Abstract
Food contamination by Salmonella can lead to serious foodborne diseases that constantly threaten public health. Innovative and effective strategies are needed to control foodborne pathogenic contamination since the incidence of foodborne diseases has increased gradually. In the present study, two broad-spectrum phages named Salmonella phage PSE-D1 and Salmonella phage PST-H1 were isolated from sewage in China. Phages PSE-D1 and PST-H1 were obtained by enrichment with Salmonella enterica subsp. enterica serovar Enteritidis (S. Enteritidis) CVCC1806 and Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) CVCC3384, respectively. They were able to lyse Salmonella, E. coli and K. pneumoniae and exhibited broad host range. Further study demonstrated that PSE-D1 and PST-H1 showed high pH and thermal tolerances. Phage PSE-D1 belongs to the Jiaodavirus genus, Tevenvirinae subfamily, while phage PST-H1 belongs to the Jerseyvirus genus, Guernseyvirinae subfamily according to morphology and phylogeny. The results of genome analysis showed that PSE-D1 and PST-H1 lack virulence and drug-resistance genes. The effects of PSE-D1 and PST-H1 on controlling S. Enteritidis CVCC1806 and S. Typhimurium CVCC3384 contamination in three kinds of foods (eggshells, sausages and milk) were further investigated, respectively. Our results showed that, compared to phage-free groups, PSE-D1 and PST-H1 inhibited the growth of their host strain significantly. A significant reduction of host bacteria titers (1.5 and 1.9 log10 CFU/sample, p < 0.001) on eggshells was observed under PSE-D1 and PST-H1 treatments, respectively. Furthermore, administration of PSE-D1 and PST-H1 decreased the counts of bacteria by 1.1 and 1.2 log10 CFU/cm2 (p < 0.001) in sausages as well as 1.5 and 1.8 log10 CFU/mL (p < 0.001) in milk, respectively. Interesting, the bacteriostasis efficacy of both phages exhibited more significantly at 4 °C than that at 28 °C in eggshells and milk and sausages. In sum, the purpose of our research was evaluating the counteracting effect of phage PSE-D1 and PST-H1 on the spread of Salmonella on contaminated foods products. Our results suggested that these two phage-based biocontrol treatments are promising strategies for controlling pathogenic Salmonella contaminated food.
1. Introduction
The primary cause of foodborne illnesses is bacterial contamination in food. Salmonella accounts for the highest proportion of bacterial food-borne contamination in the U.S.A, causing 1.4 million cases of Salmonella food-borne diseases each year [1]. During 2017, it was reported in Britain that S. Typhimurium and S. Enteritidis were two of the top five most common serotypes in bacterial infections [2]. Fresh-cut produce that is easily consumed, uncooked and under-cooked meat, poultry, and eggs contaminated with Salmonella are further risk factors for infection [1]. Salmonella contamination was very common in eggs, meat, and meat products [3].
Effective and innovative methods to preserve food are critical for food quality and safety. Bacterial food contamination can be controlled by chemical, physical, and biological techniques. Nevertheless, some traditional procedures cause adverse effects on food quality [4]. Thermal treatment, the most widely used procedure for microbial inactivation in foods, usually causes unwanted side effects in the sensory and nutritional qualities of food [5]. In the germicidal process, irradiation can change the flavor and color of meat products in addition to producing a distinctive odor. Although antimicrobials like potassium benzoate and sodium lactate are frequently used for increasing shelf-life, the safety of foods cannot be guaranteed if they are used alone [6]. In order to reduce the occurrence of food-borne salmonellosis and inhibit the contamination of Salmonella in foods, appropriate approaches are urgently required.
Phage-based biological control strategies are considered potential methods to control bacterial contamination of food gradually. Phages are the most abundant microorganisms found on earth and are widespread on foods of various origins [7]. Compared to conventional antimicrobials, bacteriophages are widespread, easily available, and harmless to the public. Phages have high specificity and an effective ability to lyse targeted pathogenic bacteria. They are natural killers of bacteria. Until now, some studies have reported the application of bacteriophages on bacterial contamination in dairy foods. Huang et al. showed that broad-spectrum Salmonella bacterio phage LPST10 had the potential to reduce titers of S. Typhimurium and S. Enteritidis by 0.92 to 5.12 log10 CFU/sample in lettuce and sausages [8]. Another similar study certified that, in both liquid egg white and yolk, considerable bacteria inhibition in two MDR Salmonella strains were observed by phage D1-2 at different temperatures [9]. Furthermore, Hudson et al. found that, compared to phage-free groups, phages could significantly inhibit the growth of E. coli O157:H7 by 4 log10 CFU/cm2 on cooked and raw beef at two different temperatures [10]. These studies suggested that phages have the potential to control bacterial food-borne contamination.
Therefore, our paper aimed to investigate the effectiveness of bacteriophages on inhibiting S. Enteritidis CVCC1806 and S. Typhimurium CVCC3384 contamination in eggshells, chicken sausages, and milk at different temperatures to explore their use in these and other food products.
2. Materials and Methods
2.1. Bacteria Strains and Culture Medium
Our study involved seventy bacteria strains, including 6 Salmonella strains, 48 E. coli strains, 15 K. pneumoniae strains, and 1 P. aeruginosa strain. Salmonella enterica subsp. enterica serovar Enteritidis (S. Enteritidis) CVCC1806, Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) CVCC3384, Salmonella enterica subsp. enterica serovar Pullorum (S. Pullorum) SX-1014, and SF-0923 were bought from the China Veterinary Culture Collection Center (CVCC). The remaining strains came from poultry and pig farms or clinics. In our lab, all bacteria strains were stored at −80 °C in LB broth contained with 20% v/v glycerol.
