African Swine Fever Virus Exhibits Distinct Replication Defects in Different Cell Types
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses, Cells and Reagents
2.2. Anti-ASFV p30/p72 Monoclonal Antibodies Preparation
2.3. Virus Passage Assay
2.4. Virus Attachment Detection
2.5. Virus Entry Detection
2.6. Virus Genomic DNA Replication Detection
2.7. Viral Early and Late Genes Expressions Detection
2.8. Viral Genes Transcription Quantification
2.9. Virus Assembly Detection
2.10. Virus Release Detection
2.11. Indirect Immunofluorescence Assay
2.12. SG Effect on ASFV Replication
2.13. Biosafety Statement and Facilities
3. Results
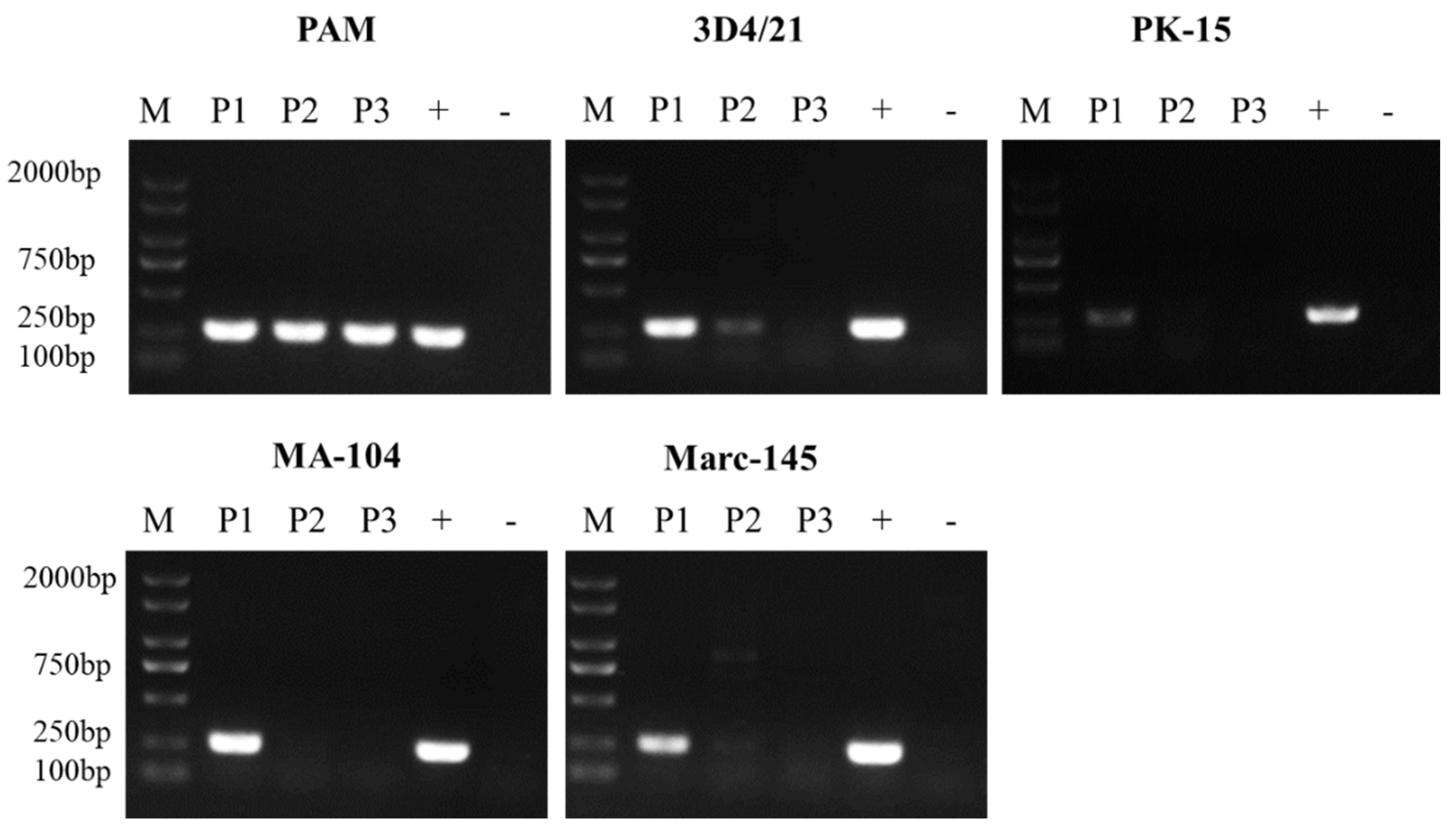
3.1. ASFV Passage in Different Cells
3.2. ASFV Attachment on Different Cells
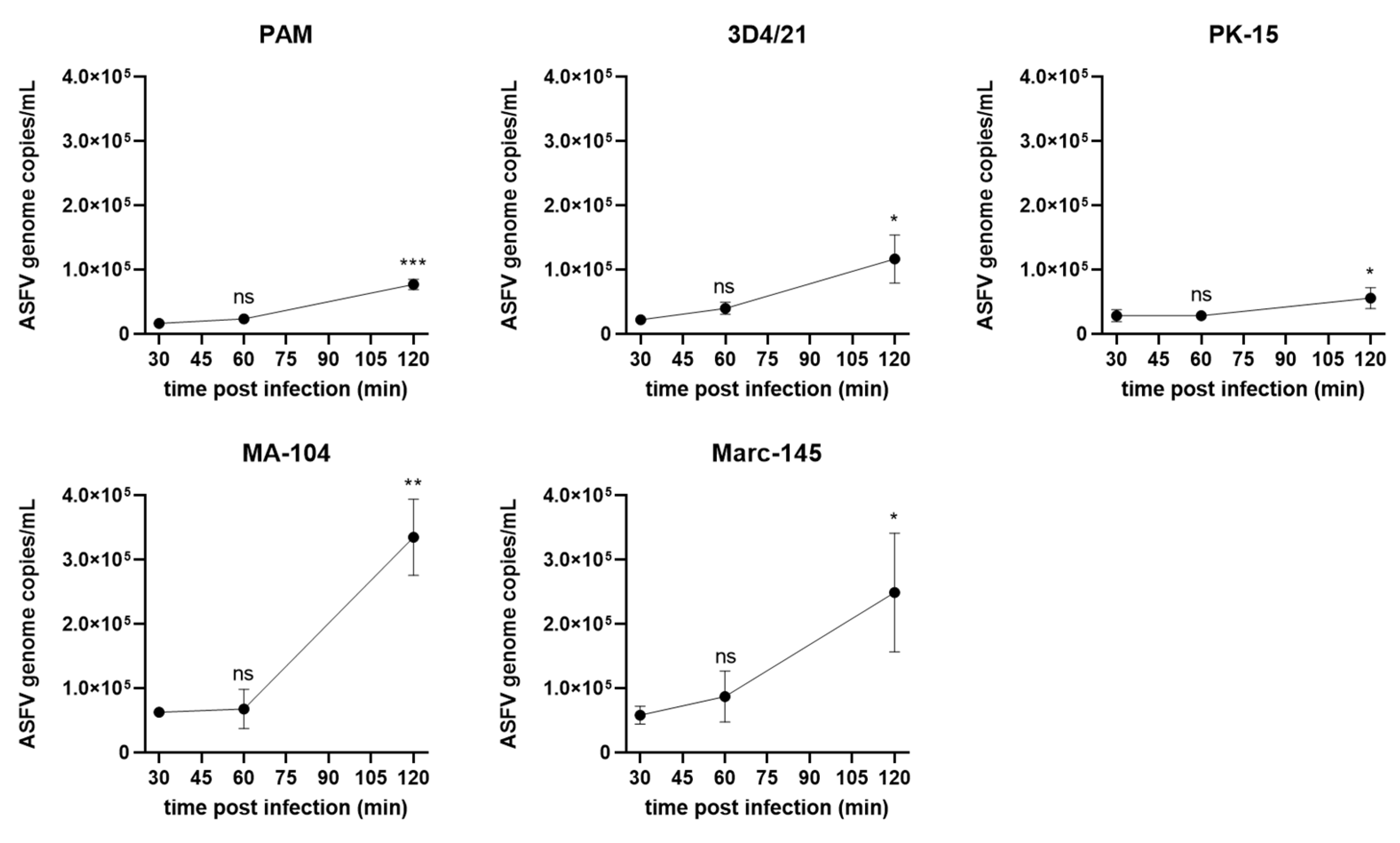
3.3. ASFV Entry into in Different Cells
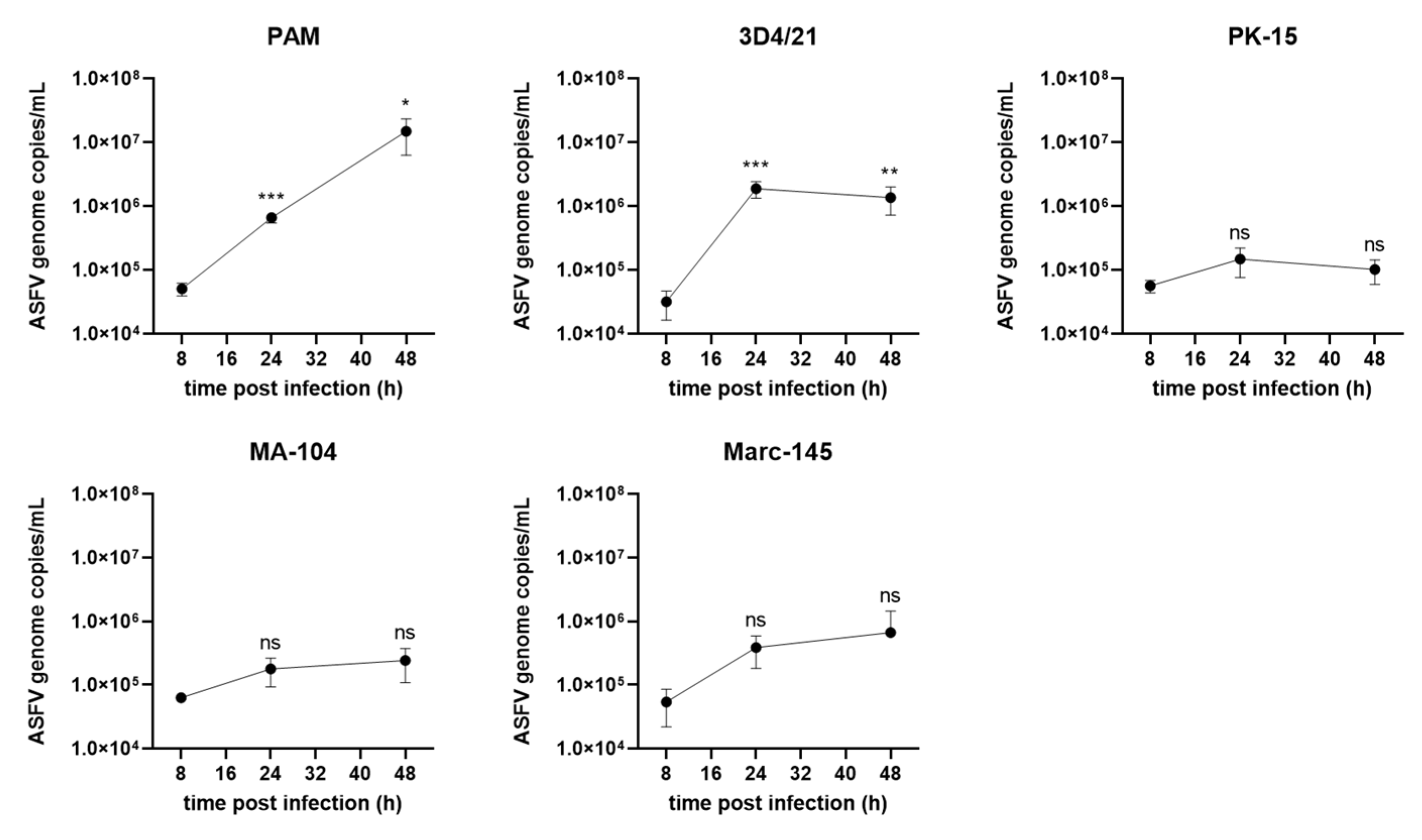
3.4. ASFV Genome Replication in Different Cells
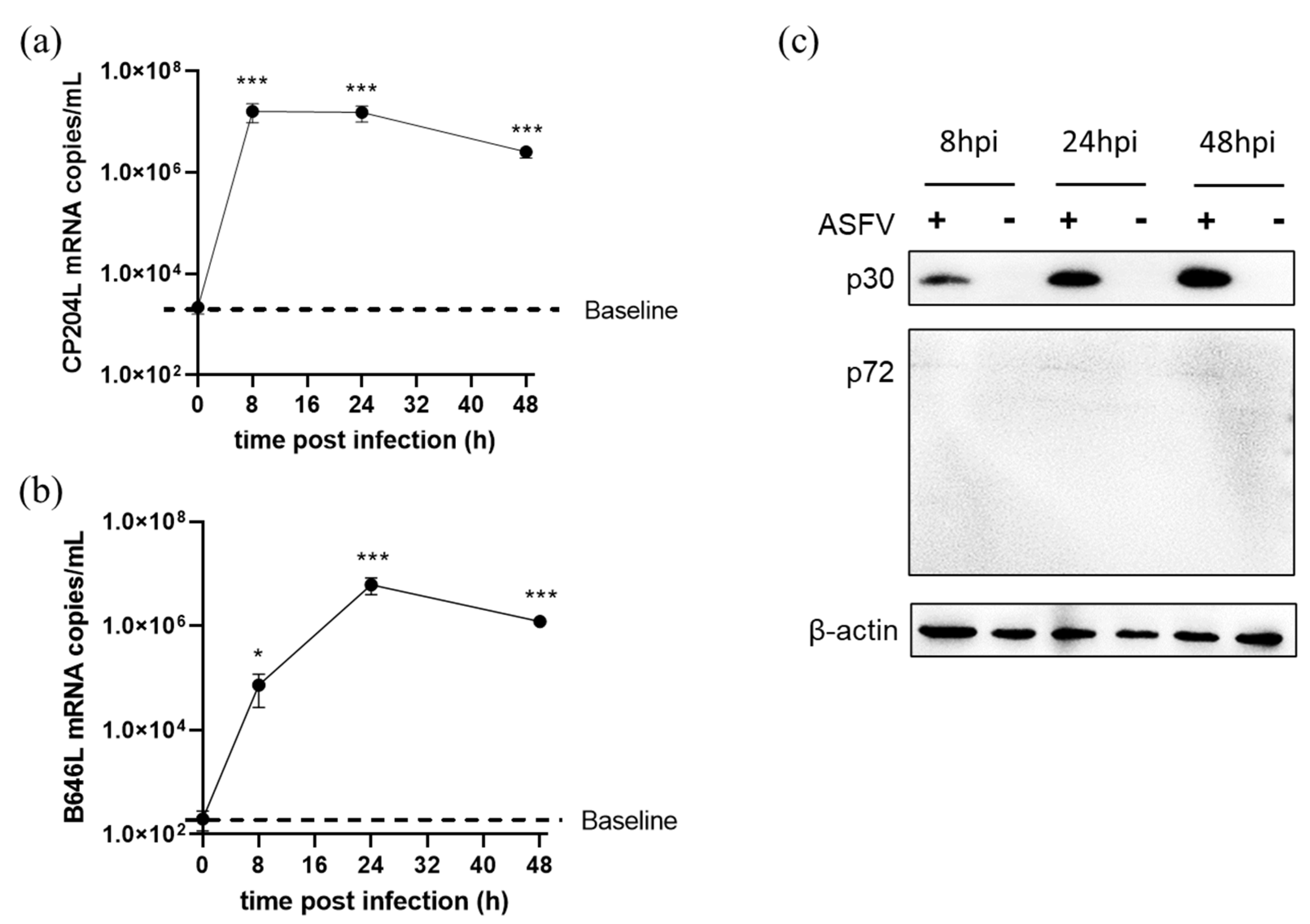
3.5. ASFV Early and Late Gene Expression in Different Cells
3.6. ASFV Early and Late Genes Transcription in 3D4/21 Cells
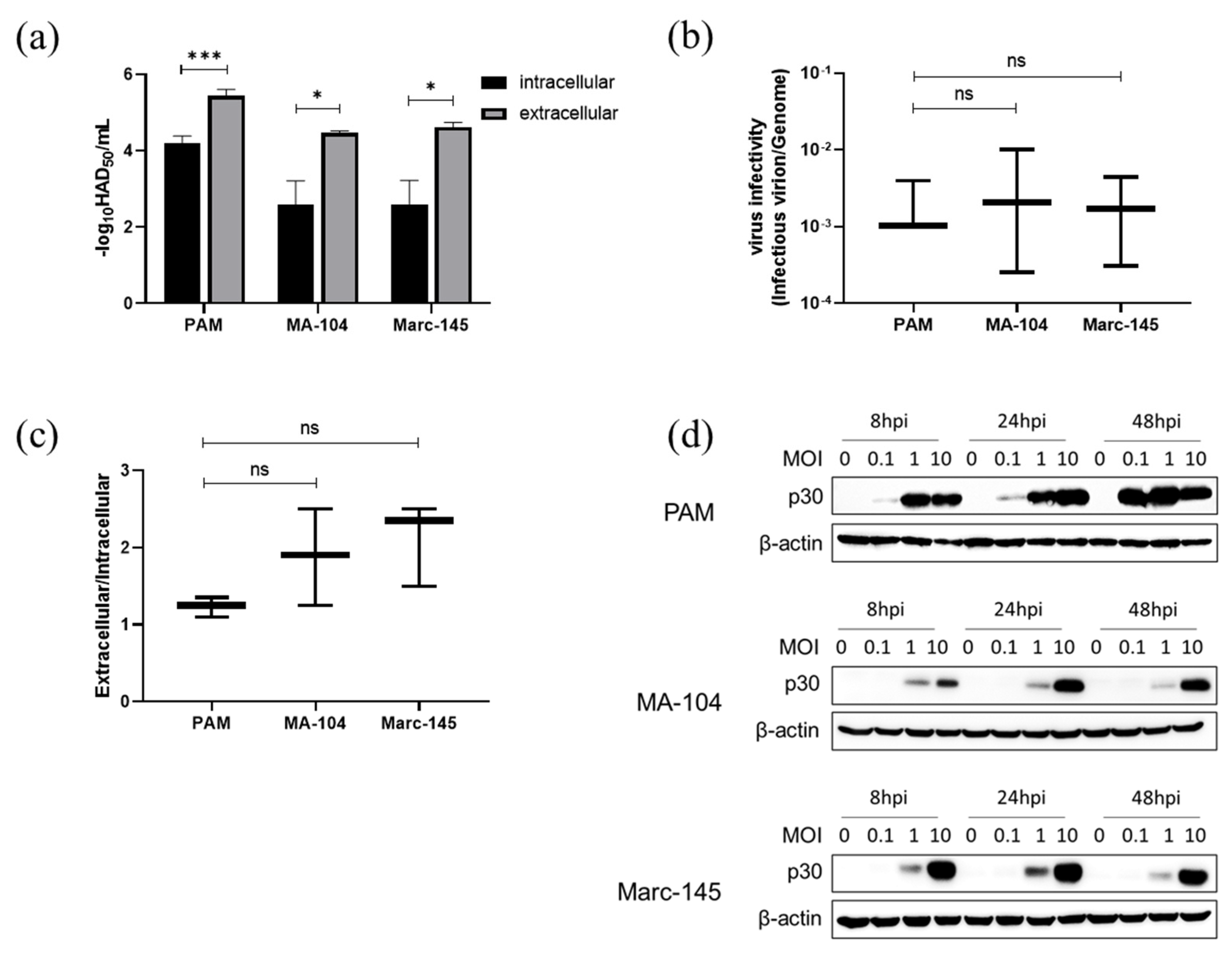
3.7. ASFV Infectious Virions Production and Release in PAMs, MA-104 and Marc-145 Cells
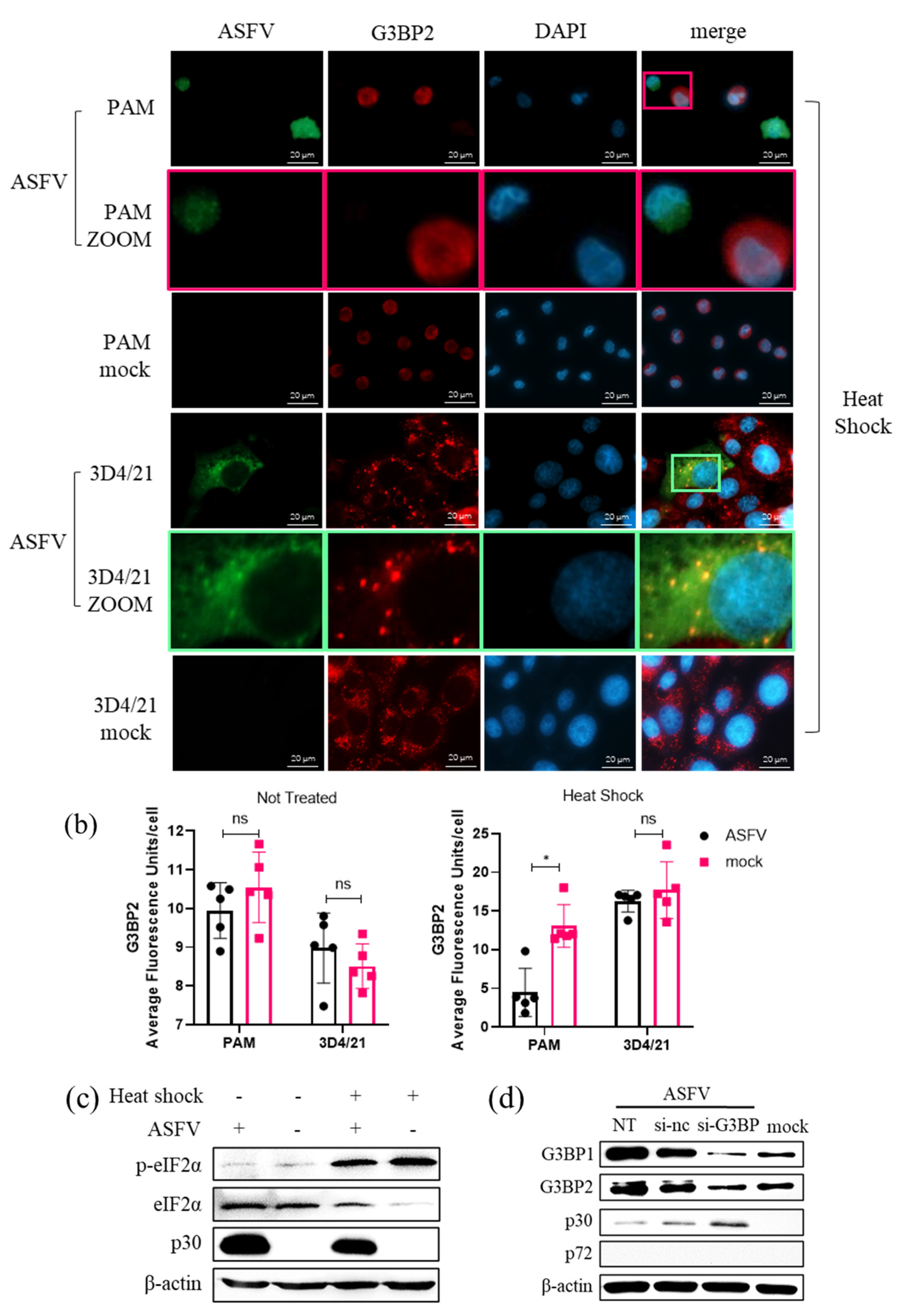
3.8. Role of SG Formation during ASFV Replication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef] [PubMed]
