Enhancing Anti-G Antibody Induction by a Live Single-Cycle Prefusion F—Expressing RSV Vaccine Improves In Vitro and In Vivo Efficacy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Antibodies
2.2. Construction of Recombinant RSV cDNAs
2.3. Recovery of Viruses from cDNA and Production of Virus Stocks
2.4. Generation of VLP-G
2.5. Cell ELISA to Measure preF and G Levels on the Surface of Infected Cells
2.6. Western Blot Analysis
2.7. Mouse Ethics Statement
2.8. Vaccination Protocol and Serum Ab Collection
2.9. Challenge Studies (Histopathology)
2.10. ELISA to Measure Anti-preF and Anti-G Ab Levels in Mouse Sera
2.11. In Vitro Neutralization
3. Results
3.1. Overview of Approaches Aimed at Enhancing Serum Anti-G Ab Levels Induced by a Single-Cycle Pref-Expressing Virus
3.2. A mutation That Blocks CatL Cleavage Does Not Raise G Ab Levels in Mice Vaccinated with RSV-preF∆CT
3.3. The Impact of Re-Introducing sG on Anti-G Ab Levels in Vaccinated Mice
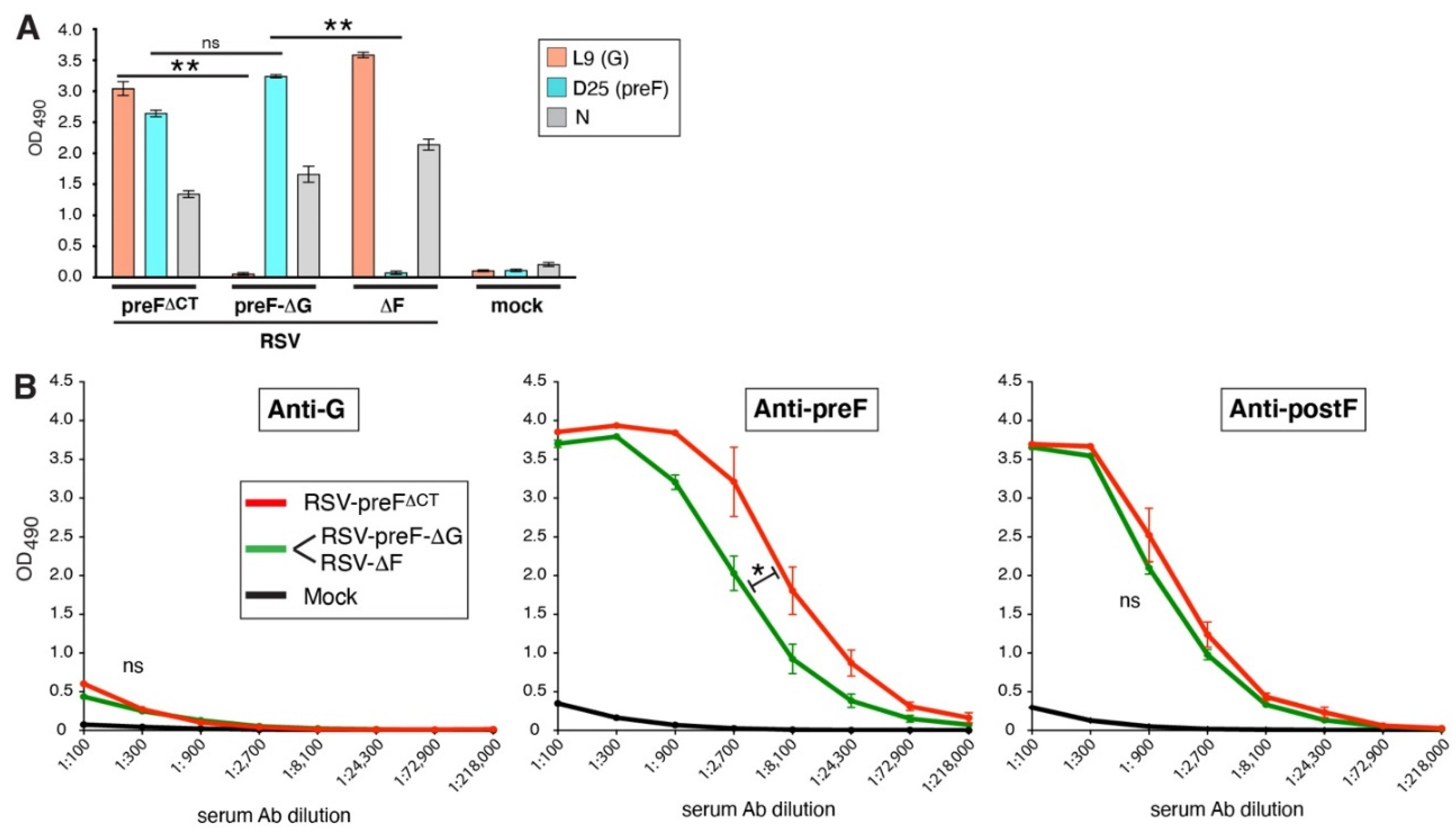
3.4. Presenting preF and G on Separate Vaccine Particles Does Not Improve G or F Ab Levels in Mice
3.5. Addition of Exogenous Input G Protein Increases Anti-G Ab Levels in Vaccinated Mice
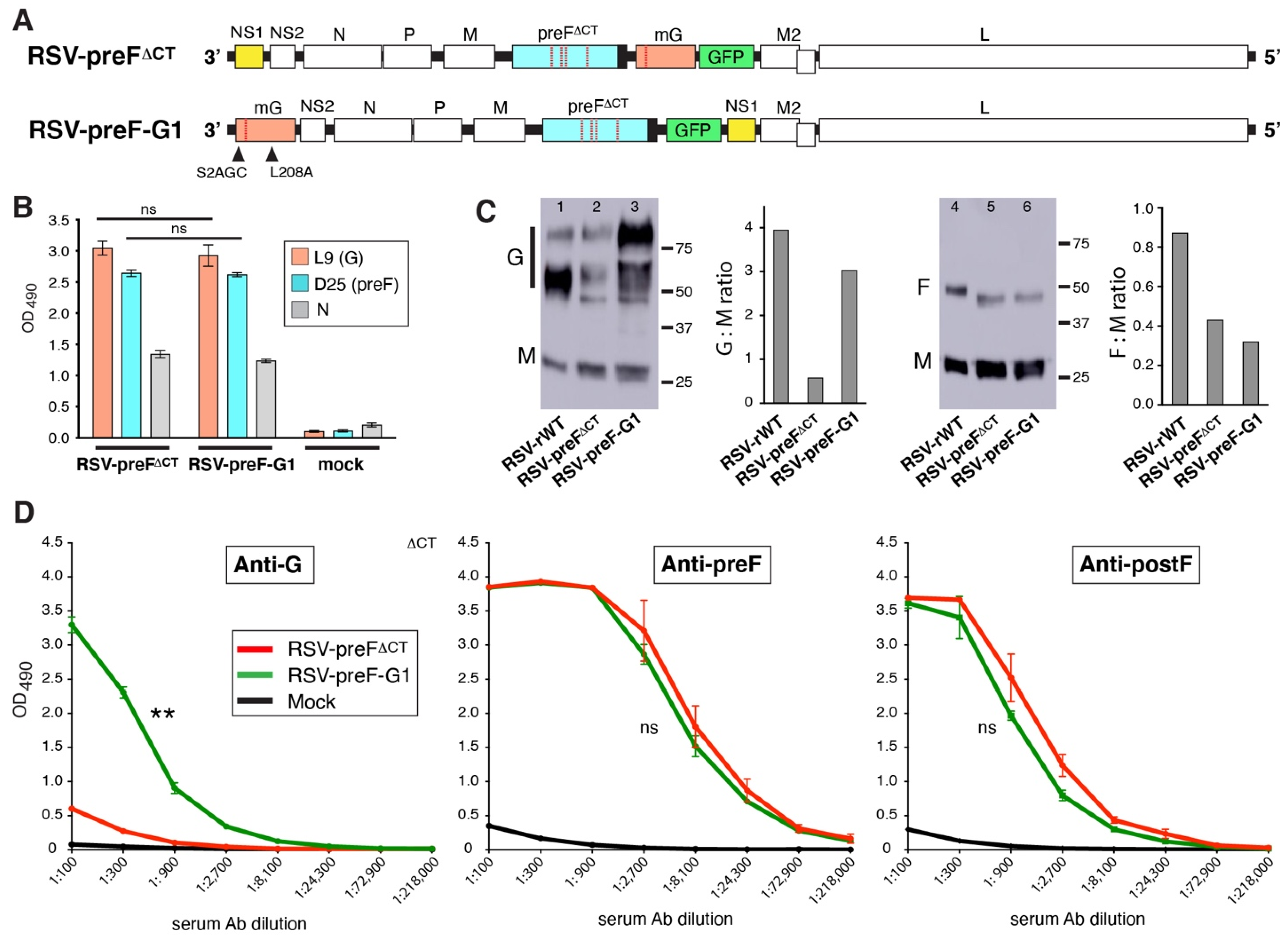
3.6. The Impact of Moving G to the First Position in the Genome
3.7. Parameters Contributing to Enhanced Anti-G Ab Levels Induced by RSV-preF-G1
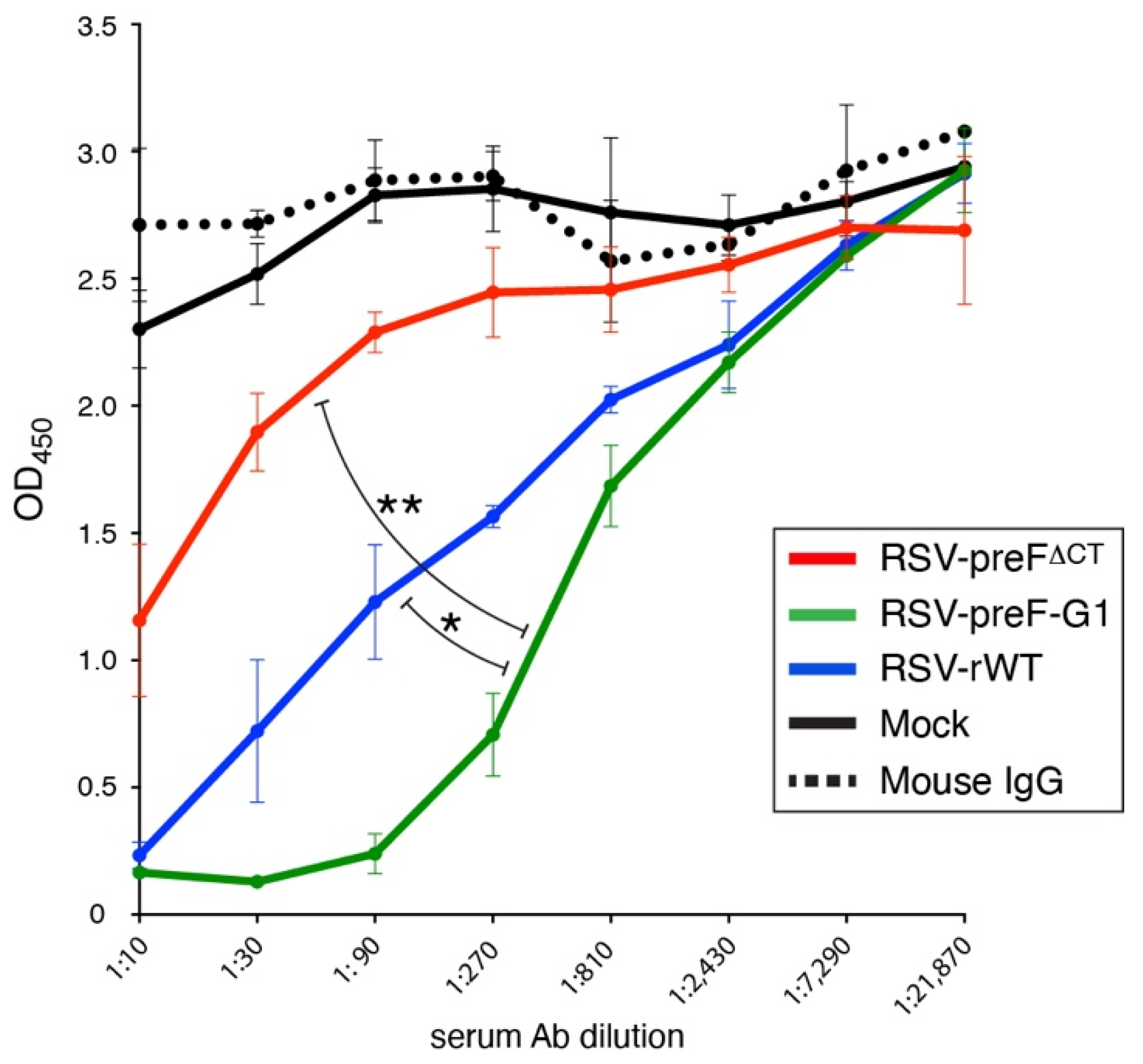
3.8. In Vitro Neutralization
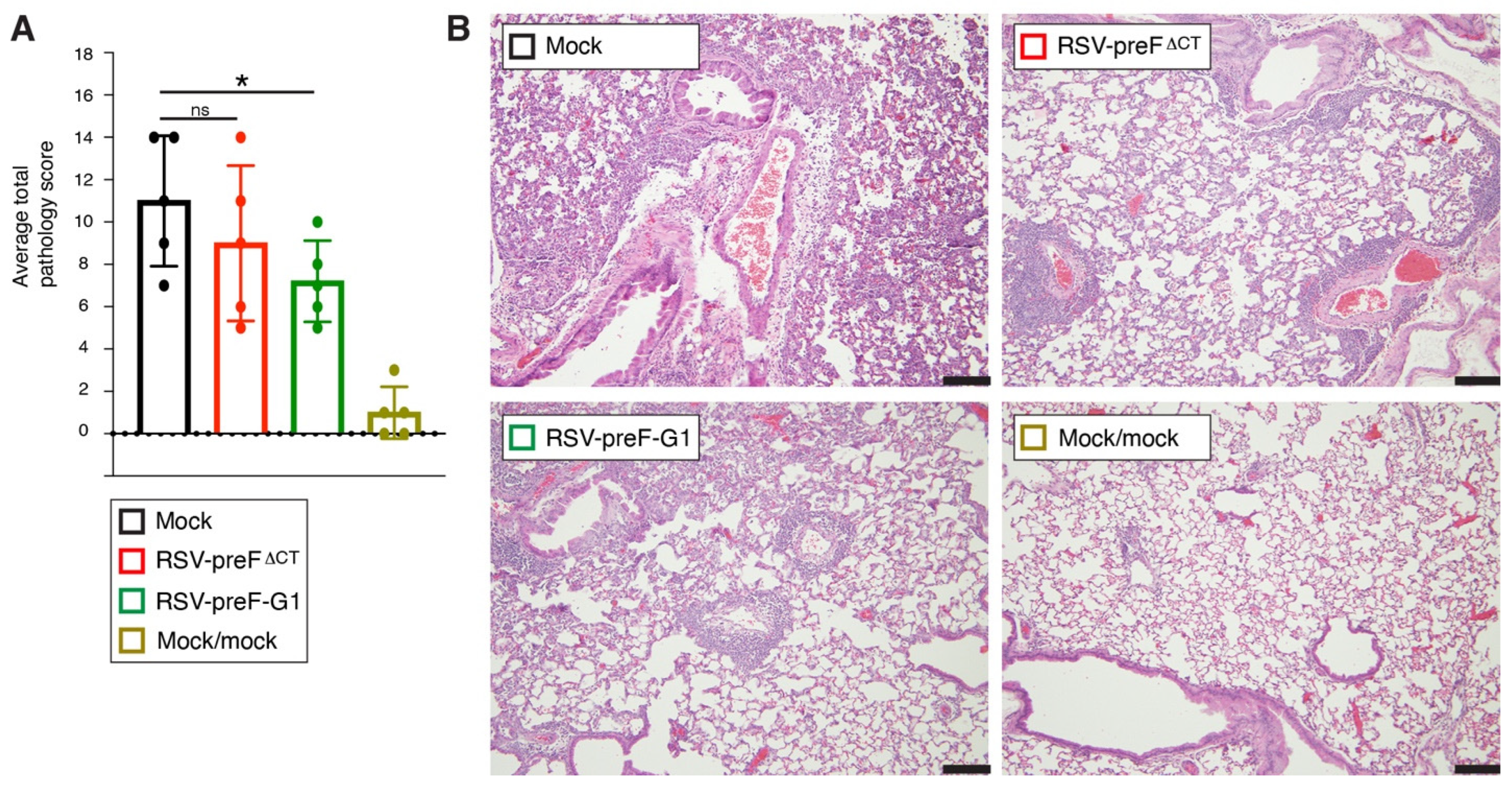
3.9. Protection from Lung Pathology after Challenge
4. Discussion
4.1. Study Objective
4.2. Several Modifications to RSV-preFΔCT Failed to Enhance the Induction of Serum Anti-G Abs in Mice