2.2. Isolation and Purification of Bacteriophages
Two phages named Salmonella phage PSE-D1 and Salmonella phage PST-H1 were isolated with S. Enteritidis CVCC1806 and S. Typhimurium CVCC3384 as host bacteria, respectively. PSE-D1 and PST-H1 were separated from cattle farm sewage in Guangxi, China. Purification and amplification of the two phages were then performed by using host strains, and 10mL of LB broth was added into the mixture of 100 μL samples and 100 μL log-phase bacteria and subsequently cultured for 4–5 h at 37 °C. The supernatant was collected by filtering through a micro-porous membrane of 0.22 µm after centrifuging at 12,000 rpm for 5 min. The morphology of phage plaque and titer were investigated by using the double-layer agar method. The presence of plaque would be observed after incubating for 4–5 h at 37 °C in an incubator [11,12]. The purified phage suspension was obtained after purification three times. Finally, phages would be preserved at −80 °C into a SM buffer containing 20% v/v glycerol in our laboratory.
2.3. Host Range
The host spectrum of phage PSE-D1 and PST-H1 were investigated against 6 Salmonella strains, 48 E. coli strains, 15 K. pneumoniae strains, and 1 P. aeruginosa strain through a spot test [13]. The bottom layer consisted of LB broth and 1.0% agar (10 mL in total). The mixture of LB contained 0.5% agar (2–3 mL), and the bacterial culture of the tested bacterial strains (100 µL) was overburden. The overlay was added after solidification of the bottom layer. Phage suspensions (4–5 µL, 109 PFU/mL) were then dropped onto the previously mentioned plates and incubated at 37 °C overnight.
2.4. Characterization of Phages
2.4.1. One Step Growth
A one-step growth curve experiment of phages was determined with some modifications [14]. A mixture of log-phase bacteria and a phage was incubated for 15 min at 37 °C. The supernatant fluid was then taken out after centrifuging at 12,000 rpm for 1 min and resuspended with LB broth. Then the suspension was transferred into 10 mL LB broth and incubated at 37 °C. Sampling was done at 0, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, and 120 min and used the double-layer method to assess phage titers. Burst size was calculated according to standard methodology [15].
2.4.2. Lytic Capacity of Phages under Different Multiplicity of Infection (MOI)
Culture S. Enteritidis CVCC1806 and S. Typhimurium CVCC3384 strains were incubated under the above conditions [16]. The culture was then loaded into glass test tubes at 1 × 106 CFU/mL, and phages were added under MOIs of 100, 10, 1, 0.1, or 0.01 (PFU/CFU). Incubated for 15h at 37 °C in a table concentrator at 180 rpm and the cellular density was evaluated at OD450nm every hour.
2.4.3. Thermal and pH Stability of Phages
The thermal and pH stability of the phages were determined using a modification of the previously described methods [17,18,19]. For temperature sensitivity testing, a purified phage was incubated at refrigeration temperature (4 °C), room temperature (25 °C), incubation temperature (37 °C), and other temperatures (50, 60, 70, 80 °C), respectively. Sampling was performed at 30 and 60 min, and phage titers were assessed by double-layer method. The purified phages were incubated at different pH values (2–13) for 2 h at 37 °C in order to evaluate the pH sensitivity of the phages.
2.4.4. Electron Microscopy
After being resuspended in ammonium acetate (0.1 mol/L), the phage suspension (1010 PFU/mL) was centrifuged at 4 °C with 40,000 rpm for 1 h. The immersed phages were suspended in the copper grid for 10 min, followed by staining with a 2% volume fraction of phosphotungstic acid solution (pH = 7) for 10 min. The morphology of the phages was observed by transmission electron microscopy (TEM) (Hitachi High-Tech Co., Ltd., Tokyo, Japan).
2.4.5. Extraction and Analysis of Phage Genome Sequence
A suspension with a high titer (1010 PFU/mL) of phages was used to extract genomic DNA with modification of the previously described methods [20]. Beijing University of Chemical Technology carried out the control of sequencing data quality and the construction of a sequencing library. A RAST server 2.0 was used to forecast and annotate the coding DNA sequences (CDSs). The Virulence Factor Database and Comprehensive Antibiotic Resistance Database were used to screen the hypothetical virulence factors and drug-resistance genes, respectively [21,22]. A CGView Server was used to depict a comparative circular genome map of the phage genome. The phylogenetic trees were constructed by using the ClustalW program based on the whole genome in MEGA-X [23].
2.5. Assays on the Surface of Egg Shells, Chicken Sausage and in Fresh Milk
2.5.1. Sample Preparation
Salmonella CVCC1806 and CVCC3384 inoculum were prepared as mentioned above. Eggs, chicken sausages, and fresh milk were bought from a supermarket and stored at 4 °C. To eliminate bacteria on the surface of the eggs, the surface was washed with 75% ethanol and exposed to ultraviolet radiation for 40 min. Packed chicken sausages were aseptically cut into slices with a diameter of 2cm and a thickness of 1cm in a biological safety cabinet. To reduce the indigenous bacteria level before inoculation, the pieces of chicken sausages were irradiated with UV light for 1h in a safety cabinet (30 min on each side).
2.5.2. Salmonella and Phage Preparation
The host strain cultures were inoculated and spread evenly on the surface of the eggshells (100 µL, 1.0 × 108 CFU/mL) and sausages (25 µL, 4.0 × 105 CFU/mL). Before phage treatment, all samples were dried for 10 min (eggs) or 15–20 min (sausages) in a biological safety cabinet. The Salmonella suspension (100 µL, 1.0 × 105 CFU/mL) was artificially inoculated into the fresh milk.
Samples were inoculated with a phage lysate at a MOI of 100 and 10, respectively at 4 °C and 28 °C for phage treatment. The above phage lysate was suspended in Tris-SM buffer, and sterile Tris-SM buffers were added into the control samples. The microbiological analysis of eggshells and sausages were carried out at 2, 4, 6, 24, and 48h [24,25], and the milk was carried out at 2, 4, 6, and 24h [26,27]. The eggshells were washed with physiological saline [28], and sausages were ground before counting. Plating on Salmonella Shigella agar (SS agar) was used to calculate bacterial counts (CFU/mL).