- Mulumba-Mfumu, L.K.; Saegerman, C.; Dixon, L.K.; Madimba, K.C.; Kazadi, E.; Mukalakata, N.T.; Oura, C.A.L.; Chenais, E.; Masembe, C.; Ståhl, K.; et al. African swine fever: Update on Eastern, Central and Southern Africa. Transbound. Emerg. Dis. 2019, 66, 1462–1480. [Google Scholar] [CrossRef] [PubMed]
- Karger, A.; Pérez-Núñez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.-F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Graham, S.P.; Giudici, S.D.; Bonelli, P.; Pilo, G.; Anfossi, A.G.; Pittau, M.; Nicolussi, P.S.; Laddomada, A.; Oggiano, A. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Veter. Microbiol. 2017, 198, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.G.; Riera, E.; Nogal, M.; Gallardo, C.; Fernández, P.; Bello-Morales, R.; López-Guerrero, J.A.; Chitko-McKown, C.G.; Richt, J.A.; Revilla, Y. Phenotyping and susceptibility of established porcine cells lines to African Swine Fever Virus infection and viral production. Sci. Rep. 2017, 7, 10369. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Cuesta-Geijo, M.A.; Hlavova, K.; Muñoz-Moreno, R.; Barrado-Gil, L.; Dominguez, J.; Alonso, C. African swine fever virus infects macrophages, the natural host cells, via clathrin- and cholesterol-dependent endocytosis. Virus Res. 2015, 200, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Alcamí, A.; Carrascosa, A.L.; Viñuela, E. Interaction of African swine fever virus with macrophages. Virus Res. 1990, 17, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Carrascosa, A.L.; Bustos, M.J.; Galindo, I.; Viñuela, E. Virus-specific cell receptors are necessary, but not sufficient, to confer cell susceptibility to African swine fever virus. Arch. Virol. 1999, 144, 1309–1321. [Google Scholar] [CrossRef]
