 2009, 106, 329–334. [Google Scholar] [CrossRef]
- Vorou, R.M.; Papavassiliou, V.G.; Pierroutsakos, I.N. Cowpox virus infection: An emerging health threat. Curr. Opin. Infect. Dis. 2008, 21, 153–156. [Google Scholar] [CrossRef]
- Campe, H.; Zimmermann, P.; Glos, K.; Bayer, M.; Bergemann, H.; Dreweck, C.; Graf, P.; Weber, B.K.; Meyer, H.; Büttner, M.; et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg. Infect. Dis. 2009, 15, 777–780. [Google Scholar] [CrossRef]
- Ladnyj, I.D.; Ziegler, P.; Kima, E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972, 46, 593–597. [Google Scholar] [PubMed]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.; et al. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, A.; Aarons, E.; Astbury, J.; Balasegaram, S.; Beadsworth, M.; Beck, C.R.; Chand, M.; O’Connor, C.; Dunning, J.; Ghebrehewet, S.; et al. Two cases of monkeypox imported to the United Kingdom, September 2018. Eurosurveillance 2018, 23, 1800509. [Google Scholar] [CrossRef] [PubMed]
- Henderson, D.A. The looming threat of bioterrorism. Science 1999, 283, 1279–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, K.L.; Smith, G.L. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008, 16, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L.; Vanderplasschen, A.; Law, M. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 2002, 83 Pt 12, 2915–2931. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus cell entry: How many proteins does it take? Viruses 2012, 4, 688–707. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L.; Murphy, B.J.; Law, M. Vaccinia virus motility. Annu. Rev. Microbiol. 2003, 57, 323–342. [Google Scholar] [CrossRef] [Green Version]
- Hiller, G.; Weber, K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 1985, 55, 651–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geada, M.M.; Galindo, I.; Lorenzo, M.M.; Perdiguero, B.; Blasco, R. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 2001, 82 Pt 11, 2747–2760. [Google Scholar] [CrossRef]
- Payne, L.G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 1980, 50, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.F.; Janeczko, R.; Esteban, M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J. Virol. 1985, 56, 482–488. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, J.C.; Chung, C.S.; Chang, W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 1999, 73, 8750–8761. [Google Scholar] [CrossRef] [Green Version]
- Davies, D.H.; McCausland, M.M.; Valdez, C.; Huynh, D.; Hernandez, J.E.; Mu, Y.; Hirst, S.; Villarreal, L.; Felgner, P.L.; Crotty, S. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 2005, 79, 11724–11733. [Google Scholar] [CrossRef] [Green Version]
- Ichihashi, Y.; Oie, M. Neutralizing epitope on penetration protein of vaccinia virus. Virology 1996, 220, 491–494. [Google Scholar] [CrossRef] [Green Version]
- Wolffe, E.J.; Vijaya, S.; Moss, B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 1995, 211, 53–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaever, T.; Matho, M.H.; Meng, X.; Crickard, L.; Schlossman, A.; Xiang, Y.; Crotty, S.; Peters, B.; Zajonc, D.M. Linear Epitopes in Vaccinia Virus A27 Are Targets of Protective Antibodies Induced by Vaccination against Smallpox. J. Virol. 2016, 90, 4334–4345. [Google Scholar] [CrossRef] [Green Version]
- Matho, M.H.; Schlossman, A.; Gilchuk, I.M.; Miller, G.; Mikulski, Z.; Hupfer, M.; Wang, J.; Bitra, A.; Meng, X.; Xiang, Y.; et al. Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8. J. Biol. Chem. 2018, 293, 390–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matho, M.H.; Schlossman, A.; Meng, X.; Benhnia, M.R.; Kaever, T.; Buller, M.; Doronin, K.; Parker, S.; Peters, B.; Crotty, S.; et al. Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer. PLoS Pathog. 2015, 11, e1005148. [Google Scholar] [CrossRef] [Green Version]
- Moss, B. Smallpox vaccines: Targets of protective immunity. Immunol. Rev. 2011, 239, 8–26. [Google Scholar] [CrossRef]
- Benhnia, M.R.; McCausland, M.M.; Moyron, J.; Laudenslager, J.; Granger, S.; Rickert, S.; Koriazova, L.; Kubo, R.; Kato, S.; Crotty, S. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J. Virol. 2009, 83, 1201–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiller, G.; Eibl, H.; Weber, K. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J. Virol. 1981, 39, 903–913. [Google Scholar] [CrossRef] [Green Version]
- Hirt, P.; Hiller, G.; Wittek, R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 1986, 58, 757–764. [Google Scholar] [CrossRef] [Green Version]