2.6. Statistical Analysis
All the experiments were conducted three times. SPSS statistical software was used for analysis statistical. The significance level (p < 0.05) between each group was determined by a one-way ANOVA with a confidence interval of 95%.
3. Results
3.1. Isolation and Purification of Bacteriophages
Two broad-spectrum phages named Salmonella phage PSE-D1 and Salmonella phage PST-H1 were isolated from sewage. Phage PSE-D1 and PST-H1 were obtained by enrichment with S. Enteritidis CVCC1806 and S. Typhimurium CVCC3384, respectively. The plaque of these bacteriophages on the double-layer agar plate was bright and clean, with clear edges. (Figure 1).
Figure 1.
Morphology on double−layer agar plates. (a) phage PSE-D1. (b) phage PST-H1.
3.2. Host Range
A total of 70 strains were used to test the hosts spectrum of phage PSE-D1 and phage PST-H1, respectively, including 6 Salmonella strains, 48 E. coli strains, 15 K. pneumoniae strains, and 1 P. aeruginosa strain (Table 1). Phage PSE-D1 could infect 3 Salmonella strains, 13 E. coli strains, and 8 K. pneumoniae strains. Phage PST-H1 could infect 3 Salmonella strains, 10 E. coli strains, and 11 K. pneumoniae strains.

Table 1.
Host strains information of phage PST-H1 and PSE-D1.
3.3. Biological Characterization of Phages
3.3.1. Electron Microscopy
According to the International Committee on Taxonomy of Viruses (ICTV) and structural analysis by transmission electron microscopy, PSE-D1 belongs to the Tevenvirinae subfamily. The head of PSE-D1 is elongated, icosahedral-shaped, and the diameter is about 69.2 nm. The tail of PSE-D1 is contractile with a 94.5 nm length approximately (Figure 2a). Phage PST-H1 belongs to the Guernseyvirinae subfamily. PST-H1 has a typical regular icosahedral-shaped head, and the diameter is about 51.0 nm. The tail of PST-H1 is long and non-contractile with a 125.1 nm length approximately (Figure 2b).
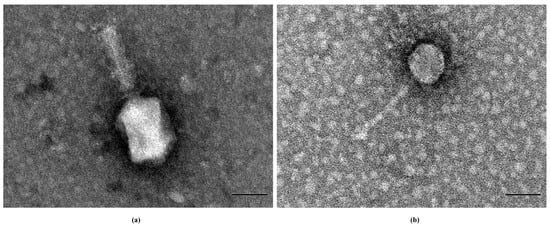
Figure 2.
TEM of phages. (a) phage PSE-D1. (b) phage PST-H1.
3.3.2. One Step Growth
The infection potential of the phages was analyzed through one-step growth curves (Figure 3). PSE-D1 had a latent period of 10 min approximately, according to the growth curve (Figure 3a). Over the next 60 min, the phage titer of PSE-D1 significantly increased, and PSE-D1 then grew stably until 120 min. The burst size of PSE-D1 was around 110 PFU/CFU. PST-H1 had a latent period of 20 min approximately (Figure 3b) and had a burst size of 183 PFU/CFU.
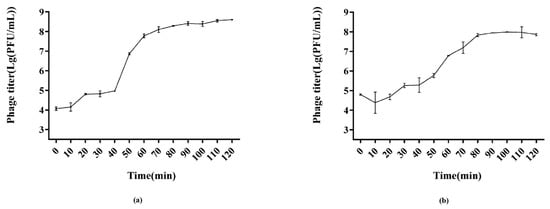
Figure 3.
One step growth curves of phages. (a) phage PSE-D1. (b) phage PST-H1.
3.3.3. Lytic Capacity of Phages against Host at Different MOI
The growth of the host (OD450 nm) was measured to determine the lytic capacity of the phage against strain. As shown in Figure 4, proliferation of the host strains could consistently be inhibited by phages PSE-D1 and PST-H1 for 15 hours, respectively. The viable counts of host strains increased at 1–15 h in the positive control groups. The efficacy at the MIOs of 100 and 10 were better than other treatment groups with lower-titer phages, revealing that the antibacterial effect of PSE-D1 was MOI dependent. PST-H1 had an obvious strong inhibitory effect of CVCC3384 within 5 h. The strain counts of phage-treatment groups reduced significantly at 1-15 h compared to the phage-free control (Figure 4b).
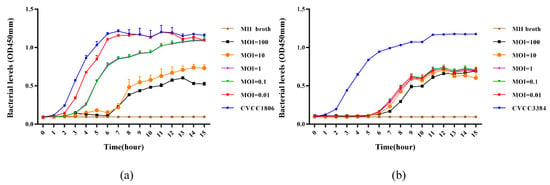
Figure 4.
(a) Lytic capacity of phage PSE-D1 on S. Enteritidis CVCC1806 in LB broth medium. (b) Inhibitory effect of phage PST-H1 on S. Typhimurium CVCC3384 in LB broth medium. Phage and host strain of each group were added at the same time.
3.3.4. Thermal and pH Stability of Phages
Phages PSE-D1 and PST-H1 were stable at 4–60 °C in a thermal stability test, revealing high thermal tolerance, and phage titers were undetectable after 60 min at 80 °C. At 70 °C, phage titers kept decreasing (Figure 5a,b). For the pH stability test, PSE-D1 and PST-H1 showed obvious stability at pH 3-11 (Figure 5c,d). At pH 2, pH 12, or pH 13, PSE-D1 and PST-H1 remained inactivated.
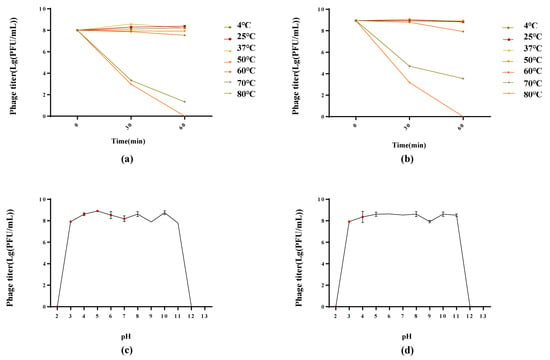
Figure 5.