- Reineke, L.C.; Lloyd, R.E. Diversion of stress granules and P-bodies during viral infection. Virology 2013, 436, 255–267. [Google Scholar] [CrossRef]
- Malinowska, M.; Niedźwiedzka-Rystwej, P.; Tokarz-Deptuła, B.; Deptuła, W. Stress granules (SG) and processing bodies (PB) in viral infections. Acta Biochim. Pol. 2016, 63, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Legros, S.; Boxus, M.; Gatot, J.S.; Van Lint, C.; Kruys, V.; Kettmann, R.; Twizere, J.C.; Dequiedt, F. The HTLV-1 Tax protein inhibits formation of stress granules by interacting with histone deacetylase 6. Oncogene 2011, 30, 4050–4062. [Google Scholar] [CrossRef] [PubMed]
- Khaperskyy, D.A.; Hatchette, T.F.; McCormick, C. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 2011, 26, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Finnen, R.L.; Hay, T.J.M.; Dauber, B.; Smiley, J.R.; Banfield, B.W. The Herpes Simplex Virus 2 Virion-Associated Ribonuclease vhs Interferes with Stress Granule Formation. J. Virol. 2014, 88, 12727–12739. [Google Scholar] [CrossRef]
- Lindquist, M.E.; Lifland, A.W.; Utley, T.J.; Santangelo, P.J.; Crowe, J.E. Respiratory Syncytial Virus Induces Host RNA Stress Granules To Facilitate Viral Replication. J. Virol. 2010, 84, 12274–12284. [Google Scholar] [CrossRef]