4.3. Addition of Exogenous (Input) G and Moving G to the First Genome Position of RSV-preF∆CT Each Substantially Improve Serum Anti-G Ab Levels in Mice
4.4. In Summary
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, T.; McAllister, D.A.; O’Brien, K.L.; Simoes, E.A.F.; Madhi, S.A.; Gessner, B.D.; Polack, F.P.; Balsells, E.; Acacio, S.; Aguayo, C.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet 2017, 390, 946–958. [Google Scholar] [CrossRef]
- Stockman, L.J.; Curns, A.T.; Anderson, L.J.; Fischer-Langley, G. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006. Pediatr. Infect. Dis. J 2012, 31, 5–9. [Google Scholar] [CrossRef]
- McLellan, J.S.; Chen, M.; Joyce, M.G.; Sastry, M.; Stewart-Jones, G.B.E.; Yang, Y.; Zhang, B.; Chen, L.; Srivatsan, S.; Zheng, A.; et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 2013, 342, 592–598. [Google Scholar] [CrossRef]
- Ngwuta, J.O.; Chen, M.; Modjarrad, K.; Joyce, M.G.; Kanekiyo, M.; Kumar, A.; Yassine, H.M.; Moin, S.M.; Killikelly, A.M.; Chuang, G.Y.; et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci. Transl. Med. 2015, 7, 309ra162. [Google Scholar] [CrossRef]
- Karron, R.A.; Buonagurio, D.A.; Georgiu, A.F.; Whitehead, S.S.; Adamus, J.E.; Clements-Mann, M.L.; Harris, D.O.; Randolph, V.B.; Udem, S.A.; Murphy, B.R.; et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: Clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 1997, 94, 13961–13966. [Google Scholar] [CrossRef]
- Chirkova, T.; Lin, S.; Oomens, A.G.P.; Gaston, K.A.; Boyoglu-Barnum, S.; Meng, J.; Stobart, C.C.; Cotton, C.U.; Hartert, T.V.; Moore, M.L.; et al. CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. J. Gen. Virol. 2015, 96, 2543–2556. [Google Scholar] [CrossRef]