Biological characterization of phages. (a) Stability at different pH of PSE-D1. (b) Stability at different pH of PST-H1. (c) Stability at different temperatures of PSE-D1. (b) Stability at different temperatures of PST-H1.
3.4. Genomic Analysis of Phages
The Gen Bank accession of PSE-D1 was no. ON550260, and that of PST-H1 was no. ON454038. The two phages were both double-stranded DNA. PSE-D1 had 166,604 bp and the GC content was 39.59% (Figure 6a). PST-H1 consisted of 42,036 bp, and the GC content was 49.95% (Figure 6b). While 115 CDSs had been predicted to encode functional proteins, there were 266 CDSs in the PSE-D1 genome totally. A total of 62 CDSs were associated with DNA replication and regulation; 48 CDSs participated in packing and morphogenesis proteins; 5 CDSs encoded proteins of host lysis among 115 CDSs (Supplementary Table S1). A total of 61 CDSs were founded in PST-H1 genomes according to protein coding genes prediction, of which 16 CDSs were predicted to encode functional proteins. Among 16 CDSs, 6 CDSs encoded the replication and regulation of DNA, 1 CDS encoded lysis protein, and 9 CDSs encoded packing and morphogenesis proteins (Supplementary Table S2).
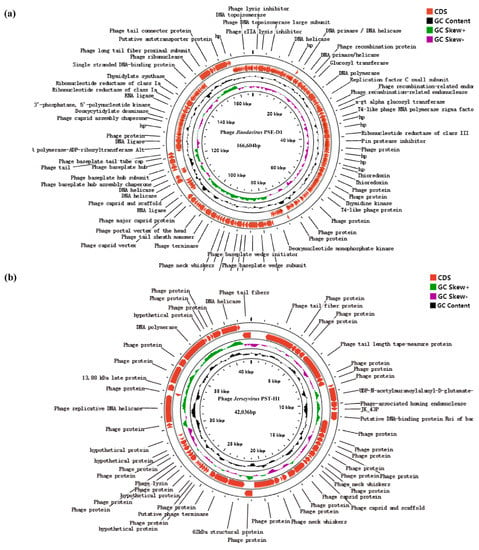
Figure 6.
Comparative circular genome maps of phages. (a) phage PSE-D1. (b) phage PST-H1. Black represents GC content; green and purple represent positive and negative GC skew.
The plaques of phage PSE-D1and PST-H1 had typical characteristics of lytic bacteriophages. Furthermore, gene clusters like Cro, CI, C2, C3, N, and Q were absent from the phage genome, which related to lysogeny, indicating that PSE-D1and PST-H1 were lytic bacteriophages [29]. No known genes associated with virulence and antibiotic-resistance were found in the genome sequence of phages, indicating that PSE-D1 and PST-H1 are safe for use in phage therapy.
PSE-D1 encoded 15 CDSs associated with tail fiber proteins, and PST-H1encoded 3 CDSs related to tail fiber proteins, which related to bind with bacterial receptors. SDS 162 of PSE-D1 encoded straight tail fiber (short tail fiber), mediating the adsorption of the phage and host strain. However, SDS162 had no specificity for bacteria adsorption [30]. Long tail fibers contributed to the host cell specificity, while CDS 244 of PSE-D1 encoded a long tail fiber proximal subunit, which related to adhesion between bacteria and bacteriophages [30]. Complete genome sequence analysis revealed that phage PSE-D1 was highly homologous with Klebsiella phage Vb-KpnM-FRZ284 (98% coverage), Klebsiella phage JD18 (98% coverage), and Klebsiella phage KP1 (97% coverage). PST-H1 was highly homologous with the following phages: Salmonella phage S55 (93% coverage), Salmonella phage LP31 (93% coverage), and Salmonella phage CKT1 (91% coverage) according to BLAST analysis.
Based on the whole genome, phylogenetic trees were created to further analyze the relationship between PSE-D1 and PST-H1 and other phages belonging to the Caudovirales order, respectively (Figure 7). The results of phylogenetic relationships revealed that the Klebsiella phage KP1 which belongs to the Jiaodavirus genus and Tevenvirinae subfamily, was the closest to phage PSE-D1. Phage PST-H1 shared a close relationship to Salmonella phage S55, which belongs to the Jerseyvirus genus, Guernseyvirinae subfamily.
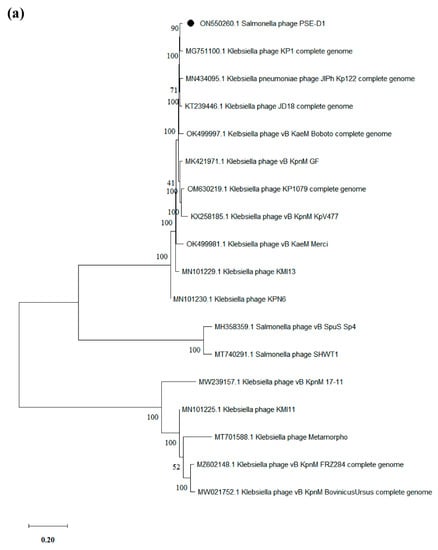
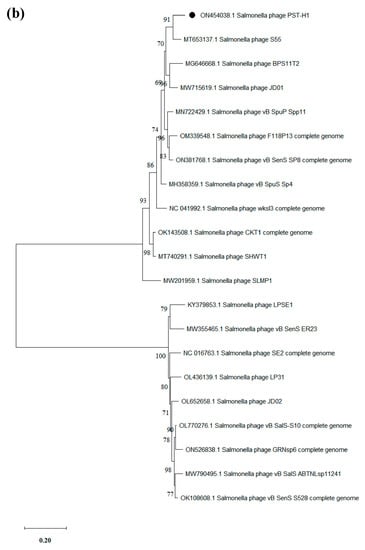
Figure 7.
