Detection of Torquetenovirus and Redondovirus DNA in Saliva Samples from SARS-CoV-2-Positive and -Negative Subjects
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Specimens
2.2. SARS-CoV-2 Detection
2.3. TTV DNA Detection
2.4. ReDoV DNA Detection
2.5. ReDoV Genetic Characterization
2.6. Statistical Analysis
3. Results
3.1. SARS-CoV-2 Presence in Saliva
3.2. TTV Prevalence and Loads in Saliva
3.3. ReDoV Prevalence and Loads in Saliva
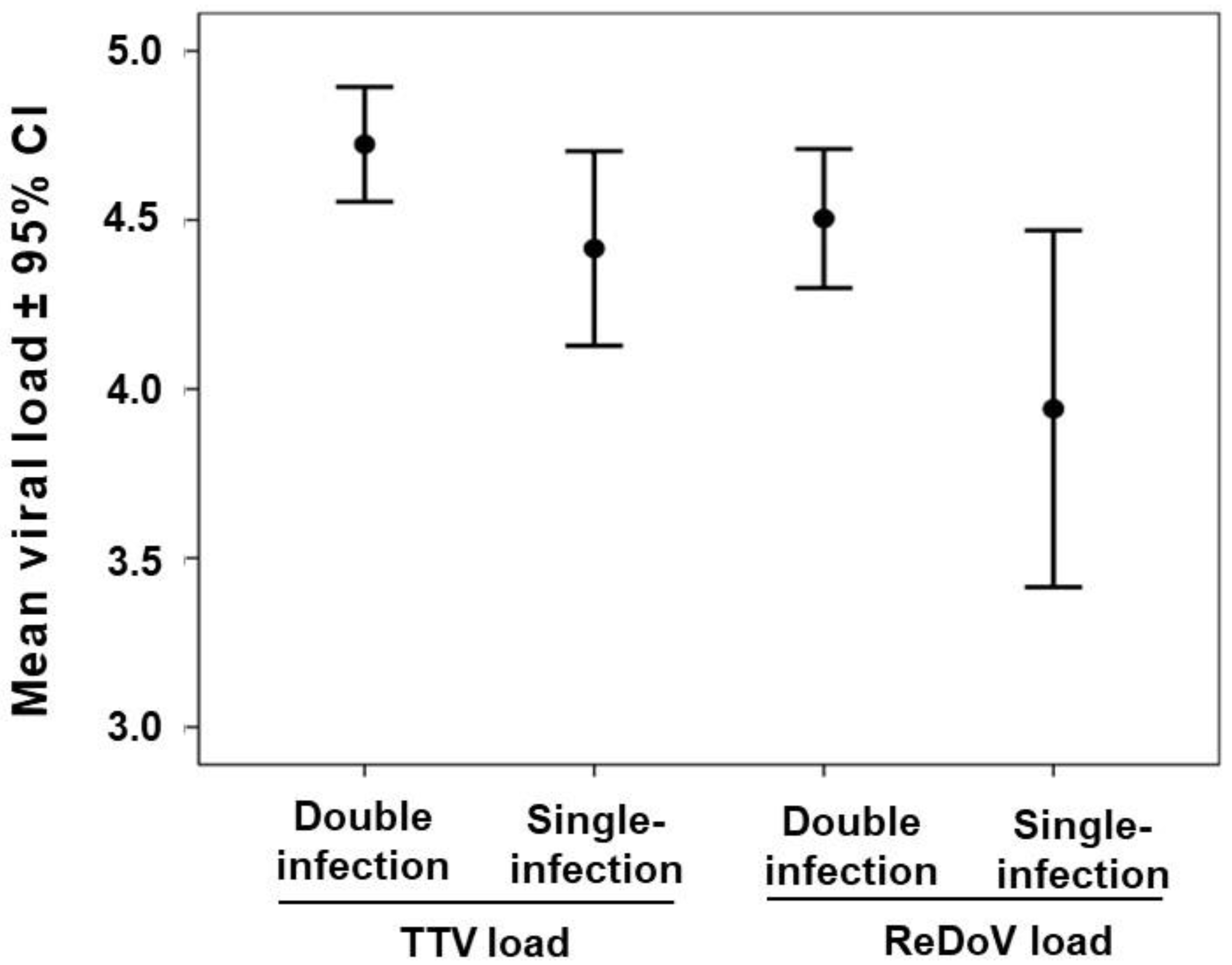
3.4. TTV and ReDoV Loads in Simultaneous Saliva and Plasma Samples
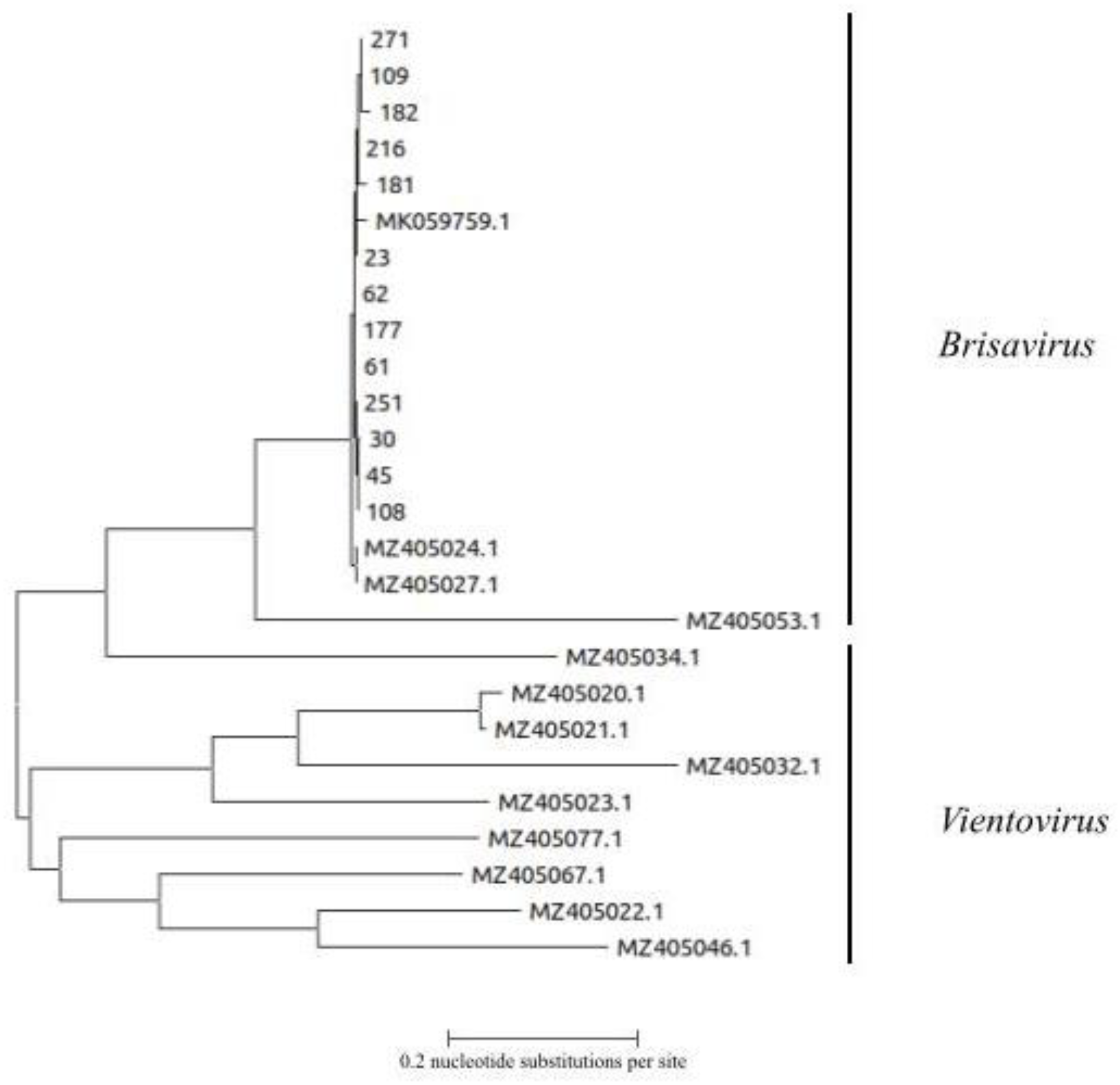
3.5. Genetic Analysis of ReDoV Isolates
4. Discussion
5. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krupovic, M.; Varsani, A.; Kazlauskas, D.; Breitbart, M.; Delwart, E.; Rosario, K.; Yutin, N.; Wolf, Y.I.; Harrach, B.; Zerbini, F.M.; et al. Cressdnaviricota: A Virus Phylum Unifying Seven Families of Rep-Encoding Viruses with Single-Stranded, Circular DNA Genomes. J. Virol. 2020, 94, e00582-20. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Taylor, L.J.; Collman, R.G.; Bushman, F.D.; Ictv Report, C. ICTV Virus Taxonomy Profile: Redondoviridae. J. Gen. Virol. 2021, 102, jgv001526. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Antonelli, G.; Pistello, M.; Maggi, F. Torquetenovirus: The human virome from bench to bedside. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016, 22, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.J.; Keeler, E.L.; Bushman, F.D.; Collman, R.G. The enigmatic roles of Anelloviridae and Redondoviridae in humans. Curr. Opin. Virol. 2022, 55, 101248. [Google Scholar] [CrossRef]
- Spezia, P.G.; Macera, L.; Mazzetti, P.; Curcio, M.; Biagini, C.; Sciandra, I.; Turriziani, O.; Lai, M.; Antonelli, G.; Pistello, M.; et al. Redondovirus DNA in human respiratory samples. J. Clin. Virol. 2020, 131, 104586. [Google Scholar] [CrossRef]