- Johnson, S.M.; McNally, B.A.; Ioannidis, I.; Flano, E.; Teng, M.N.; Oomens, A.G.; Walsh, E.E.; Peeples, M.E. Respiratory Syncytial Virus Uses CX3CR1 as a Receptor on Primary Human Airway Epithelial Cultures. PLoS Pathog. 2015, 11, e1005318. [Google Scholar] [CrossRef]
- Kwilas, S.; Liesman, R.M.; Zhang, L.; Walsh, E.; Pickles, R.J.; Peeples, M.E. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J. Virol. 2009, 83, 10710–10718. [Google Scholar] [CrossRef]
- Techaarpornkul, S.; Barretto, N.; Peeples, M.E. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 2001, 75, 6825–6834. [Google Scholar] [CrossRef]
- Teng, M.N.; Whitehead, S.S.; Collins, P.L. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 2001, 289, 283–296. [Google Scholar] [CrossRef]
- Corry, J.; Johnson, S.M.; Cornwell, J.; Peeples, M.E. Preventing Cleavage of the Respiratory Syncytial Virus Attachment Protein in Vero Cells Rescues the Infectivity of Progeny Virus for Primary Human Airway Cultures. J. Virol. 2016, 90, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Vanover, D.; Smith, D.V.; Blanchard, E.L.; Alonas, E.; Kirschman, J.L.; Lifland, A.W.; Zurla, C.; Santangelo, P.J. RSV glycoprotein and genomic RNA dynamics reveal filament assembly prior to the plasma membrane. Nat. Commun. 2017, 8, 667. [Google Scholar] [CrossRef]
- Bukreyev, A.; Yang, L.; Collins, P.L. The secreted G protein of human respiratory syncytial virus antagonizes antibody-mediated restriction of replication involving macrophages and complement. J. Virol. 2012, 86, 10880–10884. [Google Scholar] [CrossRef] [PubMed]