Phylogenetic trees of phages. (a) phage PSE-D1. (b) phage PST-H1.
3.5. Application of Phages in Different Foods
3.5.1. Salmonella Reduction on Egg Shells
At both 4 °C and 28 °C, the viable counts of S. Enteritidis CVCC1806 with PSE-D1 treatment reduced significantly (p < 0.05, Figure 8a,b) after 48 h in eggshells. At 4 °C, compared with the positive control, PSE-D1 significantly inhibited the growth of CVCC1806 on eggshells at a MOI of 100 (1.5 log10 CFU/sample, p < 0.001) and a MOI of 10 (1.1 log10 CFU/sample, p < 0.05) at 48h (Figure 8a). Compared to the positive control, PSE-D1 showed a significant inhibition effect on the growth of CVCC1806 on eggshells at a MOI of 100 (1.0 log10 CFU/sample) and a MOI of 10 (0.9 log10 CFU/sample) at 48 h at 28 °C (p < 0.05, Figure 8b). At 4 °C and 28 °C, the group with PSE-D1 at a MOI of 100 showed better bactericidal efficiency than the group at a MOI of 10. At 4 °C, PST-H1 achieved a 1.9 log10 CFU/sample (p < 0.001) reduction of S. Typhimurium CVCC3384 at a MOI of 100 and 1.4 log10 CFU/sample (p < 0.001) reduction when at a MOI of 10 on 48 h, respectively (Figure 8c). At 28 °C, the CVCC3384 was reduced by more than 1.4 log10 CFU/sample at 48h at both a MOI of 100 and 10 (p < 0.001, Figure 8d), indicating a strong bactericidal effect of PST-H1 against CVCC3384 on eggshells. No Salmonella was detected from the negative control samples.
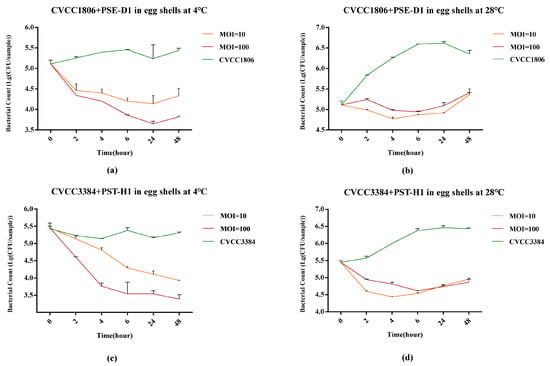
Figure 8.
Determination of the antibacterial effect of phages in eggshells at different temperatures. (a) CVCC1806+PSE-D1 in eggshells at 4 °C. (b) CVCC1806+PSE-D1 in eggshells at 28 °C. (c) CVCC3384+PST-H1 in eggshells at 4 °C. (d) CVCC3384+PST-H1 in eggshells at 28 °C.
3.5.2. Salmonella Reduction on Sausages
At 4 °C, compared to the phage-free group, PSE-D1 significantly inhibited the growth of CVCC1806 on sausages at a MOI of 100 (1.1 log10 CFU/cm2, p < 0.001) and a MOI of 10 (1.1 log10 CFU/cm2, p < 0.001) at 48h (Figure 9a). At 28 °C, PSE-D1 achieved 0.6 log10 CFU/cm2 (p < 0.001, Figure 9b) reduction on CVCC1806 with a MOI of 100 at 48 h. At 4 °C, the viable counts of CVCC3384 decreased (0.9 log10 CFU/cm2, 1.0 log10 CFU/cm2) with PST-H1 treatment at a MOI of 100 and 10, respectively (p < 0.001, Figure 9c). PST-H1 at a MOI of 100 and 10 achieved reduction (0.8 log10 CFU/cm2, 0.7 log10 CFU/cm2, p < 0.001) in CVCC3384 at 48 h at 28 °C, respectively (Figure 9b). Within 48 h, the viable counts of CVCC3384 decreased (1.2 log10 CFU/cm2, 0.9 log10 CFU/cm2, p < 0.001) with PST-H1 treatment at 4 °C and at 28 °C, respectively (Figure 9c,d). No Salmonella was detected from the negative control samples.
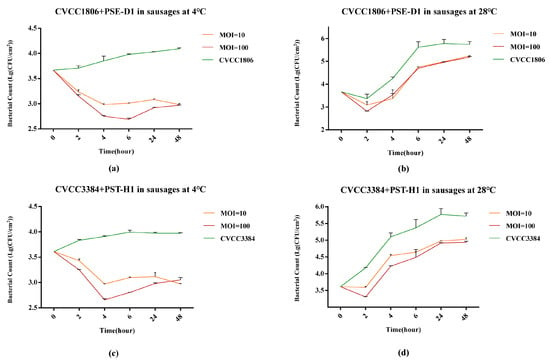
Figure 9.
Determination of the antibacterial effect of phages in sausages at different temperatures. (a) CVCC1806+PSE-D1 in sausages at 4 °C. (b) CVCC1806+PSE-D1 in sausages at 28 °C. (c) CVCC3384+PST-H1 in sausages at 4 °C. (d) CVCC3384+PST-H1 in sausages at 28 °C.
3.5.3. Salmonella Reduction in Milk
At 4 °C, compared to the positive control, PSE-D1 had a remarkable inhibitory effect on CVCC1806 growth (1.5 log10 CFU/mL, 1.2 log10 CFU/mL, p < 0.001) when at a MOI of 100 and 10 in 24 h in milk, respectively (Figure 10a). At 28 °C, the viable counts of CVCC1806 decreased to a low level with PSE-D1 at a MOI of 100 and 10 (reduction of 1.0 log10 CFU/mL, 0.7 log10 CFU/mL, p < 0.001, Figure 10b). After 24 h of incubation at both 4 °C and 28 °C, treatment with PST-H1 showed remarkable reductions of CVCC3384 counts in contaminated milk (p < 0.001, Figure 10c,d). PST-H1 obviously inhibited CVCC3384 at 4 °C and 28 °C (reduction of 1.8 log10 CFU/mL, 1.6 log10 CFU/mL, p < 0.001) after 24 h incubation, respectively (Figure 10c,d). No Salmonella was detected from the negative control samples.