- Tu, N.T.K.; Deng, X.; Hong, N.T.T.; Ny, N.T.H.; Phuc, T.M.; Tam, P.T.T.; Han, D.A.; Ha, L.T.T.; Thwaites, G.; Doorn, H.R.V.; et al. Redondoviridae: High Prevalence and Possibly Chronic Shedding in Human Respiratory Tract, But No Zoonotic Transmission. Viruses 2021, 13, 533. [Google Scholar] [CrossRef]
- Taylor, L.J.; Dothard, M.I.; Rubel, M.A.; Allen, A.A.; Hwang, Y.; Roche, A.M.; Graham-Wooten, J.; Fitzgerald, A.S.; Khatib, L.A.; Ranciaro, A.; et al. Redondovirus Diversity and Evolution on Global, Individual, and Molecular Scales. J. Virol. 2021, 95, e0081721. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, C.; Feng, X.; Chen, X.; Zhang, W. Redondoviridae and periodontitis: A case-control study and identification of five novel redondoviruses from periodontal tissues. Virus Evol. 2021, 7, veab033. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, C.; Zhu, C.; Ye, W.; Gu, Q.; Shu, C.; Feng, X.; Chen, X.; Zhang, W.; Shan, T. Redondoviridae infection regulates circRNAome in periodontitis. J. Med. Virol. 2022, 94, 2537–2547. [Google Scholar] [CrossRef]
- Abbas, A.A.; Taylor, L.J.; Dothard, M.I.; Leiby, J.S.; Fitzgerald, A.S.; Khatib, L.A.; Collman, R.G.; Bushman, F.D. Redondoviridae, a Family of Small, Circular DNA Viruses of the Human Oro-Respiratory Tract Associated with Periodontitis and Critical Illness. Cell Host Microbe 2019, 25, 719–729.e714. [Google Scholar] [CrossRef]
- Swets, M.C.; Russell, C.D.; Harrison, E.M.; Docherty, A.B.; Lone, N.; Girvan, M.; Hardwick, H.E.; ISARIC4C Investigators; Visser, L.G.; Openshaw, P.J.M.; et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet 2022, 399, 1463–1464. [Google Scholar] [CrossRef]