- Grosenbach, D.W.; Hruby, D.E. Analysis of a vaccinia virus mutant expressing a nonpalmitylated form of p37, a mediator of virion envelopment. J. Virol. 1998, 72, 5108–5120. [Google Scholar] [CrossRef] [Green Version]
- Grosenbach, D.W.; Ulaeto, D.O.; Hruby, D.E. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. J. Biol. Chem. 1997, 272, 1956–1964. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, C.; Rindisbacher, L.; Galmiche, M.C.; Wittek, R. Biochemical analysis of the major vaccinia virus envelope antigen. Virology 1995, 213, 19–27. [Google Scholar] [CrossRef]
- Borrego, B.; Lorenzo, M.M.; Blasco, R. Complementation of P37 (F13L gene) knock-out in vaccinia virus by a cell line expressing the gene constitutively. J. Gen. Virol. 1999, 80 Pt 2, 425–432. [Google Scholar] [CrossRef]
- Husain, M.; Moss, B. Vaccinia virus F13L protein with a conserved phospholipase catalytic motif induces colocalization of the B5R envelope glycoprotein in post-Golgi vesicles. J. Virol. 2001, 75, 7528–7542. [Google Scholar] [CrossRef] [Green Version]
- Breman, J.G.; Henderson, D.A. Poxvirus dilemmas—Monkeypox, smallpox, and biologic terrorism. N. Engl. J. Med. 1998, 339, 556–559. [Google Scholar] [CrossRef]
- Grosenbach, D.W.; Jordan, R.; Hruby, D.E. Development of the small-molecule antiviral ST-246 as a smallpox therapeutic. Future Virol. 2011, 6, 653–671. [Google Scholar] [CrossRef] [Green Version]
- Schmaljohn, C.; Cui, Y.; Kerby, S.; Pennock, D.; Spik, K. Production and characterization of human monoclonal antibody Fab fragments to vaccinia virus from a phage-display combinatorial library. Virology 1999, 258, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Farajnia, S.; Ahmadzadeh, V.; Tanomand, A.; Veisi, K.; Khosroshahi, S.A.; Rahbarnia, L. Development trends for generation of single-chain antibody fragments. Immunopharmacol. Immunotoxicol. 2014, 36, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Czerny, C.P.; Mahnel, H. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J. Gen. Virol. 1990, 71 Pt 10, 2341–2352. [Google Scholar] [CrossRef] [PubMed]
- Joklik, W.K. The purification fo four strains of poxvirus. Virology 1962, 18, 9–18. [Google Scholar] [CrossRef]
- Czerny, C.P.; Johann, S.; Holzle, L.; Meyer, H. Epitope detection in the envelope of intracellular naked orthopox viruses and identification of encoding genes. Virology 1994, 200, 764–777. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Galfre, G.; Milstein, C. Preparation of monoclonal antibodies: Strategies and procedures. Methods Enzymol. 1981, 73 Pt B, 3–46. [Google Scholar]
- Schmelz, M.; Sodeik, B.; Ericsson, M.; Wolffe, E.J.; Shida, H.; Hiller, G.; Griffiths, G. Assembly of vaccinia virus: The second wrapping cisterna is derived from the trans Golgi network. J. Virol. 1994, 68, 130–147. [Google Scholar] [CrossRef] [Green Version]
- Diesterbeck, U.S.; Ahsendorf, H.P.; Frenzel, A.; Sharifi, A.R.; Schirrmann, T.; Czerny, C.P. Characterization of an In Vivo Neutralizing Anti-Vaccinia Virus D8 Single-Chain Fragment Variable (scFv) from a Human Anti-Vaccinia Virus-Specific Recombinant Library. Vaccines 2021, 9, 1308. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Brochet, X.; Lefranc, M.P.; Giudicelli, V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008, 36, W503–W508. [Google Scholar] [CrossRef] [Green Version]
- Giudicelli, V.; Brochet, X.; Lefranc, M.P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011, 2011, 695–715. [Google Scholar] [CrossRef]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Michaelis, L.; Menten, M.L. Die Kinetik der Invertinwirkung. Biochem. Z 1913, 49, 333–369. [Google Scholar]
- Frank, R. Spot-Synthesis—an Easy Technique for the Positionally Addressable, Parallel Chemical Synthesis on a Membrane Support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Beutling, U.; Stading, K.; Stradal, T.; Frank, R. Large-scale analysis of protein-protein interactions using cellulose-bound peptide arrays. Protein–Protein Interact. 2008, 110, 115–152. [Google Scholar]
- Ahsendorf, H.P.; Gan, L.L.; Eltom, K.H.; Abd El Wahed, A.; Hotop, S.K.; Roper, R.L.; Beutling, U.; Broenstrup, M.; Stahl-Hennig, C.; Hoelzle, L.E.; et al. Species-Specific Conservation of Linear Antigenic Sites on Vaccinia Virus A27 Protein Homologs of Orthopoxviruses. Viruses 2019, 11, 493. [Google Scholar] [CrossRef] [Green Version]