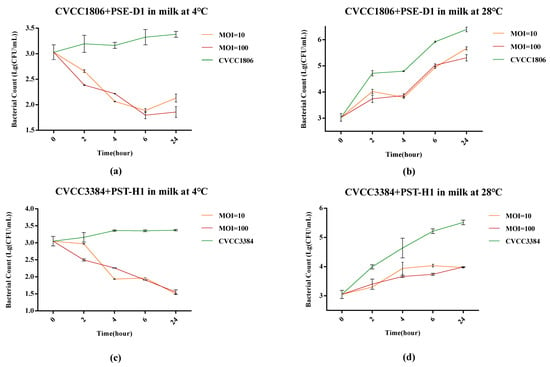
Figure 10.
Determination of the antibacterial effect of phages in milk at different temperatures. (a) CVCC1806+PSE-D1 in milk at 4 °C. (b) CVCC1806+PSE-D1 in milk at 28 °C. (c) CVCC3384+PST-H1 in milk at 4 °C. (d) CVCC3384+PST-H1 in milk at 28 °C.
4. Discussion
In this study, two phages named Salmonella phage PSE-D1 and Salmonella phage PST-H1 were isolated from cattle farm sewage in Guangxi, China. Phage PSE-D1 could infect 3 of 6 Salmonella strains, 13 of 48 E. coli strains, and 8 of 15 K. pneumoniae strains tested in this paper. A total of 3 Salmonella strains, 10 E. coli strains, and 11 K. pneumoniae strains were all lysed by phage PST-H1. Salmonella and E. coli are common bacterial foodborne pathogens, which are often found in contaminated dairy products, eggs, chicken, and chicken products. S. Typhimurium and S. Enteritidis collections were chosen to evaluate the host spectrum because they are the most commonly reported serovars that affect the food and veterinary industry (specifically poultry), suggesting that the two phages have the potential to inhibit Salmonella from contaminating food.
One-step growth curves reveal the infection potential of each phage. The latent period of PSE-D1 was around 10 min, and PST-H1 had a latent period of 20 min approximately. Their latent periods were shorter than that of many other Salmonella phages that have been reported [31,32,33]. The previous study revealed that shorter latent periods can substantially reduce phage generation times, making them more suitable for biocontrol applications. The burst size of PSE-D1 is 110 PFU/CFU, while that of PST-H1 is 183 PFU/CFU, which is higher than many other reported phages [8,31,34].
PSE-D1 and PST-H1 are highly stable at 60 °C for 1h, implying that they could be utilized to pasteurize dairy products. In line with the previous studies, PSE-D1 and PST-H1 exhibited high pH tolerance by remaining active under pH ranging from 3 to 11. The two phages can be employed in food with varying pH, since they are stable under a wide range of acidic and alkaline circumstances. The high stability of phage PSE-D1 and PST-H1 at temperature and pH might allow PSE-D1 and PST-H1 to be stored in food for a longer time.
For simulating the storage conditions of food products at different temperatures, we chose 4 °C and 28 °C to represent cold storage and room temperature, respectively. Salmonella counts with the phage treatment decreased significantly (p < 0.05) on the surface of egg shells (1.9 log10 CFU/sample), sausages (1.2 log10 CFU/cm2), and in milk (1.8 log10 CFU/mL) compared to phage-free groups after 48h at two different temperatures (4 °C and 28 °C). Temperature largely influenced the effectiveness of phage PSE-D1 and PST-H1. It is reported that phages had greater bactericidal capacity when refrigerated [8,9,35]. A similar result was observed in this study. At 4 °C, PSE-D1 and PST-H1 showed a stronger inhibitory effect on Salmonella growth than that at 28 °C. PSE-D1 and PST-H1 can maintain high activity for a long time at 4 °C. The phage titer in Chinese cabbage was dramatically lower after incubation at 25 °C for 24 hours than that at 4 °C in the previous study, demonstrating that storage at room temperature may also impair the stability of the phage [36]. Salmonella activity was relatively lower at 4 °C than that at 28 °C, indicating that the phages had a better bacteriostasis effect at 4 °C.
The quality and safety of milk are highly valued in the dairy industry. Milk might be an important medium for Salmonella transmission to humans [37]. In raw milk, Salmonella is one of the most common bacterial foodborne pathogens especially in developing countries [35]. The existence of Salmonella is the lead cause of contamination after pasteurization in dairy products [38]. Therefore, controlling the contamination after milk processing is a key factor to prevent the transmission of Salmonella into dairy products. Because Salmonella is widely spread in chickens, it is important to reduce the incidence of Salmonella contamination in chicken and chicken products during their production, transportation and processing [39]. Consumption of shell eggs has been associated with S. Enteritidis infections in humans in the U.S.A. With the reduction of pathogens on eggshells, the possibility of cross-shell contamination becomes less [40]. The application of salmonella bacteriophage products in meat and poultry products has been authorized [41]. There are few reports on the direct application of phages on eggshells, while phages PSE-D1 and PST-H1 showed 1.5 log10 CFU/sample reduction of Salmonella on the surface of eggshells with significant difference (p < 0.001) occurring in 48h. It is reported that phages had good applications in chicken and milk [37,38,39]. In this study, phages PSE-D1 and PST-H1 showed good bactericidal effect in chicken sausages and milk with significant difference (p < 0.001), indicating that the two phages had potentiality for being used in controlling Salmonella contaminated food.