- Bukreyev, A.; Yang, L.; Fricke, J.; Cheng, L.; Ward, J.M.; Murphy, B.R.; Collins, P.L. The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J. Virol. 2008, 82, 12191–12204. [Google Scholar] [CrossRef]
- Shingai, M.; Azuma, M.; Ebihara, T.; Sasai, M.; Funami, K.; Ayata, M.; Ogura, H.; Tsutsumi, H.; Matsumoto, M.; Seya, T. Soluble G protein of respiratory syncytial virus inhibits Toll-like receptor 3/4-mediated IFN-beta induction. Int. Immunol. 2008, 20, 1169–1180. [Google Scholar] [CrossRef]
- Maher, C.F.; Hussell, T.; Blair, E.; Ring, C.J.; Openshaw, P.J. Recombinant respiratory syncytial virus lacking secreted glycoprotein G is attenuated, non-pathogenic but induces protective immunity. Microbes. Infect. 2004, 6, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Kabatova, B.; Kabat, J.; Dorward, D.W.; Liu, X.; Surman, S.; Liu, X.; Moseman, A.P.; Buchholz, U.J.; Collins, P.L.; et al. Effects of Alterations to the CX3C Motif and Secreted Form of Human Respiratory Syncytial Virus (RSV) G Protein on Immune Responses to a Parainfluenza Virus Vector Expressing the RSV G Protein. J. Virol. 2019, 93, e02043-18. [Google Scholar] [CrossRef]
- Lamichhane, P.; Schmidt, M.E.; Terhüja, M.; Varga, S.M.; Snider, T.A.; Rostad, C.A.; Oomens, A.G.P. A live single-cycle RSV vaccine expressing prefusion F protein. Virology 2022, 577, 51–64. [Google Scholar] [CrossRef]
- Oomens, A.G.; Bevis, K.P.; Wertz, G.W. The cytoplasmic tail of the human respiratory syncytial virus F protein plays critical roles in cellular localization of the F protein and infectious progeny production. J. Virol. 2006, 80, 10465–10477. [Google Scholar] [CrossRef]