- Zhou, Y.; Fang, L.; Wang, D.; Cai, K.; Chen, H.; Xiao, S. Porcine Reproductive and Respiratory Syndrome Virus Infection Induces Stress Granule Formation Depending on Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK) in MARC-145 Cells. Front. Cell. Infect. Microbiol. 2017, 7, 111. [Google Scholar] [CrossRef]
- Okonski, K.M.; Samuel, C.E. Stress Granule Formation Induced by Measles Virus Is Protein Kinase PKR Dependent and Impaired by RNA Adenosine Deaminase ADAR1. J. Virol. 2013, 87, 756–766. [Google Scholar] [CrossRef]
- Katoh, H.; Okamoto, T.; Fukuhara, T.; Kambara, H.; Morita, E.; Mori, Y.; Kamitani, W.; Matsuura, Y. Japanese Encephalitis Virus Core Protein Inhibits Stress Granule Formation through an Interaction with Caprin-1 and Facilitates Viral Propagation. J. Virol. 2013, 87, 489–502. [Google Scholar] [CrossRef]
- Scholte, F.E.M.; Tas, A.; Albulescu, I.C.; Žusinaite, E.; Merits, A.; Snijder, E.J.; van Hemert, M.J. Stress Granule Components G3BP1 and G3BP2 Play a Proviral Role Early in Chikungunya Virus Replication. J. Virol. 2015, 89, 4457–4469. [Google Scholar] [CrossRef]
- Gotte, B.; Panas, M.D.; Hellström, K.; Liu, L.; Samreen, B.; Larsson, O.; Ahola, T.; McInerney, G.M. Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery. PLoS Pathog. 2019, 15, e1007842. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Bustos, M.J.; de Leon, P. Methods for Growing and Titrating African Swine Fever Virus: Field and Laboratory Samples. Curr. Protoc. Cell Biol. 2011, 53, 14–26. [Google Scholar] [CrossRef]