- Cong, B.; Deng, S.; Wang, X.; Li, Y. The role of respiratory co-infection with influenza or respiratory syncytial virus in the clinical severity of COVID-19 patients: A systematic review and meta-analysis. J. Glob. Health 2022, 12, 5040. [Google Scholar] [CrossRef] [PubMed]
- Maggi, F.; Pifferi, M.; Fornai, C.; Andreoli, E.; Tempestini, E.; Vatteroni, M.; Presciuttini, S.; Marchi, S.; Pietrobelli, A.; Boner, A.; et al. TT virus in the nasal secretions of children with acute respiratory diseases: Relations to viremia and disease severity. J. Virol. 2003, 77, 2418–2425. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; He, X.; Ma, J.; Hong, W.; Hu, F.; Zhao, L.; Li, Q.; Zhang, J.; Zhang, C.; et al. Gemykibivirus genome in lower respiratory tract of elderly woman with unexplained acute respiratory distress syndrome. Clin. Infect. Dis. 2019, 69, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Forqué, L.; Albert, E.; Giménez, E.; Torres, I.; Carbonell, N.; Ferreres, J.; Blasco, M.L.; Navarro, D. Monitoring of Torque Teno virus DNAemia in critically ill COVID-19 patients: May it help to predict clinical outcomes? J. Clin. Virol. 2022, 148, 105082. [Google Scholar] [CrossRef]
- Solis, M.; Gallais, F.; Garnier-Kepka, S.; Lefebvre, N.; Benotmane, I.; Ludes, P.O.; Castelain, V.; Meziani, F.; Caillard, S.; Collange, O.; et al. Combining predictive markers for severe COVID-19: Torquetenovirus DNA load and SARS-CoV-2 RNAemia. J. Clin. Virol. 2022, 148, 105120. [Google Scholar] [CrossRef]
- Stincarelli, M.A.; Baj, A.; Guidotti, B.; Spezia, P.G.; Novazzi, F.; Lucenteforte, E.; Tillati, S.; Focosi, D.; Maggi, F.; Giannecchini, S. Plasma Torquetenovirus (TTV) microRNAs and severity of COVID-19. Virol. J. 2022, 19, 79. [Google Scholar] [CrossRef]
- Mendes-Correa, M.C.; Tozetto-Mendoza, T.R.; Freire, W.S.; Palao, H.G.O.; Ferraz, A.B.C.; Mamana, A.C.; Ferreira, N.E.; de Paula, A.V.; Felix, A.C.; Romano, C.M.; et al. Torquetenovirus in saliva: A potential biomarker for SARS-CoV-2 infection? PLoS ONE 2021, 16, e0256357. [Google Scholar] [CrossRef]