- Schmidt, M.E.; Varga, S.M. Modulation of the host immune response by respiratory syncytial virus proteins. J. Microbiol. 2017, 55, 161–171. [Google Scholar] [CrossRef]
- Baviskar, P.S.; Hotard, A.L.; Moore, M.L.; Oomens, A.G. The respiratory syncytial virus fusion protein targets to the perimeter of inclusion bodies and facilitates filament formation by a cytoplasmic tail-dependent mechanism. J. Virol. 2013, 87, 10730–10741. [Google Scholar] [CrossRef] [PubMed]
- Oomens, A.G.; Wertz, G.W. Trans-Complementation allows recovery of human respiratory syncytial viruses that are infectious but deficient in cell-to-cell transmission. J. Virol. 2004, 78, 9064–9072. [Google Scholar] [CrossRef]
- Anderson, L.J.; Jadhao, S.J.; Paden, C.R.; Tong, S. Functional Features of the Respiratory Syncytial Virus G Protein. Viruses 2021, 13, 1214. [Google Scholar] [CrossRef]
- Boyoglu-Barnum, S.; Gaston, K.A.; Todd, S.O.; Boyoglu, C.; Chirkova, T.; Barnum, T.R.; Jorquera, P.; Haynes, L.M.; Tripp, R.A.; Moore, M.L.; et al. A respiratory syncytial virus (RSV) anti-G protein F(ab’)2 monoclonal antibody suppresses mucous production and breathing effort in RSV rA2-line19F-infected BALB/c mice. J. Virol. 2013, 87, 10955–10967. [Google Scholar] [CrossRef] [PubMed]
- Boyoglu-Barnum, S.; Todd, S.O.; Chirkova, T.; Barnum, T.R.; Gaston, K.A.; Haynes, L.M.; Tripp, R.A.; Moore, M.L.; Anderson, L.J. An anti-G protein monoclonal antibody treats RSV disease more effectively than an anti-F monoclonal antibody in BALB/c mice. Virology 2015, 483, 117–125. [Google Scholar] [CrossRef]