- Rai, A.; Pruitt, S.; Ramirez-Medina, E.; Vuono, E.A.; Silva, E.; Velazquez-Salinas, L.; Carrillo, C.; Borca, M.V.; Gladue, D.P. Identification of a Continuously Stable and Commercially Available Cell Line for the Identification of Infectious African Swine Fever Virus in Clinical Samples. Viruses 2020, 12, 820. [Google Scholar] [CrossRef] [PubMed]
- Weingartl, H.; Sabara, M.; Pasick, J.; van Moorlehem, E.; Babiuk, L. Continuous porcine cell lines developed from alveolar macrophages: Partial characterization and virus susceptibility. J. Virol. Methods 2002, 104, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Urbano, A.C.; Ferreira, F. African swine fever control and prevention: An update on vaccine development. Emerg. Microbes Infect. 2022, 11, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G.; Terpstra, C.; Pol, J.M.A.; Ter Laak, E.A.; Bloemraad, M.; De Kluyver, E.P.; Kragten, C.; Van Buiten, L.; den Besten, A.; Wagenaar, F.; et al. Mystery swine disease in the Netherlands: The isolation of Lelystad virus. Veter. Q. 1991, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.K.; Singh, A.; Mukherjee, K.; Panda, A.K. Refolding and purification of recombinant L-asparaginase from inclusion bodies of E. coli into active tetrameric protein. Front. Microbiol. 2014, 5, 486. [Google Scholar] [CrossRef]
- Groth, S.D.S.; Scheidegger, D. Production of monoclonal antibodies: Strategy and tactics. J. Immunol. Methods 1980, 35, 1–21. [Google Scholar] [CrossRef]