The bacteriostatic effects of PSE-D1 and PST-H1 were found to be more effective with phages with higher titers. At 4 °C, PSE-D1 treatment with different counts (1010 PFU/sample, 109 PFU/sample) lead to a significant decrease of bacteria counts (1.5 log10 CFU/sample, 1.1 log10 CFU/sample, p < 0.05) in CVCC1806 contaminated eggshells. At 4 °C, the viable counts of CVCC3384 decreased (1.9 log10 CFU/mL, 1.4 log10 CFU/mL, p < 0.001) with phages treatment with different titers (1010 PFU/mL, 109 PFU/mL) in eggshells. The previous study revealed that direct contact between bacteria and bacteriophages is more likely to occur at higher concentrations [14,42]. High concentrations of phage increased the probability of encountering Salmonella cells [24,25].
5. Conclusions
Our study described two broad-spectrum phages, PSE-D1 and PST-H1, isolated from cattle farm sewage, exhibiting a strong lytic effect on S. Enteritidis CVCC1806 and S. Typhimurium CVCC3384. They showed high pH and thermal tolerances as well as short latent periods. Additionally, PSE-D1 and PST-H1 showed considerable inhibition of growth of S. Typhimurium and S. Enteritidis in LB broth and three different foods, respectively. No known virulence factors or antibiotic-resistance related genes were found in PSE-D1 and PST-H1 genomes, indicating they are a promising bactericide during food preservation.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/v14122647/s1, Table S1. CDS function prediction of phage PSE-D1; Table S2. CDS function prediction of phage PST-H1.
Author Contributions
Conceptualization, X.L. and X.W.; Data curation, Y.C., X.M., Y.W., K.H., L.L. and Y.Z.; Formal analysis, Y.C. and R.M.; Funding acquisition, X.L. and X.W.; Investigation, L.W.; Methodology, Y.C. and R.M.; Software, Z.L., X.M., Y.L., Y.W., and X.W.; Supervision, X.L. and X.W.; Validation, D.M.; Visualization, Y.C.; Writing—original draft, Y.C.; Writing—review & editing, Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Guangxi Province (2021GXNSFAA196058), the Key Research and Development Program of Nanning (20212023), the Key R&D Program of Liangqing District (202109), the Major Science and Technology Project of Liangqing District (202118), the Key R&D Program of Fangchenggang City (AB21014016), the Key Research and Development of Wuming District (20210102), and the Jiangnan Key Research and Development (20220620-2).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequence of Salmonella phage PSE-D1 and Salmonella phage PST-H1 has been submitted to the NCBI Gene Bank and can be accessed by accession numbers ON550260 and ON454038, respectively. The data presented in this study are available on request from the corresponding author.
Acknowledgments
The construction of sequencing library and the control of sequencing data quality were performed by Beijing University of Chemical Technology.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Brown, E.W.; Bell, R.; Zhang, G.; Timme, R.; Zheng, J.; Hammack, T.S.; Allard, M.W. Salmonella Genomics in Public Health and Food Safety. EcoSal Plus 2021, 9, eESP00082020. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar]
- Centers for Disease Control and Prevention (CDC). Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, United States, 2005. Jama J. Am. Med. Assoc. 2006, 295, 2241–2243, reprinted in MMWR 2006, 55, 392–395. [Google Scholar] [CrossRef][Green Version]
- Liu, D.; Gu, W.; Wang, L.; Sun, J. Photodynamic inactivation and its application in food preservation. Crit. Rev. Food Sci. Nutr. 2021, 1–15. [Google Scholar] [CrossRef]
- Ramos-Vivas, J.; Elexpuru-Zabaleta, M.; Samano, M.L.; Barrera, A.P.; Forbes-Hernández, T.Y.; Giampieri, F.; Battino, M. Phages and enzybiotics in food biopreservation. Molecules 2021, 26, 5138. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.U.; Kim, I.S.; Lee, E.J. Irradiation and additive combinations on the pathogen reduction and quality of poultry meat. Poult. Sci. 2013, 92, 534–545. [Google Scholar] [CrossRef]
- Singh, V.P. Recent approaches in food bio-preservation—A review. Open Veter J. 2018, 8, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shi, J.; Ma, W.; Li, Z.; Wang, J.; Li, J.; Wang, X. Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 2018, 111, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, W.; Li, W.; Ding, Y.; Zhang, Y.; Yang, Q.; Wang, J.; Wang, X. A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 2020, 132, 109011. [Google Scholar] [CrossRef]
- Hudson, J.; Billington, C.; Wilson, T.; On, S. Effect of phage and host concentration on the inactivation of Escherichia coli O157:H7 on cooked and raw beef. Food Sci. Technol. Int. 2013, 21, 104–109. [Google Scholar] [CrossRef]
- Xing, S.; Zhang, X.; Sun, Q.; Wang, J.; Mi, Z.; Pei, G.; Huang, Y.; An, X.; Fu, K.; Zhou, L.; et al. Complete genome sequence of a novel, virulent Ahjdlikevirus bacteriophage that infects Enterococcus faecium. Arch. Virol. 2017, 162, 3843–3847. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, E.; Song, J.; Yang, L.; Wu, B. Complete Genome Sequence of a Novel T7-Like Bacteriophage from a Pasteurella multocida Capsular Type A Isolate. Curr. Microbiol. 2018, 75, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Yun, J.; Lim, J.-A.; Roh, E.; Jung, K.-S.; Chang, Y.; Ryu, S.; Heu, S. Characterization and genomic analysis of two Staphylococcus aureus bacteriophages isolated from poultry/livestock farms. J. Gen. Virol. 2013, 94, 2569–2576. [Google Scholar] [CrossRef] [PubMed]