- Caidi, H.; Harcourt, J.L.; Tripp, R.A.; Anderson, L.J.; Haynes, L.M. Combination therapy using monoclonal antibodies against respiratory syncytial virus (RSV) G glycoprotein protects from RSV disease in BALB/c mice. PLoS ONE 2012, 7, e51485. [Google Scholar] [CrossRef]
- Caidi, H.; Miao, C.; Thornburg, N.J.; Tripp, R.A.; Anderson, L.J.; Haynes, L.M. Anti-respiratory syncytial virus (RSV) G monoclonal antibodies reduce lung inflammation and viral lung titers when delivered therapeutically in a BALB/c mouse model. Antiviral. Res. 2018, 154, 149–157. [Google Scholar] [CrossRef]
- Capella, C.; Chaiwatpongsakorn, S.; Gorrell, E.; Risch, Z.A.; Ye, F.; Mertz, S.E.; Johnson, S.M.; Moore-Clingenpeel, M.; Ramilo, O.; Mejias, A.; et al. Prefusion F, Postfusion F, G Antibodies, and Disease Severity in Infants and Young Children with Acute Respiratory Syncytial Virus Infection. J. Infect. Dis. 2017, 216, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Takeda, K.; Wang, M.; Zeng, W.; Jia, Y.; Shiraishi, Y.; Okamoto, M.; Dakhama, A.; Gelfand, E.W. Effects of antig and anti-f antibodies on airway function after respiratory syncytial virus infection. Am. J. Respir. Cell Mol. Biol. 2014, 51, 143–154. [Google Scholar] [CrossRef]
- Haynes, L.M.; Caidi, H.; Radu, G.U.; Miao, C.; Harcourt, J.L.; Tripp, R.A.; Anderson, L.J. Therapeutic monoclonal antibody treatment targeting respiratory syncytial virus (RSV) G protein mediates viral clearance and reduces the pathogenesis of RSV infection in BALB/c mice. J. Infect. Dis. 2009, 200, 439–447. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, J.Y.; Park, M.H.; Kim, J.Y.; Chang, J. Monoclonal Antibody against G Glycoprotein Increases Respiratory Syncytial Virus Clearance In Vivo and Prevents Vaccine-Enhanced Diseases. PLoS ONE 2017, 12, e0169139. [Google Scholar] [CrossRef]