- Wang, P.; Bai, J.; Liu, X.; Wang, M.; Wang, X.; Jiang, P. Tomatidine inhibits porcine epidemic diarrhea virus replication by targeting 3CL protease. Vet. Res. 2020, 51, 136. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Geijo, M.A.; Galindo, I.; Hernáez, B.; Quetglas, J.I.; Dalmau-Mena, I.; Alonso, C. Endosomal Maturation, Rab7 GTPase and Phosphoinositides in African Swine Fever Virus Entry. PLoS ONE 2012, 7, e48853. [Google Scholar] [CrossRef]
- Alonso, C.; Miskin, J.; Hernáez, B.; Fernandez-Zapatero, P.; Soto, L.; Cantó, C.; Rodríguez-Crespo, I.; Dixon, L.; Escribano, J.M. African Swine Fever Virus Protein p54 Interacts with the Microtubular Motor Complex through Direct Binding to Light-Chain Dynein. J. Virol. 2001, 75, 9819–9827. [Google Scholar] [CrossRef]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, e01293-18. [Google Scholar] [CrossRef] [PubMed]
- Matos, A.P.A.; Carvalho, Z.G. African swine fever virus interaction with microtubules. Biol. Cell 1993, 78, 229–234. [Google Scholar] [CrossRef]
- Breese, S.S.; DeBoer, C.J. Electron microscope observations of African swine fever virus in tissue culture cells. Virology 1966, 28, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Andrés, G.; Garcıa-Escudero, R.; Viñuela, E.; Salas, M.L.; Rodríguez, J.M. African Swine Fever Virus Structural Protein pE120R Is Essential for Virus Transport from Assembly Sites to Plasma Membrane but Not for Infectivity. J. Virol. 2001, 75, 6758–6768. [Google Scholar] [CrossRef]