- Husain, M.; Weisberg, A.; Moss, B. Topology of epitope-tagged F13L protein, a major membrane component of extracellular vaccinia virions. Virology 2003, 308, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Roos, N.; Cyrklaff, M.; Cudmore, S.; Blasco, R.; Krijnse-Locker, J.; Griffiths, G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996, 15, 2343–2355. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.M.; Wilkins, T.D. Localization of two epitopes recognized by monoclonal antibody PCG-4 on Clostridium difficile toxin A. Infect. Immun. 1992, 60, 2488–2492. [Google Scholar] [CrossRef] [Green Version]
- Krawczyk, A.; Krauss, J.; Eis-Hubinger, A.M.; Daumer, M.P.; Schwarzenbacher, R.; Dittmer, U.; Schneweis, K.E.; Jager, D.; Roggendorf, M.; Arndt, M.A.E.; et al. Impact of valency of a glycoprotein B-specific monoclonal antibody on neutralization of herpes simplex virus. J. Virol. 2011, 85, 1793–1803. [Google Scholar] [CrossRef] [Green Version]
- Matho, M.H.; de Val, N.; Miller, G.M.; Brown, J.; Schlossman, A.; Meng, X.; Crotty, S.; Peters, B.; Xiang, Y.; Hsieh-Wilson, L.C.; et al. Murine anti-vaccinia virus D8 antibodies target different epitopes and differ in their ability to block D8 binding to CS-E. PLoS Pathog. 2014, 10, e1004495. [Google Scholar] [CrossRef] [Green Version]
- Petersen, B.; Lundegaard, C.; Petersen, T.N. NetTurnP--neural network prediction of beta-turns by use of evolutionary information and predicted protein sequence features. PLoS ONE 2010, 5, e15079. [Google Scholar] [CrossRef] [Green Version]
- Chou, P.Y.; Fasman, G.D. Prediction of beta-turns. Biophys. J. 1979, 26, 367–383. [Google Scholar] [CrossRef] [Green Version]
- McCausland, M.M.; Benhnia, M.R.; Crickard, L.; Laudenslager, J.; Granger, S.W.; Tahara, T.; Kubo, R.; Koriazova, L.; Kato, S.; Crotty, S. Combination therapy of vaccinia virus infection with human anti-H3 and anti-B5 monoclonal antibodies in a small animal model. Antivir. Ther. 2010, 15, 661–675. [Google Scholar] [CrossRef] [Green Version]
- Benhnia, M.R.; McCausland, M.M.; Laudenslager, J.; Granger, S.W.; Rickert, S.; Koriazova, L.; Tahara, T.; Kubo, R.T.; Kato, S.; Crotty, S. Heavily isotype-dependent protective activities of human antibodies against vaccinia virus extracellular virion antigen B5. J. Virol. 2009, 83, 12355–12367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, J.C.; Tapia, E.; Esteban, M. Administration to mice of a monoclonal antibody that neutralizes the intracellular mature virus form of vaccinia virus limits virus replication efficiently under prophylactic and therapeutic conditions. J. Gen. Virol. 2002, 83 Pt 5, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Benhnia, M.R.; Maybeno, M.; Blum, D.; Aguilar-Sino, R.; Matho, M.; Meng, X.; Head, S.; Felgner, P.L.; Zajonc, D.M.; Koriazova, L.; et al. Unusual features of vaccinia virus extracellular virion form neutralization resistance revealed in human antibody responses to the smallpox vaccine. J. Virol. 2013, 87, 1569–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005, 23, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Milenic, D.E.; Whitlow, M.; Schlom, J. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms. Cancer Res. 1992, 52, 3402–3408. [Google Scholar] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahsendorf, H.P.; Diesterbeck, U.S.; Hotop, S.-K.; Winkler, M.; Brönstrup, M.; Czerny, C.-P. Characterisation of an Anti-Vaccinia Virus F13 Single Chain Fragment Variable from a Human Anti-Vaccinia Virus-Specific Recombinant Immunoglobulin Library. Viruses 2022, 14, 197. https://doi.org/10.3390/v14020197
Ahsendorf HP, Diesterbeck US, Hotop S-K, Winkler M, Brönstrup M, Czerny C-P. Characterisation of an Anti-Vaccinia Virus F13 Single Chain Fragment Variable from a Human Anti-Vaccinia Virus-Specific Recombinant Immunoglobulin Library. Viruses. 2022; 14(2):197. https://doi.org/10.3390/v14020197
Chicago/Turabian StyleAhsendorf, Henrike P., Ulrike S. Diesterbeck, Sven-Kevin Hotop, Michael Winkler, Mark Brönstrup, and Claus-Peter Czerny. 2022. "Characterisation of an Anti-Vaccinia Virus F13 Single Chain Fragment Variable from a Human Anti-Vaccinia Virus-Specific Recombinant Immunoglobulin Library" Viruses 14, no. 2: 197. https://doi.org/10.3390/v14020197
APA StyleAhsendorf, H. P., Diesterbeck, U. S., Hotop, S.-K., Winkler, M., Brönstrup, M., & Czerny, C.-P. (2022). Characterisation of an Anti-Vaccinia Virus F13 Single Chain Fragment Variable from a Human Anti-Vaccinia Virus-Specific Recombinant Immunoglobulin Library. Viruses, 14(2), 197. https://doi.org/10.3390/v14020197