- Merenstein, C.; Liang, G.; Whiteside, S.A.; Cobián-Güemes, A.G.; Merlino, M.S.; Taylor, L.J.; Glascock, A.; Bittinger, K.; Tanes, C.; Graham-Wooten, J.; et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. mBio 2021, 12, e0177721. [Google Scholar] [CrossRef]
- Honorato, L.; Witkin, S.S.; Mendes-Correa, M.C.; Conde Toscano, A.L.C.; Linhares, I.M.; de Paula, A.V.; Paiao, H.G.O.; de Paula, V.S.; Lopes, A.O.; Lima, S.H.; et al. The Torque Teno virus titer in saliva reflects the level of circulating CD4+ T lymphocytes and HIV in individuals undergoing antiretroviral maintenance therapy. Front. Med. 2022, 8, 809312. [Google Scholar] [CrossRef]
- Batista, A.M.; Caetano, M.W.; Stincarelli, M.A.; Mamana, A.C.; Zerbinati, R.M.; Sarmento, D.J.S.; Gallottini, M.; Caixeta, R.A.V.; Medina-Pestana, J.; Hasseus, B.; et al. Quantification of torque teno virus (TTV) DNA in saliva and plasma samples in patients at short time before and after kidney transplantation. J. Oral Microbiol. 2021, 14, 2008140. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Spezia, P.G.; Macera, L.; Salvadori, S.; Navarro, D.; Lanza, M.; Antonelli, G.; Pistello, M.; Maggi, F. Assessment of prevalence and load of torquetenovirus viraemia in a large cohort of healthy blood donors. Clin. Microbiol. Infect. 2020, 26, 1406–1410. [Google Scholar] [CrossRef] [PubMed]
- Albert, E.; Torres, I.; Talaya, A.; Giménez, E.; Piñana, J.L.; Hernández-Boluda, J.C.; Focosi, D.; Macera, L.; Maggi, F.; Solano, C.; et al. Kinetics of torque teno virus DNA load in saliva and plasma following allogeneic hematopoietic stem cell transplantation. J. Med. Virol. 2018, 90, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