- Miao, C.; Radu, G.U.; Caidi, H.; Tripp, R.A.; Anderson, L.J.; Haynes, L.M. Treatment with respiratory syncytial virus G glycoprotein monoclonal antibody or F(ab’)2 components mediates reduced pulmonary inflammation in mice. J. Gen. Virol. 2009, 90, 1119–1123. [Google Scholar] [CrossRef]
- Stott, E.J.; Taylor, G.; Ball, L.A.; Anderson, K.; Young, K.K.; King, A.M.; Wertz, G.W. Immune and histopathological responses in animals vaccinated with recombinant vaccinia viruses that express individual genes of human respiratory syncytial virus. J. Virol. 1987, 61, 3855–3861. [Google Scholar] [CrossRef]
- Walsh, E.E.; Karron, R.A.; Belshe, R.B.; Shi, J.R.; Randolph, V.B.; Collins, P.L.; O’Shea, P.F.; Gruber, W.C.; Murphy, B.R. Comparison of antigenic sites of subtype-specific respiratory syncytial virus attachment proteins. J. Gen. Virol. 1989, 70, 2953–2961. [Google Scholar] [CrossRef]
- Mitra, R.; Baviskar, P.; Duncan-Decocq, R.R.; Patel, D.; Oomens, A.G. The human respiratory syncytial virus matrix protein is required for maturation of viral filaments. J. Virol. 2012, 86, 4432–4443. [Google Scholar] [CrossRef]
- Schmidt, M.E.; Oomens, A.G.P.; Varga, S.M. Vaccination with a Single-Cycle Respiratory Syncytial Virus Is Immunogenic and Protective in Mice. J. Immunol. 2019, 202, 3234–3245. [Google Scholar] [CrossRef]