- Dauber, B.; Pelletier, J.; Smiley, J.R. The Herpes Simplex Virus 1 vhs Protein Enhances Translation of Viral True Late mRNAs and Virus Production in a Cell Type-Dependent Manner. J. Virol. 2011, 85, 5363–5373. [Google Scholar] [CrossRef]
- Gómez-Villamandos, J.C.; Bautista, M.J.; Sánchez-Cordón, P.J.; Carrasco, L. Pathology of African swine fever: The role of monocyte-macrophage. Virus Res. 2013, 173, 140–149. [Google Scholar] [CrossRef]
- Gómez-Villamandos, J.C.; Hervás, J.; Méndez, A.; Carrasco, L.; Mulas, J.M.D.L.; Villeda, C.J.; Wilkinson, P.J.; Sierra, M.A. Experimental African swine fever: Apoptosis of lymphocytes and virus replication in other cells. J. Gen. Virol. 1995, 76 Pt 9, 2399–2405. [Google Scholar] [CrossRef]
- Carrasco, L.; Sierra, M.A.; Hervas, J.; Gomez-Villamandos, J.C.; Mendez, A. The lesional changes and pathogenesis in the kidney in african swine fever. Vet. Res. Commun. 1996, 20, 285–299. [Google Scholar] [CrossRef]
- Sánchez, E.G.; Quintas, A.; Pérez-Núñez, D.; Nogal, M.; Barroso, S.; Carrascosa, L.; Revilla, Y. African Swine Fever Virus Uses Macropinocytosis to Enter Host Cells. PLoS Pathog. 2012, 8, e1002754. [Google Scholar] [CrossRef]



| Cell | Organism | Tissue | Cell Type |
|---|---|---|---|
| PAM | Sus scrofa | Lung | Primary macrophage, alveolar |
| 3D4/21 | Sus scrofa | Lung | Macrophage, alveolar |
| PK-15 | Sus scrofa | Kidney | epithelial |
| MA-104 | Cercopithecus aethiops | Kidney | epithelial |
| Marc-145 | Cercopithecus aethiops | Kidney | epithelial |