| Parameter | SARS-CoV-2 Negative | SARS-CoV-2 Positive | Total |
|---|---|---|---|
| Saliva samples examined (No.) | 326 | 122 | 448 |
| TTV DNA positive (%) | 238 (73) | 87 (71) | 325 (72) |
| ReDoV DNA positive (%) | 167 (65) a,b | 63 (52) c | 230 (61) d,e |
| TTV DNA load (Log copies/mL ± 95% CI) | 4.6 (4.4–4.8) | 4.7 (4.3–4.8) | 4.6 (4.4–4.7) |
| ReDoV DNA load (Log copies/mL ± 95% CI) | 4.4 (4.1–4.6) | 4.5 (4.0–4.7) | 4.4 (4.2–4.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spezia, P.G.; Baj, A.; Drago Ferrante, F.; Boutahar, S.; Azzi, L.; Genoni, A.; Dalla Gasperina, D.; Novazzi, F.; Dentali, F.; Focosi, D.; et al. Detection of Torquetenovirus and Redondovirus DNA in Saliva Samples from SARS-CoV-2-Positive and -Negative Subjects. Viruses 2022, 14, 2482. https://doi.org/10.3390/v14112482
Spezia PG, Baj A, Drago Ferrante F, Boutahar S, Azzi L, Genoni A, Dalla Gasperina D, Novazzi F, Dentali F, Focosi D, et al. Detection of Torquetenovirus and Redondovirus DNA in Saliva Samples from SARS-CoV-2-Positive and -Negative Subjects. Viruses. 2022; 14(11):2482. https://doi.org/10.3390/v14112482
Chicago/Turabian StyleSpezia, Pietro Giorgio, Andreina Baj, Francesca Drago Ferrante, Sara Boutahar, Lorenzo Azzi, Angelo Genoni, Daniela Dalla Gasperina, Federica Novazzi, Francesco Dentali, Daniele Focosi, and et al. 2022. "Detection of Torquetenovirus and Redondovirus DNA in Saliva Samples from SARS-CoV-2-Positive and -Negative Subjects" Viruses 14, no. 11: 2482. https://doi.org/10.3390/v14112482
APA StyleSpezia, P. G., Baj, A., Drago Ferrante, F., Boutahar, S., Azzi, L., Genoni, A., Dalla Gasperina, D., Novazzi, F., Dentali, F., Focosi, D., & Maggi, F. (2022). Detection of Torquetenovirus and Redondovirus DNA in Saliva Samples from SARS-CoV-2-Positive and -Negative Subjects. Viruses, 14(11), 2482. https://doi.org/10.3390/v14112482