- Meshram, C.D.; Baviskar, P.S.; Ognibene, C.M.; Oomens, A.G. The Respiratory Syncytial Virus Phosphoprotein, Matrix Protein, and Fusion Protein Carboxy-Terminal Domain Drive Efficient Filamentous Virus-Like Particle Formation. J. Virol. 2016, 90, 10612–10628. [Google Scholar] [CrossRef]
- Kozak, M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef]
- Hotard, A.L.; Lee, S.; Currier, M.G.; Crowe, J.E.; Sakamoto, K., Jr.; Newcomb, D.C.; Peebles, R.S., Jr.; Plemper, R.K.; Moore, M.L. Identification of residues in the human respiratory syncytial virus fusion protein that modulate fusion activity and pathogenesis. J. Virol. 2015, 89, 512–522. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamichhane, P.; Terhüja, M.; Snider, T.A.; Oomens, A.G.P. Enhancing Anti-G Antibody Induction by a Live Single-Cycle Prefusion F—Expressing RSV Vaccine Improves In Vitro and In Vivo Efficacy. Viruses 2022, 14, 2474. https://doi.org/10.3390/v14112474
Lamichhane P, Terhüja M, Snider TA, Oomens AGP. Enhancing Anti-G Antibody Induction by a Live Single-Cycle Prefusion F—Expressing RSV Vaccine Improves In Vitro and In Vivo Efficacy. Viruses. 2022; 14(11):2474. https://doi.org/10.3390/v14112474
Chicago/Turabian StyleLamichhane, Pramila, Megolhubino Terhüja, Timothy A. Snider, and Antonius G. P. Oomens. 2022. "Enhancing Anti-G Antibody Induction by a Live Single-Cycle Prefusion F—Expressing RSV Vaccine Improves In Vitro and In Vivo Efficacy" Viruses 14, no. 11: 2474. https://doi.org/10.3390/v14112474
APA StyleLamichhane, P., Terhüja, M., Snider, T. A., & Oomens, A. G. P. (2022). Enhancing Anti-G Antibody Induction by a Live Single-Cycle Prefusion F—Expressing RSV Vaccine Improves In Vitro and In Vivo Efficacy. Viruses, 14(11), 2474. https://doi.org/10.3390/v14112474