| Target Detection | Primers | Sequences |
|---|---|---|
| PCR | B646L-F | 5′-AGTTATGGGAAACCCGACCC-3′ |
| B646L-R | 5′-CCCTGAATCGGAGCATCCT-3′ | |
| qPCR for ASFV genomic DNA /RT-qPCR for B646L mRNA | B646L-qPCR-F | 5′-CGGAGAACGACTTTATGA-3′ |
| B646L-qPCR-R | 5′-CCTGGGATGCAAAATTTG-3′ | |
| B646L-qPCR-probe | FAM-CAAGCGTTGTGACATCCGAACTATATT-TAMRA | |
| RT-qPCR for CP204L mRNA | CP204L-qPCR-F | 5′-AGGAGACGGAATCCTCAGCA-3′ |
| CP204L-qPCR-R | 5′-GGGCTCTTGCTCAAACAACG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Xia, T.; Bai, J.; Zhang, L.; Jiang, X.; Yang, X.; Zhang, K.; Jiang, P. African Swine Fever Virus Exhibits Distinct Replication Defects in Different Cell Types. Viruses 2022, 14, 2642. https://doi.org/10.3390/v14122642
Gao Y, Xia T, Bai J, Zhang L, Jiang X, Yang X, Zhang K, Jiang P. African Swine Fever Virus Exhibits Distinct Replication Defects in Different Cell Types. Viruses. 2022; 14(12):2642. https://doi.org/10.3390/v14122642
Chicago/Turabian StyleGao, Yanni, Tingting Xia, Juan Bai, Lujie Zhang, Xiaolin Jiang, Xing Yang, Keshan Zhang, and Ping Jiang. 2022. "African Swine Fever Virus Exhibits Distinct Replication Defects in Different Cell Types" Viruses 14, no. 12: 2642. https://doi.org/10.3390/v14122642
APA StyleGao, Y., Xia, T., Bai, J., Zhang, L., Jiang, X., Yang, X., Zhang, K., & Jiang, P. (2022). African Swine Fever Virus Exhibits Distinct Replication Defects in Different Cell Types. Viruses, 14(12), 2642. https://doi.org/10.3390/v14122642