- Hagens, S.; Loessner, M.J. Bacteriophage for biocontrol of foodborne pathogens: Calculations and considerations. Curr. Pharm. Biotechnol. 2010, 11, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. Physiological aspects of bacteriophage genetics. Adv. Virus Res. 1959, 6, 137–158. [Google Scholar]
- Aguilera, M.; Martínez, S.; Tello, M.; Gallardo, M.J.; García, V. Use of Cocktail of Bacteriophage for Salmonella Typhimurium Control in Chicken Meat. Foods 2022, 11, 1164. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Ramesh, N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS ONE 2018, 13, e0206278. [Google Scholar] [CrossRef] [PubMed]
- Kokkari, C.; Sarropoulou, E.; Bastías, R.; Mandalakis, M.; Katharios, P. Isolation and characterization of a novel bacteriophage infecting Vibrio alginolyticus. Arch. Microbiol. 2018, 200, 707–718. [Google Scholar] [CrossRef]
- Lam, H.Y.P.; Peng, S.Y.; Paramita, P.; Wu, W.J.; Chen, L.K.; Chao, H.J.; Lai, M.J.; Chang, K.C. Biological and genomic characterization of two newly isolated Elizabethkingia anophelis bacteriophages. J. Microbiol. Immunol. Infect. 2022, 55, 634–642. [Google Scholar]
- Lu, S.; Le, S.; Tan, Y.; Zhu, J.; Li, M.; Rao, X.; Zou, L.; Li, S.; Wang, J.; Jin, X.; et al. Genomic and Proteomic Analyses of the Terminally Redundant Genome of the Pseudomonas aeruginosa Phage PaP1: Establishment of Genus PaP1-Like Phages. PLoS ONE 2013, 8, e62933. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2018, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.; Billington, C.; Cornelius, A.; Wilson, T.; On, S.; Premaratne, A.; King, N. Use of a bacteriophage to inactivate Escherichia coli O157:H7 on beef. Food Microbiol. 2013, 36, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Q.; Zhang, C.; Yang, J.; Lu, Z.; Lu, F.; Bie, X. Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 2017, 236, 14–23. [Google Scholar] [CrossRef]
- McLean, S.K.; Dunn, L.A.; Palombo, E.A. Phage Inhibition of Escherichia coli in Ultrahigh-Temperature-Treated and Raw Milk. Foodborne Pathog. Dis. 2013, 10, 956–962. [Google Scholar] [CrossRef]
- Luo, D.D.; Li, C.S.; Wu, Q.P.; Ding, Y.; Yang, M.Y.; Hu, Y.D.; Zeng, H.; Zhang, J. Isolation and characterization of new phage vB_CtuP_A24 and application to control Cronobacter spp. in infant milk formula and lettuce. Food Res. Int. 2021, 141, 110109. [Google Scholar] [CrossRef]
- Sritha, K.S.; Bhat, S.G. In vitro efficiency evaluation of phage cocktail for biocontrol of Salmonella spp. in food products. Arch. Microbiol. 2021, 203, 5445–5452. [Google Scholar]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in Bacteriophage Lambda Development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef]
- Burda, M.R.; Hindennach, I.; Miller, S. Stability of Bacteriophage T4 Short Tail Fiber. Biol. Chem. 2000, 381, 255–258. [Google Scholar] [CrossRef]
- Feng, X.; Yan, W.; Wang, A.; Ma, R.; Chen, X.; Lin, T.-H.; Chen, Y.-L.; Wei, S.; Jin, T.; Jiao, N.; et al. A Novel Broad Host Range Phage Infecting Alteromonas. Viruses 2021, 13, 987. [Google Scholar] [CrossRef]
- Naknaen, A.; Suttinun, O.; Surachat, K.; Khan, E.; Pomwised, R. A Novel Jumbo Phage PhiMa05 Inhibits Harmful Microcystis sp. Front. Microbiol. 2021, 12, 660351. [Google Scholar] [CrossRef]
- Wang, R.; Xing, S.; Zhao, F.; Li, P.; Mi, Z.; Shi, T.; Liu, H.; Tong, Y. Characterization and genome analysis of novel phage vB_EfaP_IME195 infecting Enterococcus faecalis. Virus Genes 2018, 54, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Q.; Fan, J.; Yan, T.; Zhang, H.; Yang, J.; Deng, D.; Liu, C.; Wei, T.; Ma, Y. Characterization and Genomic Analysis of ValSw3-3, a New Siphoviridae Bacteriophage Infecting Vibrio alginolyticus. J. Virol. 2020, 94, e00066-20. [Google Scholar] [CrossRef] [PubMed]
- Grace, D.; Wu, F.; Havelaar, A.H. MILK Symposium review: Foodborne diseases from milk and milk products in developing countries-Review of causes and health and economic implications. J. Dairy Sci. 2020, 103, 9715–9729. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Zhang, P.; Zhang, H.; Zhou, Y.; Zhang, L.; Wang, R. Bio-Control of Salmonella Enteritidis in Foods Using Bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef] [PubMed]
- Phongtang, W.; Choi, G.-P.; Chukeatirote, E.; Ahn, J. Bacteriophage control of Salmonella Typhimurium in milk. Food Sci. Biotechnol. 2018, 28, 297–301. [Google Scholar] [CrossRef]
- Olsen, S.J.; Ying, M.; Davis, M.F.; Deasy, M.; Holland, B.; Iampietro, L.; Baysinger, C.M.; Sassano, F.; Polk, L.D.; Gormley, B.; et al. Multidrug-resistant Salmonella Typhimurium Infection from Milk Contaminated after Pasteurization. Emerg. Infect. Dis. 2004, 10, 932–935. [Google Scholar] [CrossRef]
- Wessels, K.; Rip, D.; Gouws, P. Salmonella in Chicken Meat: Consumption, Outbreaks, Characteristics, Current Control Methods and the Potential of Bacteriophage Use. Foods 2021, 10, 1742. [Google Scholar] [CrossRef]
- Messens, W.; Grijspeerdt, K.; Herman, L. Eggshell characteristics and penetration by Salmonella enterica serovar Enteritidis through the production period of a layer flock. Br. Poult Sci. 2005, 46, 694–700. [Google Scholar] [CrossRef]
- Sharma, M. Lytic bacteriophages: Potential interventions against enteric bacterial pathogens on produce. Bacteriophage 2013, 3, e25518. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, H.J.; Jung, S.J.; Mizan, M.F.R.; Park, S.H.; Ha, S.D. Characterization of Salmonella spp.-specific bacteriophages and their biocontrol application in chicken breast meat. J. Food Sci. 2020, 85, 526–534. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).